Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations
Abstract
:1. Introduction
2. Long Noncoding RNAs Mediating Complementary Target-RNA Degradation
2.1. LncRNAs Involved in Discordant Regulation
2.2. LncRNAs as Precursors of Small Regulatory RNAs
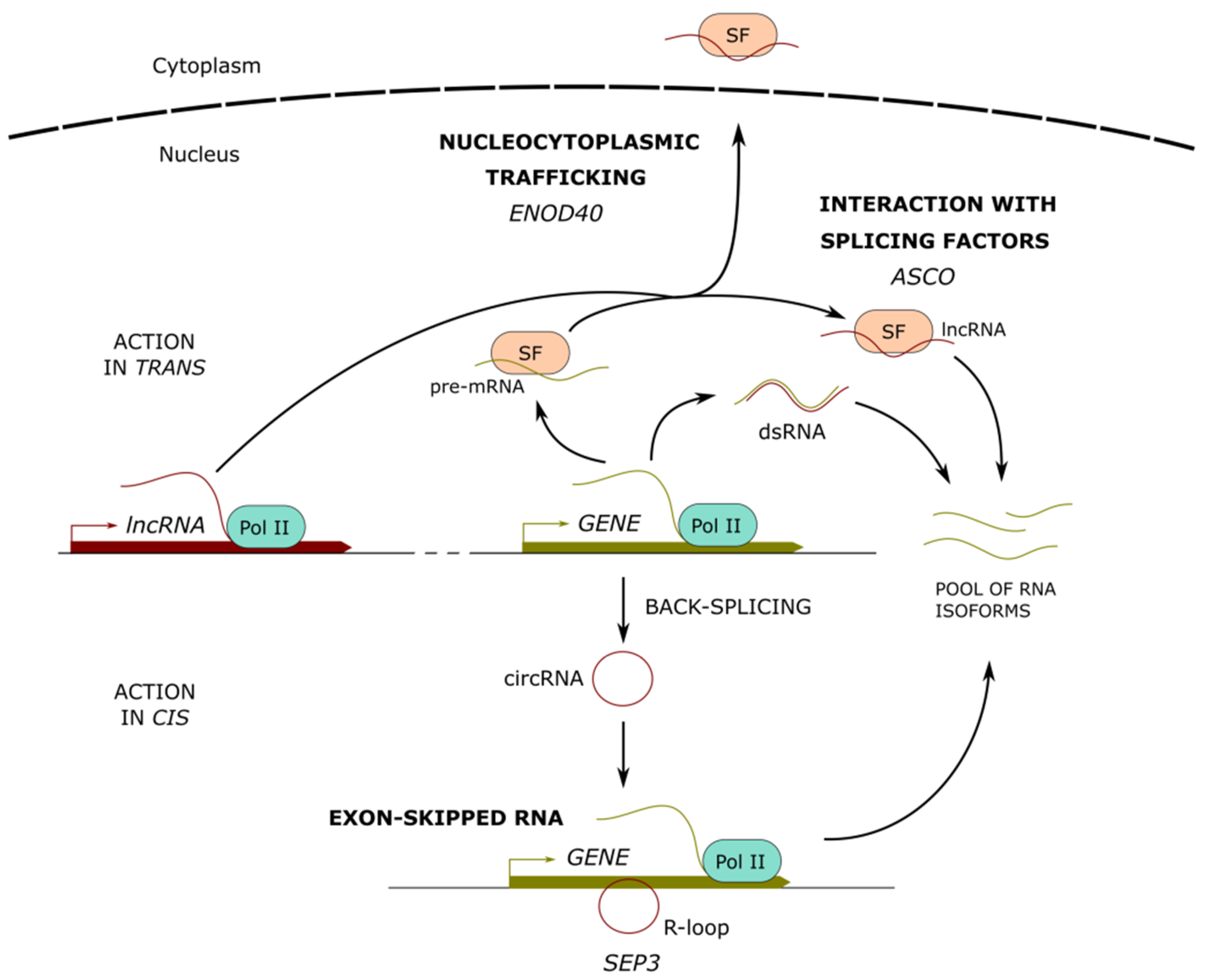
3. Long Noncoding RNAs Involved in the Regulation of Alternative Splicing
3.1. LncRNAs Interacting with Splicing Factors
3.2. LncRNAs Regulating Splicing Through Chromatin Remodeling
3.3. LncRNA-RNA Duplexes Regulating Alternative Splicing
4. Long Noncoding RNAs as Molecular Cargos for Protein Re-Localization
5. Long Noncoding RNAs Promoting Translation
5.1. LncRNA-mRNA Pairs into Polysomes
5.2. LncRNAs as Target Mimics for miRNAs
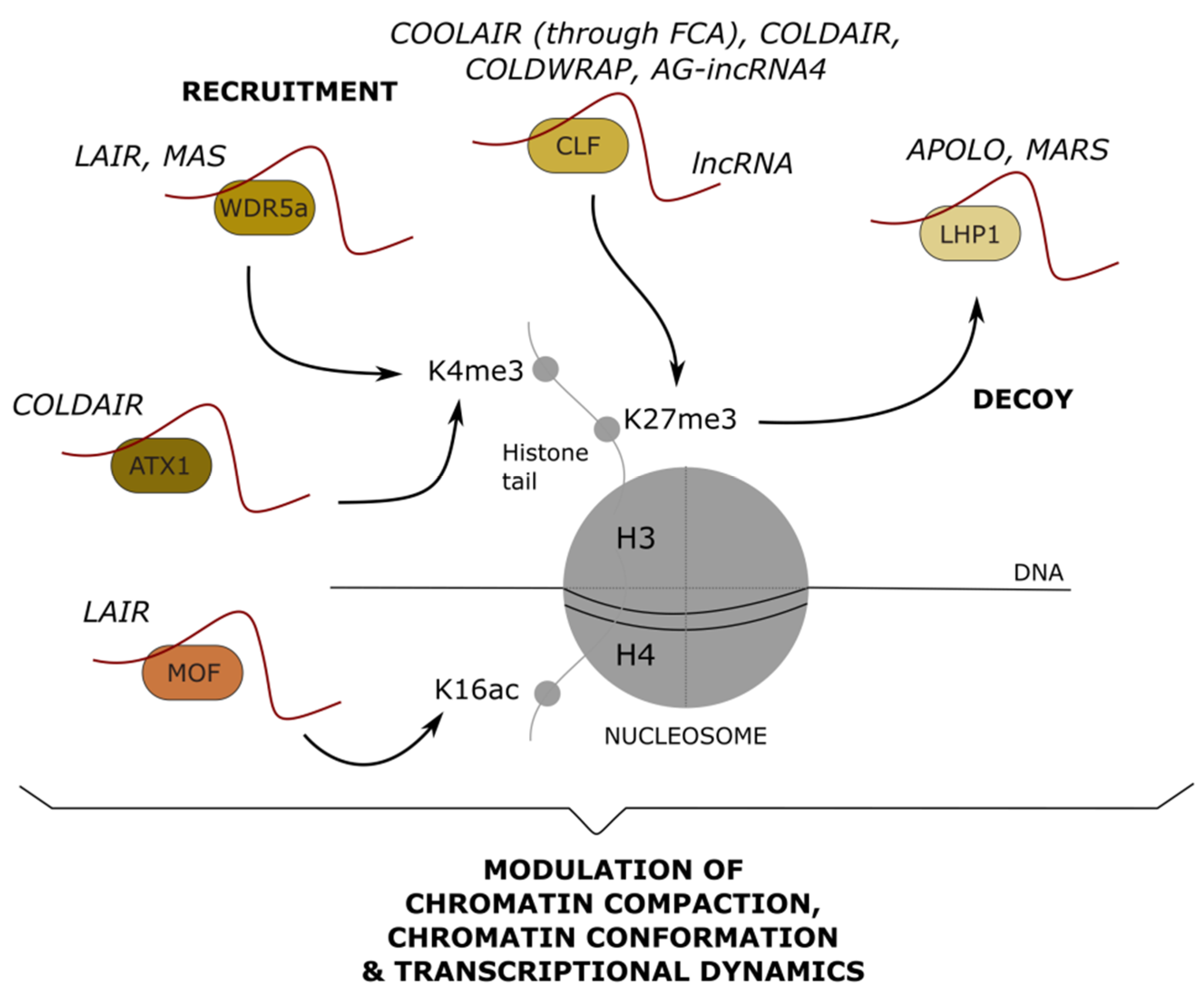
6. Long Noncoding RNAs Mediating Post-Translational Modifications: Impact on Chromatin Remodeling and Transcription
7. Conclusions and Future Perspectives
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corradi, N.; Pombert, J.-F.; Farinelli, L.; Didier, E.S.; Keeling, P.J. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 2010, 1, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellicer, J.; Fay, M.F.; Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010, 164, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C. The Genetic Organization of Chromosomes. Annu. Rev. Genet. 1971, 5, 237–256. [Google Scholar] [CrossRef]
- Eddy, S.R. The C-value paradox, junk DNA and ENCODE. Curr. Biol. 2012, 22, R898–R899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant. Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, Z.; Krause, H.M. Long Noncoding RNAs and Repetitive Elements: Junk or Intimate Evolutionary Partners? Trends Genet. 2019, 35, 892–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comings, D.E. The Structure and Function of Chromatin. Adv. Hum. Genet. 1972, 3, 237–431. [Google Scholar] [CrossRef]
- Ohno, S. So much “junk” DNA in our genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 1–12. [Google Scholar] [CrossRef]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucero, L.; Ferrero, L.; Fonouni-Farde, C.; Ariel, F. Functional classification of plant long noncoding RNAs: A transcript is known by the company it keeps. New Phytol. 2021, 229, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Lucero, L.; Fonouni-Farde, C.; Crespi, M.; Ariel, F. Long noncoding RNAs shape transcription in plants. Transcription 2020, 11, 160–171. [Google Scholar] [CrossRef]
- Lapidot, M.; Pilpel, Y. Genome-wide natural antisense transcription: Coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006, 7, 1216–1222. [Google Scholar] [CrossRef] [Green Version]
- Britto-Kido, S.D.A.; Ferreira Neto, J.R.C.; Pandolfi, V.; Marcelino-Guimarães, F.C.; Nepomuceno, A.L.; Vilela Abdelnoor, R.; Benko-Iseppon, A.M.; Kido, E.A. Natural antisense transcripts in plants: A re-view and identification in soybean infected with phakopsora pachyrhizi supersage library. Sci. World J. 2013, 2013, 219798. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-J.; Gaasterland, T.; Chua, N.-H. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005, 6, R30. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-Y.; Qin, L.; Guo, Z.-M.; Liu, L.; Xu, H.; Hao, P.; Su, J.; Shi, Y.; He, W.-Z.; Li, Y.-X. In silico discovery of human natural antisense transcripts. BMC Bioinform. 2006, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Osato, N.; Yamada, H.; Satoh, K.; Ooka, H.; Yamamoto, M.; Suzuki, K.; Kawai, J.; Carninci, P.; Ohtomo, Y.; Murakami, K.; et al. Antisense transcripts with rice full-length cDNAs. Genome Biol. 2003, 5, R5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jen, C.-H.; Michalopoulos, I.; Westhead, D.R.; Meyer, P. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 2005, 6, R51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Chua, N.-H.; Wang, X.-J. Prediction of trans-antisense transcripts in Arabidopsis thaliana. Genome Biol. 2006, 7, R92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Sunkar, R.; Jin, H.; Zhu, J.K.; Zhang, W. Genome-wide identification and analysis of small RNAs origi-nated from natural antisense transcripts in Oryza sativa. Genome Res. 2009, 19, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Zhu, C.; Lu, G.; Guo, Y.; Zhou, Y.; Zhang, Z.; Zhao, Y.; Li, W.; Lu, Y.; Tang, W.; et al. Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genom. 2012, 13, 721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Chung, P.J.; Liu, J.; Jang, I.C.; Kean, M.J.; Xu, J.; Chua, N.H. Genome-wide identification of long non-coding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2004, 24, 444–453. [Google Scholar] [CrossRef] [Green Version]
- Wunderlich, M.; Gross-Hardt, R.; Schöffl, F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014, 85, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Fedak, H.; Palusinska, M.; Krzyczmonik, K.; Brzezniak, L.; Yatusevich, R.; Pietras, Z.; Kaczanowski, S.; Swiezewski, S. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 2016, 113, E7846–E7855. [Google Scholar] [CrossRef] [Green Version]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.-F.; Chua, N.-H. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, D.; Zhang, D.; Yin, D.; Zhao, Y.; Ji, C.; Zhao, X.; Li, X.; He, Q.; Chen, R.; et al. A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 2018, 218, 774–788. [Google Scholar] [CrossRef] [Green Version]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous siRNAs Derived from a Pair of Natural cis-Antisense Transcripts Regulate Salt Tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katiyar-Agarwal, S.; Morgan, R.; Dahlbeck, D.; Borsani, O.; Villegas, A.; Zhu, J.-K.; Staskawicz, B.J.; Jin, H. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 2006, 103, 18002–18007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ron, M.; Saez, M.A.; Eshed-Williams, L.; Fletcher, J.C.; McCormick, S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 2010, 24, 1010–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Held, M.A.; Penning, B.; Brandt, A.S.; Kessans, S.A.; Yong, W.; Scofield, S.R.; Carpita, N.C. Small-interfering RNAs from natural antisense transcripts derived from a cellulose synthase gene modulate cell wall biosynthesis in barley. Proc. Natl. Acad. Sci. USA 2008, 105, 20534–20539. [Google Scholar] [CrossRef] [Green Version]
- Zubko, E.; Meyer, P. A natural antisense transcript of the Petunia hybrida Sho gene suggests a role for an anti-sense mechanism in cytokinin regulation. Plant J. 2007, 52, 1131–1139. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Luo, Q.-J.; Samanta, M.P.; Koksal, F.; Janda, J.; Galbraith, D.W.; Richardson, C.R.; Ou-Yang, F.; Rock, C.D. Evidence for Antisense Transcription Associated with MicroRNA Target mRNAs in Arabidopsis. PLoS Genet. 2009, 5, e1000457. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Feurtado, J.A.; Smith, M.A.; Flatman, L.K.; Koh, C.; Cutler, A.J. Long noncoding miRNA gene repres-ses wheat β-diketone waxes. Proc. Natl. Acad. Sci. USA 2017, 114, E3149–E3158. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, W. Why genes in pieces? Nat. Cell Biol. 1978, 271, 501. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.; Liu, P.; Wu, C.-A.; Yang, G.-D.; Xu, R.; Guo, Q.-H.; Huang, J.-G.; Zheng, C.-C. Stress-Induced Alternative Splicing Provides a Mechanism for the Regulation of MicroRNA Processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Palusa, S.G.; Reddy, A.S.N. Differential recruitment of splice variants from SR Pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015, 56, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Tran, V.D.T.; Souiai, O.; Romero-Barrios, N.; Crespi, M.; Gautheret, D. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016, 13, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Bazin, J.; Romero-Barrios, N.; Rigo, R.; Charon, C.; Blein, T.; Ariel, F.; Crespi, M. Nuclear Speckle RNA Binding Proteins Remodel Alternative Splicing and the Non-coding Arabidopsis Transcriptome to Regulate a Cross-Talk Between Auxin and Immune Responses. Front. Plant. Sci. 2018, 9, 1209. [Google Scholar] [CrossRef] [PubMed]
- Rigo, R.; Bazin, J.; Romero-Barrios, N.; Moison, M.; Lucero, L.; Christ, A.; Benhamed, M.; Blein, T.; Huguet, S.; Charon, C.; et al. The Arabidopsis lnc RNA ASCO modulates the transcriptome through interaction with splicing factors. EMBO Rep. 2020, 21, e48977. [Google Scholar] [CrossRef]
- Grainger, R.J.; Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA 2005, 11, 533–557. [Google Scholar] [CrossRef] [Green Version]
- Elvira-Matelot, E.; Bardou, F.; Ariel, F.; Jauvion, V.; Bouteiller, N.; Le Masson, I.; Cao, J.; Crespi, M.D.; Vaucheret, H. The Nuclear Ribonucleoprotein SmD1 Interplays with Splicing, RNA Quality Control, and Posttranscriptional Gene Silencing in Arabidopsis. Plant Cell 2016, 28, 426–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Severing, E.I.; Van Dijk, A.D.J.; Morabito, G.; Busscher-Lange, J.; Immink, R.G.H.; Van Ham, R.C.H.J. Predicting the Impact of Alternative Splicing on Plant MADS Domain Protein Function. PLoS ONE 2012, 7, e30524. [Google Scholar] [CrossRef] [Green Version]
- Maclntosh, G.; Wilkerson, C.; Green, P. Identification and analysis of Arabidopsis expressed sequence tags cha-racteristic of non-coding RNAs. Plant Physiol. 2001, 3, 765–776. [Google Scholar] [CrossRef]
- Lindsey, K.; Casson, S.; Chilley, P. Peptides: New signalling molecules in plants. Trends Plant Sci. 2002, 7, 78–83. [Google Scholar] [CrossRef]
- Crespi, M.; Jurkevitch, E.; Poiret, M.; D’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef] [PubMed]
- Compaan, B.; Yang, W.-C.; Bisseling, T.; Franssen, H. ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil 2001, 230, 1–8. [Google Scholar] [CrossRef]
- Röhrig, H.; Schmidt, J.; Miklashevichs, E.; Schell, J.; John, M. Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 1915–1920. [Google Scholar] [CrossRef] [Green Version]
- Asad, S.; Fang, Y.; Wycoff, K.L.; Hirsch, A.M. Isolation and characterization of cDNA and genomic clones of MsENOD40; transcripts are detected in meristematic cells of alfalfa. Protoplasma 1994, 183, 10–23. [Google Scholar] [CrossRef]
- Lucero, L.; Bazin, J.; Rodriguez Melo, J.; Ibañez, F.; Crespi, M.D.; Ariel, F. Evolution of the small family of alter-native splicing modulators nuclear speckle RNA-binding proteins in plants. Genes 2020, 11, 207. [Google Scholar] [CrossRef] [Green Version]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a short open reading frame-containing mRNA, induces cyto-plasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 2004, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, B.; Von Arnim, A.G. Translational Regulation of Cytoplasmic mRNAs. Arab. Book 2013, 11, e0165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, Y.; Meyerowitz, E.M. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010, 6, 419. [Google Scholar] [CrossRef]
- Juntawong, P.; Sorenson, R.; Bailey-Serres, J. Cold shock protein 1 chaperones mRNAs during translation inArabidopsis thaliana. Plant J. 2013, 74, 1016–1028. [Google Scholar] [CrossRef]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 2013, 153, 562–574. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Le, B.; Ma, X.; Li, S.; You, C.; Yu, Y.; Zhang, B.; Liu, L.; Gao, L.; Shi, T.; et al. Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. eLife 2016, 5, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Sorenson, R.; Bailey-Serres, J. Selective mRNA sequestration by OLIGOURIDYLATEBINDING PROTEIN 1 con-tributes to translational control during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2373–2378. [Google Scholar] [CrossRef] [Green Version]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef] [Green Version]
- Hamburger, D.; Rezzonico, E.; Petétot, J.M.-C.; Somerville, C.; Poirier, Y. Identification and Characterization of the Arabidopsis PHO1 Gene Involved in Phosphate Loading to the Xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [Green Version]
- Stefanovic, A.; Arpat, A.B.; Bligny, R.; Gout, E.; Vidoudez, C.; Bensimon, M.; Poirier, Y. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 2011, 66, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Arpat, A.B.; Magliano, P.; Wege, S.; Rouached, H.; Stefanovic, A.; Poirier, Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012, 71, 479–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabnoune, M.; Secco, D.; Lecampion, C.; Robaglia, C.; Shu, Q.; Poirier, Y. A Rice cis-Natural Antisense RNA Acts as a Translational Enhancer for Its Cognate mRNA and Contributes to Phosphate Homeostasis and Plant Fitness. Plant Cell 2013, 25, 4166–4182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deforges, J.; Reis, R.S.; Jacquet, P.; Sheppard, S.; Gadekar, V.P.; Hart-Smith, G.; Tanzer, A.; Hofacker, I.L.; Iseli, C.; Xenarios, I.; et al. Control of Cognate Sense mRNA Translation by cis-Natural Antisense RNAs. Plant Physiol. 2019, 180, 305–322. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA Targets Identified by Sequencing of the Arabidopsis Degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [Green Version]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Du, Q.; Wang, K.; Zou, C.; Xu, C.; Li, W.X. The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol. 2018, 177, 1743–1753. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen-Hao, Z.; Mysore, K.S.; Zhang, W.-H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Cui, J.; Shi, Y.; Yang, G.; Zhou, X.; Hou, X.; Meng, J.; Luan, Y. Tomato lncRNA23468 functions as a com-peting endogenous RNA to modulate NBS-LRR genes by decoying miR482b in the tomato-Phytophthora infestans interaction. Hortic. Res. 2019, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.-J.; Wang, Z.-M.; Wang, M.; Wang, X.-J. Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2005, 5, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Huang, K.; Han, Z.; Wang, P.; Fang, Y. Genome-wide identification of Arabidopsis long noncoding RNAs in response to the blue light. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.; Paszkowski, J. Regulation of rice root development by a retrotransposon acting as a microRNA sponge. eLife 2017, 6, e30038. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Patel, N.A.; Watson, J.E.; Apostolatos, H.; Kleiman, E.; Hanson, O.; Hagiwara, M.; Cooper, D.R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternati-ve splicing of PKCβJII messenger ribonucleic acid. Endocrinology 2019, 150, 2087–2097. [Google Scholar] [CrossRef]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARγ2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef] [Green Version]
- Taniue, K.; Kurimoto, A.; Sugimasa, H.; Nasu, E.; Takeda, Y.; Iwasaki, K.; Nagashima, T.; Okada-Hatakeyama, M.; Oyama, M.; Kozuka-Hata, H.; et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. USA 2016, 113, 1273–1278. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [Green Version]
- Goodrich, J.; Puangsomlee, P.; Martín, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nat. Cell Biol. 1997, 386, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Primavesi, L.; Bishopp, A.; Roberts, G.; Doonan, J.; Jenuwein, T.; Goodrich, J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006, 25, 4638–4649. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Al-Abdallat, A.; Ndamukong, I.; Alvarez-Venegas, R.; Avramova, Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 2007, 35, 6290–6296. [Google Scholar] [CrossRef] [Green Version]
- Hennig, L.; Derkacheva, M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009, 25, 414–423. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis Homolog of Trithorax, Activates Flower Homeotic Genes. Curr. Biol. 2003, 13, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a re-pressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Howard, M.; Dean, C. Antagonistic Roles for H3K36me3 and H3K27me3 in the Cold-Induced Epigenetic Switch at Arabidopsis FLC. Curr. Biol. 2014, 24, 1793–1797. [Google Scholar] [CrossRef] [Green Version]
- Pien, S.; Fleury, D.; Mylne, J.S.; Crevillen, P.; Inzé, D.; Avramova, Z.; Dean, C.; Grossniklaus, U. ARABIDOPSIS TRITHORAX1 Dynamically Regulates FLOWERING LOCUS C Activation via Histone 3 Lysine 4 Trimethylation. Plant Cell 2008, 20, 580–588. [Google Scholar] [CrossRef] [Green Version]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2010, 331, 76–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312.e4. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposi-tion at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, 7246–7270. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-W.; Zhao, N.; Su, Y.-N.; Chen, S.-S.; He, X.-J. Exogenously overexpressed intronic long noncoding RNAs activate host gene expression by affecting histone modification in Arabidopsis. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Luo, X.; Sun, F.; Hu, J.; Zha, X.; Su, W.; Yang, J. Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Sieburth, L.E.; Meyerowitz, E.M. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 1997, 9, 355–365. [Google Scholar]
- Deyholos, M.K.; Sieburth, L.E. Separable whorl-specific expression and negative regulation by enhancer ele-ments within the AGAMOUS second intron. Plant Cell 2000, 12, 1799–1810. [Google Scholar] [CrossRef] [Green Version]
- Busch, W.; Miotk, A.; Ariel, F.D.; Zhao, Z.; Forner, J.; Daum, G.; Suzaki, T.; Schuster, C.; Schultheiss, S.J.; Leibfried, A.; et al. Transcriptional Control of a Plant Stem Cell Niche. Dev. Cell 2010, 18, 841–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.-W.; Deng, S.; Xu, H.; Mao, H.-Z.; Liu, J.; Niu, Q.-W.; Wang, H.; Chua, N.-H. A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 2018, 219, 1480–1491. [Google Scholar] [CrossRef] [Green Version]
- Veluchamy, A.; Jégu, T.; Ariel, F.; Latrasse, D.; Mariappan, K.G.; Kim, S.-K.; Crespi, M.; Hirt, H.; Bergounioux, C.; Raynaud, C.; et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS ONE 2016, 11, e0158936. [Google Scholar] [CrossRef] [Green Version]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcrip-tion by alternative rna polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef] [PubMed]
- Roulé, T.; Ariel, F.; Hartmann, C.; Crespi, M.; Blein, T. The lncRNA MARS modulates the epigenetic reprogram-ming of the marneral cluster in response to ABA. BioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.08.10.236562v1.full (accessed on 26 January 2021). [CrossRef]
- Blein, T.; Balzergue, C.; Roulé, T.; Gabriel, M.; Scalisi, L.; François, T.; Sorin, C.; Christ, A.; Godon, C.; Delannoy, E.; et al. Landscape of the Noncoding Transcriptome Response of Two Arabidopsis Ecotypes to Phosphate Starvation. Plant. Physiol. 2020, 183, 1058–1072. [Google Scholar] [CrossRef]
- Kindgren, P.; Ivanov, M.; Marquardt, S. Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis. Nucleic Acids Res. 2020, 48, 2332–2347. [Google Scholar] [CrossRef] [Green Version]
- Kindgren, P.; Ard, R.; Ivanov, M.; Marquardt, S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nat. Cell Biol. 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Kortmann, J.F.; Narberhaus, F. Bacterial RNA thermometers: Molecular zippers and switches. Nat. Rev. Genet. 2012, 10, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Watters, K.E.; Yu, A.M.; Strobel, E.J.; Settle, A.H.; Lucks, J.B. Characterizing RNA structures in vitro and in vivo with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Methods 2016, 103, 34–48. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonouni-Farde, C.; Ariel, F.; Crespi, M. Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations. Non-Coding RNA 2021, 7, 12. https://doi.org/10.3390/ncrna7010012
Fonouni-Farde C, Ariel F, Crespi M. Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations. Non-Coding RNA. 2021; 7(1):12. https://doi.org/10.3390/ncrna7010012
Chicago/Turabian StyleFonouni-Farde, Camille, Federico Ariel, and Martin Crespi. 2021. "Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations" Non-Coding RNA 7, no. 1: 12. https://doi.org/10.3390/ncrna7010012
APA StyleFonouni-Farde, C., Ariel, F., & Crespi, M. (2021). Plant Long Noncoding RNAs: New Players in the Field of Post-Transcriptional Regulations. Non-Coding RNA, 7(1), 12. https://doi.org/10.3390/ncrna7010012




