The Role of LncRNAs in Translation
Abstract
1. Introduction
2. An Overview of the Characteristics of LncRNAs
3. Regulatory Functions of LncRNAs Depending on Their Subcellular Location
3.1. Cytoplasmic LncRNAs
3.2. Nuclear LncRNAs
4. Acting Mechanisms of LncRNAs in the Regulation of Translation
4.1. Overview of Protein Translation Process
4.2. Regulation of Translational Factors by LncRNAs
4.2.1. Inhibitory Roles of LncRNAs in Translation through Regulation of Translation Factors
4.2.2. LncRNAs Positively Regulate Protein Translation
4.3. LncRNAs Involved in Signaling Pathways Regulating Protein Translation
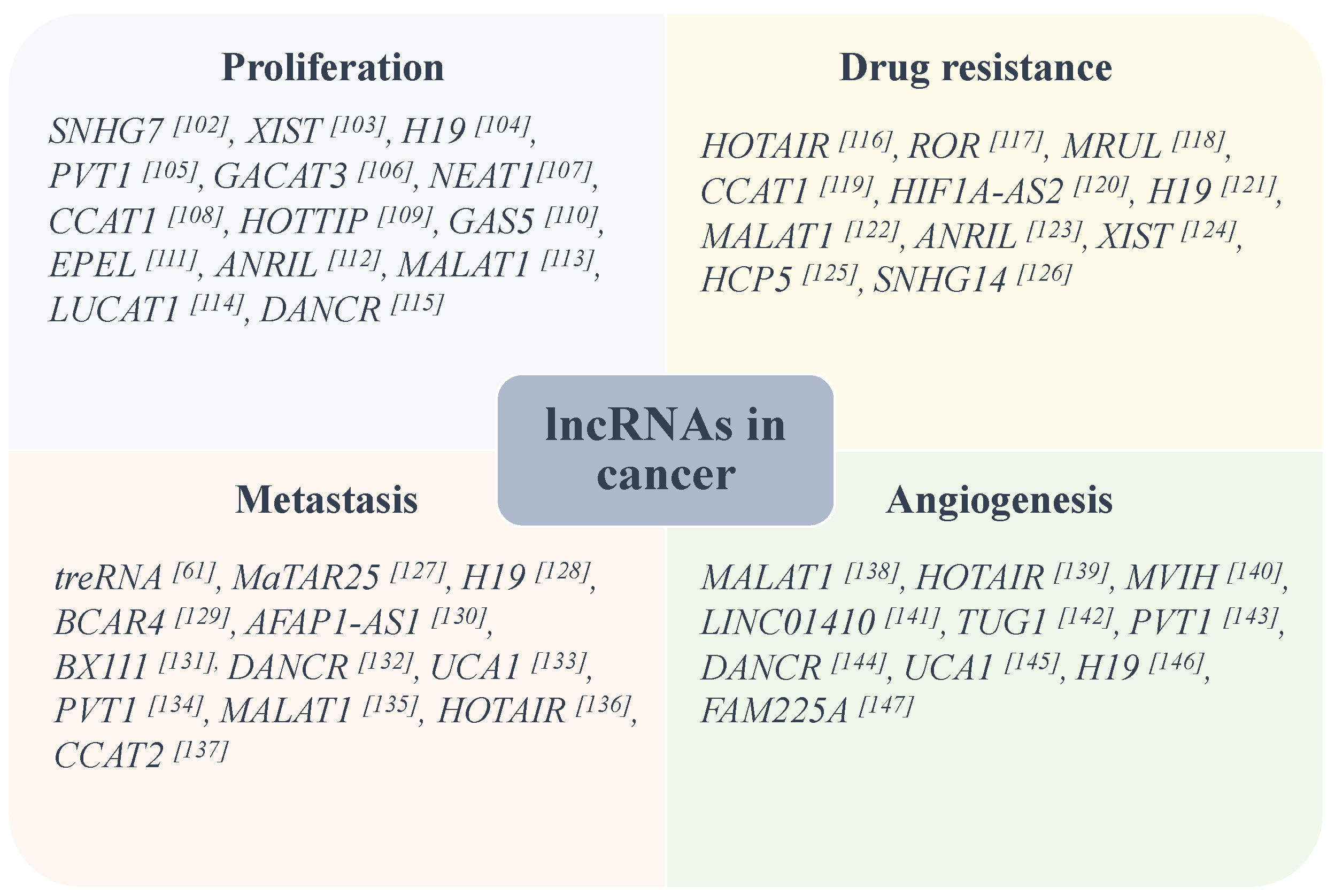
4.4. LncRNAs in Cancer
4.4.1. LncRNAs Can Contribute Hallmarks of Cancer
4.4.2. The Functions of LncRNAs in Regulating Translation of Cancer-Related Proteins
5. Conclusions
Funding
Conflicts of Interest
References
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Aillaud, M.; Schulte, L.N. Emerging Roles of Long Noncoding RNAs in the Cytoplasmic Milieu. ncRNA 2020, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Böhmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef]
- Connerty, P.; Lock, R.B.; de Bock, C.E. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front. Oncol. 2020, 10, 285. [Google Scholar] [CrossRef]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [PubMed]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef]
- Struhl, K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007, 14, 103–105. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.N.; Antonangeli, F. LncRNAs: New Players in Apoptosis Control. Int. J. Cell Biol. 2014, 2014, 473857. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H. Crosstalk of lncRNA and Cellular Metabolism and Their Regulatory Mechanism in Cancer. Int. J. Mol. Sci. 2020, 21, 2947. [Google Scholar] [CrossRef] [PubMed]
- Chew, G.L.; Pauli, A.; Rinn, J.L.; Regev, A.; Schier, A.F.; Valen, E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 2013, 140, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Aspden, J.L.; Eyre-Walker, Y.C.; Phillips, R.J.; Amin, U.; Mumtaz, M.A.; Brocard, M.; Couso, J.P. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 2014, 3, e03528. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long noncoding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef] [PubMed]
- Van Heesch, S.; van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; de Bruijn, E.; Hao, W.; Macinnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef]
- Mackowiak, S.D.; Zauber, H.; Bielow, C.; Thiel, D.; Kutz, K.; Calviello, L.; Mastrobuoni, G.; Rajewsky, N.; Kempa, S.; Selbach, M.; et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Yeasmin, F.; Yada, T.; Akimitsu, N. Micropeptides Encoded in Transcripts Previously Identified as Long Noncoding RNAs: A New Chapter in Transcriptomics and Proteomics. Front. Genet. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- López-Urrutia, E.; Bustamante Montes, L.P.; Ladrón de Guevara Cervantes, D.; Pérez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the Cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef]
- Singh, D.K.; Prasanth, K.V. Functional insights into the role of nuclear-retained long noncoding RNAs in gene expression control in mammalian cells. Chromosome Res. 2013, 21, 695–711. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St-Laurent, G., 3rd; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, L.; Li, J.; Le, T.D. LncmiRSRN: Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics 2018, 34, 4232–4240. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Li, W.; Zhai, L.; Wang, H.; Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef]
- Xiao, J.; Lin, L.; Luo, D.; Shi, L.; Chen, W.; Fan, H.; Li, Z.; Ma, X.; Ni, P.; Yang, L.; et al. Long noncoding RNA TRPM2-AS acts as a microRNA sponge of miR-612 to promote gastric cancer progression and radioresistance. Oncogenesis 2020, 9, 29. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef]
- Hu, G.; Lou, Z.; Gupta, M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef]
- Cao, L.; Zhang, P.; Li, J.; Wu, M. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife 2017, 6, e30433. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, H.; Mei, Y.; Wu, M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell. 2014, 53, 88–100. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; Huang, G.; Ye, B.; Liu, B.; Wu, J.; Du, Y.; He, L.; Fan, Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat. Struct. Mol. Biol. 2016, 23, 631–639. [Google Scholar] [CrossRef]
- Bergmann, J.H.; Spector, D.L. Long non-coding RNAs: Modulators of nuclear structure and function. Curr. Opin. Cell Biol. 2014, 26, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Quinodoz, S.; Guttman, M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014, 24, 651–663. [Google Scholar] [CrossRef]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb. Perspect. Med. 2017, 7, a026492. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Dever, T.E. Gene-specific regulation by general translation factors. Cell 2002, 108, 545–556. [Google Scholar] [CrossRef]
- Ali, M.U.; Ur Rahman, M.S.; Jia, Z.; Jiang, C. Eukaryotic translation initiation factors and cancer. Tumor Biol. 2017, 39, 1010428317709805. [Google Scholar] [CrossRef] [PubMed]
- Grifo, J.A.; Tahara, S.M.; Morgan, M.A.; Shatkin, A.J.; Merrick, W.C. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 1983, 258, 5804–5810. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011, 75, 434–467. [Google Scholar] [CrossRef] [PubMed]
- Llácer, J.L.; Hussain, T.; Saini, A.K.; Nanda, J.S.; Kaur, S.; Gordiyenko, Y.; Kumar, R.; Hinnebusch, A.G.; Lorsch, J.R.; Ramakrishnan, V. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. eLife 2018, 7, e39273. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Ramakrishnan, V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013, 82, 203–236. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; Gromadski, K.B.; Kothe, U.; Wieden, H.J. Recognition and selection of tRNA in translation. FEBS Lett. 2005, 579, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Stansfield, I.; Jones, K.M.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Poznyakovski, A.I.; Paushkin, S.V.; Nierras, C.R.; Cox, B.S.; Ter-Avanesyan, M.D.; Tuite, M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995, 14, 4365–4373. [Google Scholar] [CrossRef]
- Zhouravleva, G.; Frolova, L.; Le Goff, X.; Le Guellec, R.; Inge-Vechtomov, S.; Kisselev, L.; Philippe, M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995, 14, 4065–4072. [Google Scholar] [CrossRef]
- Alkalaeva, E.Z.; Pisarev, A.V.; Frolova, L.Y.; Kisselev, L.L.; Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 2006, 125, 1125–1136. [Google Scholar] [CrossRef]
- Salas-Marco, J.; Bedwell, D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell Biol. 2004, 24, 7769–7778. [Google Scholar] [CrossRef]
- Frolova, L.; Le Goff, X.; Zhouravleva, G.; Davydova, E.; Philippe, M.; Kisselev, L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 1996, 2, 334–341. [Google Scholar]
- Jia, X.; Shi, L.; Wang, X.; Luo, L.; Ling, L.; Yin, J.; Song, Y.; Zhang, Z.; Qiu, N.; Liu, H.; et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019, 10, 373. [Google Scholar] [CrossRef]
- Hu, G.; Witzig, T.E.; Gupta, M. Novel Long Non-Coding RNA, SNHG4 Complex With Eukaryotic Initiation Factor-4E and Regulate Aberrant Protein Translation In Mantle Cell Lymphoma: Implications For Novel Biomarker. Blood 2013, 122, 81. [Google Scholar] [CrossRef]
- Gumireddy, K.; Li, A.; Yan, J.; Setoyama, T.; Johannes, G.J.; Orom, U.A.; Tchou, J.; Liu, Q.; Zhang, L.; Speicher, D.W.; et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013, 32, 2672–2684. [Google Scholar] [CrossRef]
- Wang, H.; Iacoangeli, A.; Popp, S.; Muslimov, I.A.; Imataka, H.; Sonenberg, N.; Lomakin, I.B.; Tiedge, H. Dendritic BC1 RNA: Functional role in regulation of translation initiation. J. Neurosci. 2002, 22, 10232–10241. [Google Scholar] [CrossRef]
- Wang, H.; Iacoangeli, A.; Lin, D.; Williams, K.; Denman, R.B.; Hellen, C.U.; Tiedge, H. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 2005, 171, 811–821. [Google Scholar] [CrossRef]
- Gu, H.; Chen, J.; Song, Y.; Shao, H. Gastric Adenocarcinoma Predictive Long Intergenic Non-Coding RNA Promotes Tumor Occurrence and Progression in Non-Small Cell Lung Cancer via Regulation of the miR-661/eEF2K Signaling Pathway. Cell Physiol. Biochem. 2018, 51, 2136–2147. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.F.; Liu, J.Y.; Mao, X.Y.; He, Z.W.; Zhu, T.; Wang, Z.B.; Li, X.; Yin, J.Y.; Zhang, W.; Zhou, H.H.; et al. LncRNA FOXD1-AS1 acts as a potential oncogenic biomarker in glioma. CNS Neurosci. Ther. 2020, 26, 66–75. [Google Scholar] [CrossRef]
- Park, S.A.; Kim, L.K.; Kim, Y.T.; Heo, T.H.; Kim, H.J. Long non-coding RNA steroid receptor activator promotes the progression of endometrial cancer via Wnt/β-catenin signaling pathway. Int. J. Biol. Sci. 2020, 16, 99–115. [Google Scholar] [CrossRef]
- Guo, C.; Gong, M.; Li, Z. Knockdown of lncRNA MCM3AP-AS1 Attenuates Chemoresistance of Burkitt Lymphoma to Doxorubicin Treatment via Targeting the miR-15a/EIF4E Axis. Cancer Manag. Res. 2020, 12, 5845–5855. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Che, J.; Gu, Y.; Song, L. LncRNA SNHG12 promotes proliferation and migration of vascular smooth muscle cells via targeting miR-766-5p/EIF5A. Res. Square 2020. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef]
- Janku, F.; Yap, T.A.; Meric-Bernstam, F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018, 15, 273–291. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 2010, 1802, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef]
- Xu, F.; Na, L.; Li, Y.; Chen, L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 2020, 10, 54. [Google Scholar] [CrossRef]
- Morrison, D.K. MAP kinase pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a011254. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Heesom, K.J.; Denton, R.M. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS Lett. 1999, 457, 489–493. [Google Scholar] [CrossRef]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef]
- Browne, G.J.; Proud, C.G. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol. Cell. Biol. 2004, 24, 2986–2997. [Google Scholar] [CrossRef]
- Knebel, A.; Morrice, N.; Cohen, P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 2001, 20, 4360–4369. [Google Scholar] [CrossRef] [PubMed]
- Knebel, A.; Haydon, C.E.; Morrice, N.; Cohen, P. Stress-induced regulation of eEF2 kinase by SB203580-sensitive and -insensitive pathways. Biochem. J. 2002, 367, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wu, P.; Su, M.; Ling, H.; Khoshaba, R.; Huang, C.; Gao, H.; Zhao, Y.; Chen, J.; Liao, Q.; et al. Long non-coding RNA UASR1 promotes proliferation and migration of breast cancer cells through the AKT/mTOR pathway. J. Cancer 2019, 10, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.E.; Chen, Y.; Zhang, G.; Xu, L.; Ge, W.; Wu, B. LncRNA H19 regulates PI3K-Akt signal pathway by functioning as a ceRNA and predicts poor prognosis in colorectal cancer: Integrative analysis of dysregulated ncRNA-associated ceRNA network. Cancer Cell Int. 2019, 19, 148. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.R.; Yan, L.; Liu, Y.T.; Cao, L.; Guo, Y.H.; Zhang, Y.; Yao, H.; Cai, L.; Shang, H.B.; Rui, W.W.; et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 2018, 9, 4624. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, D.; Liu, H.; Yang, K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019, 10, 41. [Google Scholar] [CrossRef]
- Gao, X.; Wang, N.; Wu, S.; Cui, H.; An, X.; Yang, Y. Long non-coding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol. Med. Rep. 2019, 20, 182–190. [Google Scholar] [CrossRef]
- Malakar, P.; Stein, I.; Saragovi, A.; Winkler, R.; Stern-Ginossar, N.; Berger, M.; Pikarsky, E.; Karni, R. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res. 2019, 79, 2480–2493. [Google Scholar] [CrossRef]
- Jiang, W.; Kai, J.; Li, D.; Wei, Z.; Wang, Y.; Wang, W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J. Cell Physiol. 2020, 235, 7194–7203. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, J.; Hou, L.; Wang, G.; Liu, H.; Zhang, R.; Chen, X.; Zhu, J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2017, 36, 194. [Google Scholar] [CrossRef]
- Li, C.; Liang, G.; Yang, S.; Sui, J.; Wu, W.; Xu, S.; Ye, Y.; Shen, B.; Zhang, X.; Zhang, Y. LncRNA-LOC101928316 contributes to gastric cancer progression through regulating PI3K-Akt-mTOR signaling pathway. Cancer Med. 2019, 8, 4428–4440. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Z. Long noncoding RNA UCA1 facilitates cell proliferation and inhibits apoptosis in retinoblastoma by activating the PI3K/Akt pathway. Transl. Cancer Res. 2020, 9. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, B.; Mao, Y.; Zhang, H.; Hong, W. Long non-coding RNA OECC promotes cell proliferation and metastasis through the PI3K/Akt/mTOR signaling pathway in human lung cancer. Oncol. Lett. 2019, 18, 3017–3024. [Google Scholar] [CrossRef]
- Liu, J.; Chen, M.; Ma, L.; Dang, X.; Du, G. LncRNA GAS5 Suppresses the Proliferation and Invasion of Osteosarcoma Cells via the miR-23a-3p/PTEN/PI3K/AKT Pathway. Cell Transplant. 2020, 29, 963689720953093. [Google Scholar] [CrossRef]
- Wang, M.R.; Fang, D.; Di, M.P.; Guan, J.L.; Wang, G.; Liu, L.; Sheng, J.Q.; Tian, D.A.; Li, P.Y. Long non-coding RNA LINC01503 promotes the progression of hepatocellular carcinoma via activating MAPK/ERK pathway. Int. J. Med. Sci. 2020, 17, 1224–1234. [Google Scholar] [CrossRef]
- Fang, K.; Hu, C.; Zhang, X.; Hou, Y.; Gao, D.; Guo, Z.; Li, L. LncRNA ST8SIA6-AS1 promotes proliferation, migration and invasion in breast cancer through the p38 MAPK signalling pathway. Carcinogenesis 2020, 41, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.; Jin, X.; Zhang, L. LncRNA FENDRR Upregulation Promotes Hepatic Carcinoma Cells Apoptosis by Targeting miR-362-5p Via NPR3 and p38-MAPK Pathway. Cancer Biother. Radiopharm. 2020, 35, 629–639. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar] [CrossRef]
- Zhou, W.; Chen, L.; Li, C.; Huang, R.; Guo, M.; Ning, S.; Ji, J.; Guo, X.; Lou, G.; Jia, X.; et al. The multifaceted roles of long noncoding RNAs in pancreatic cancer: An update on what we know. Cancer Cell Int. 2020, 20, 41. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.K.; Peng, Y.; He, W.; Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 2020, 19, 77. [Google Scholar] [CrossRef]
- Siddiqui, H.; Al-Ghafari, A.; Choudhry, H.; Al Doghaither, H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A Review. Mol. Clin. Oncol. 2019, 11, 167–172. [Google Scholar] [CrossRef]
- Gao, J.; Wang, F.; Wu, P.; Chen, Y.; Jia, Y. Aberrant LncRNA Expression in Leukemia. J. Cancer 2020, 11, 4284–4296. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Fan, J.; Ma, Y.; Zhou, Y.; Qin, K.; Shi, M.; Yang, J. LncRNA SNHG7 promotes pancreatic cancer proliferation through ID4 by sponging miR-342-3p. Cell Biosci. 2019, 9, 28. [Google Scholar] [CrossRef]
- Wei, W.; Liu, Y.; Lu, Y.; Yang, B.; Tang, L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J. Cell Biochem. 2017, 118, 3349–3358. [Google Scholar] [CrossRef] [PubMed]
- Berteaux, N.; Lottin, S.; Monté, D.; Pinte, S.; Quatannens, B.; Coll, J.; Hondermarck, H.; Curgy, J.J.; Dugimont, T.; Adriaenssens, E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 2005, 280, 29625–29636. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Y.; Sang, Y.; Yu, B.; Lv, D.; Zhang, W.; Feng, H. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene 2018, 37, 4723–4734. [Google Scholar] [CrossRef]
- Zhong, H.; Yang, J.; Zhang, B.; Wang, X.; Pei, L.; Zhang, L.; Lin, Z.; Wang, Y.; Wang, C. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR-497/CCND2. Cancer Biomark. 2018, 22, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, W.B.; Wang, Z.W.; Wang, X.H. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1020–1026. [Google Scholar]
- You, Z.; Liu, C.; Wang, C.; Ling, Z.; Wang, Y.; Wang, Y.; Zhang, M.; Chen, S.; Xu, B.; Guan, H.; et al. LncRNA CCAT1 Promotes Prostate Cancer Cell Proliferation by Interacting with DDX5 and MIR-28-5P. Mol. Cancer Ther. 2019, 18, 2469–2479. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Gao, G.; Wang, Z.; Sun, D.; Wei, X.; Ma, Y.; Ding, Y. Long non-coding RNA HOTTIP promotes prostate cancer cells proliferation and migration by sponging miR-216a-5p. Biosci. Rep. 2018, 38, BSR20180566. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, X.; Kong, Z.; Fu, F.; Zhang, P.; Wang, D.; Wu, H.; Wan, X.; Li, Y. An androgen reduced transcript of LncRNA GAS5 promoted prostate cancer proliferation. PLoS ONE 2017, 12, e0182305. [Google Scholar] [CrossRef]
- Park, S.M.; Choi, E.Y.; Bae, D.H.; Sohn, H.A.; Kim, S.Y.; Kim, Y.J. The LncRNA EPEL Promotes Lung Cancer Cell Proliferation Through E2F Target Activation. Cell Physiol. Biochem. 2018, 45, 1270–1283. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.Q.; Sun, M.; Yang, J.S.; Xie, M.; Xu, T.P.; Xia, R.; Liu, Y.W.; Liu, X.H.; Zhang, E.B.; Lu, K.H.; et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef]
- Feng, C.; Zhao, Y.; Li, Y.; Zhang, T.; Ma, Y.; Liu, Y. LncRNA MALAT1 Promotes Lung Cancer Proliferation and Gefitinib Resistance by Acting as a miR-200a Sponge. Arch. Bronconeumol. 2019, 55, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Li, L.; Bai, Y.; Yu, B.; Xie, C.; Wu, H.; Zhang, Y.; Huang, L.; Yan, Y.; Li, X.; et al. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing Nucleolin to regulate MYC expression. Cell Death Dis. 2020, 11, 908. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Z.; Wang, N.; Feng, J.; Zhang, J.; Luan, L.; Zhao, W.; Zeng, X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp. Mol. Med. 2018, 50, 1–17. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, M.; Lu, K.; Liu, J.; Zhang, M.; Wu, W.; De, W.; Wang, Z.; Wang, R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS ONE 2013, 8, e77293. [Google Scholar]
- Pan, Y.; Chen, J.; Tao, L.; Zhang, K.; Wang, R.; Chu, X.; Chen, L. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget 2017, 8, 33144–33158. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Wu, K.; Zhao, Q.; Nie, Y.; Fan, D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol. Cell Biol. 2014, 34, 3182–3193. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, K.; Song, H.; Wang, R.; Chu, X.; Chen, L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget 2016, 7, 62474–62489. [Google Scholar] [CrossRef]
- Jiang, Y.Z.; Liu, Y.R.; Xu, X.E.; Jin, X.; Hu, X.; Yu, K.D.; Shao, Z.M. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016, 76, 2105–2114. [Google Scholar] [CrossRef]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef]
- Lan, W.G.; Xu, D.H.; Xu, C.; Ding, C.L.; Ning, F.L.; Zhou, Y.L.; Ma, L.B.; Liu, C.M.; Han, X. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol. Rep. 2016, 36, 263–270. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, R.; Yang, D.; Li, J.; Yan, X.; Jin, K.; Li, W.; Liu, X.; Zhao, J.; Shang, W.; et al. Knockdown of Long Non-Coding RNA XIST Inhibited Doxorubicin Resistance in Colorectal Cancer by Upregulation of miR-124 and Downregulation of SGK1. Cell Physiol. Biochem. 2018, 51, 113–128. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Dong, L.; Xia, L.; Zhu, H.; Li, Z.; Yu, X. Long Noncoding RNA HCP5 Regulates Pancreatic Cancer Gemcitabine (GEM) Resistance By Sponging Hsa-miR-214-3p To Target HDGF. Oncol. Ther. 2019, 12, 8207–8216. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, P.; Wang, C.; Xin, B. SNHG14 enhances gemcitabine resistance by sponging miR-101 to stimulate cell autophagy in pancreatic cancer. Biochem. Biophys. Res. Commun. 2019, 510, 508–514. [Google Scholar] [CrossRef]
- Chang, K.C.; Diermeier, S.D.; Yu, A.T.; Brine, L.D.; Russo, S.; Bhatia, S.; Alsudani, H.; Kostroff, K.; Bhuiya, T.; Brogi, E.; et al. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat. Commun. 2020, 11, 6438. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, W.; Yuan, Y.; Li, J.; Wu, J.; Yu, J.; He, Y.; Wei, Z.; Zhang, C. Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1. J. Exp. Clin. Cancer Res. 2020, 39, 141. [Google Scholar] [CrossRef]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef]
- Leng, X.; Ding, X.; Wang, S.; Fang, T.; Shen, W.; Xia, W.; You, R.; Xu, K.; Yin, R. Long noncoding RNA AFAP1-AS1 is upregulated in NSCLC and associated with lymph node metastasis and poor prognosis. Oncol. Lett. 2018, 16, 727–732. [Google Scholar] [CrossRef]
- Deng, S.J.; Chen, H.Y.; Ye, Z.; Deng, S.C.; Zhu, S.; Zeng, Z.; He, C.; Liu, M.L.; Huang, K.; Zhong, J.X.; et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene 2018, 37, 5811–5828. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, Q.; Teng, L.; Zhang, J.; Song, J.; Bo, W.; Liu, D.; He, Y.; Tan, A. LncRNA DANCR promotes proliferation and metastasis in pancreatic cancer by regulating miRNA-33b. FEBS Open Bio 2020, 10, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, Y.; Zhang, Y.; Wang, D.; Gu, S.; Feng, W.; Peng, W.; Gong, A.; Xu, M. LncRNA UCA1 promotes migration and invasion in pancreatic cancer cells via the Hippo pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1770–1782. [Google Scholar] [CrossRef]
- Sun, F.; Wu, K.; Yao, Z.; Mu, X.; Zheng, Z.; Sun, M.; Wang, Y.; Liu, Z.; Zhu, Y. Long Noncoding RNA PVT1 Promotes Prostate Cancer Metastasis by Increasing NOP2 Expression via Targeting Tumor Suppressor MicroRNAs. OncoTargets Ther. 2020, 13, 6755–6765. [Google Scholar] [CrossRef]
- Yang, M.H.; Hu, Z.Y.; Xu, C.; Xie, L.Y.; Wang, X.Y.; Chen, S.Y.; Li, Z.G. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim. Biophys. Acta 2015, 1852, 166–174. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Tee, A.E.; Liu, B.; Song, R.; Li, J.; Pasquier, E.; Cheung, B.B.; Jiang, C.; Marshall, G.M.; Haber, M.; Norris, M.D.; et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget 2016, 7, 8663–8675. [Google Scholar] [CrossRef]
- Fu, W.M.; Lu, Y.F.; Hu, B.G.; Liang, W.C.; Zhu, X.; Yang, H.D.; Li, G.; Zhang, J.F. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef]
- Yuan, S.X.; Yang, F.; Yang, Y.; Tao, Q.F.; Zhang, J.; Huang, G.; Yang, Y.; Wang, R.Y.; Yang, S.; Huo, X.S.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef]
- Zhang, J.X.; Chen, Z.H.; Chen, D.L.; Tian, X.P.; Wang, C.Y.; Zhou, Z.W.; Gao, Y.; Xu, Y.; Chen, C.; Zheng, Z.S.; et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene 2018, 37, 2660–2675. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, X.; Zheng, J.; Xue, Y.; Ma, J.; Li, Z.; Xi, Z.; Li, Z.; Bao, M.; Liu, Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene 2017, 36, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Du, P.; Cui, P.; Qin, Y.; Hu, C.; Wu, J.; Zhou, Z.; Zhang, W.; Qin, L.; Huang, G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018, 37, 4094–4109. [Google Scholar] [CrossRef]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Mol. Carcinog. 2019, 58, 2286–2296. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, X.; Yang, Y.; Chen, W.; Zhang, K.; Teng, B.; Huang, C.; Zhao, Q.; Qiu, Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol. Ther. Nucl. Acids 2020, 22, 179–195. [Google Scholar] [CrossRef]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016, 124, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Luo, Y.; Cao, J.; Wang, X.; Miao, Z.; Shao, G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020, 9, 8600–8611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, Z.; Zou, Y. LncRNA NEAT1 exacerbates non-small cell lung cancer by upregulating EIF4G2 via miR-582-5p sponging. Arch. Biol. Sci. 2020, 72, 243–252. [Google Scholar] [CrossRef]
- Fang, Q.L.; Zhou, J.Y.; Xiong, Y.; Xiong, Y.; Xie, C.R.; Wang, F.Q.; Li, Y.T.; Yin, Z.Y.; Luo, G.H. Long non-coding RNA RP11-284P20.2 promotes cell proliferation and invasion in hepatocellular carcinoma by recruiting EIF3b to induce c-met protein synthesis. Biosci. Rep. 2020, 40, BSR20200297. [Google Scholar] [CrossRef]
- Hu, M.; Yang, J. Down-regulation of lncRNA UCA1 enhances radiosensitivity in prostate cancer by suppressing EIF4G1 expression via sponging miR-331-3p. Cancer Cell Int. 2020, 20, 449. [Google Scholar] [CrossRef]
- Hansji, H.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Cameron-Smith, D.; Figueiredo, V.C.; Askarian-Amiri, M.E. ZFAS1: A long noncoding RNA associated with ribosomes in breast cancer cells. Biol. Direct. 2016, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.Y.; Tee, A.E.; Milazzo, G.; Hannan, K.M.; Maag, J.; Mondal, S.; Atmadibrata, B.; Bartonicek, N.; Peng, H.; Ho, N.; et al. The long noncoding RNA lncNB1 promotes tumorigenesis by interacting with ribosomal protein RPL35. Nat. Commun. 2019, 10, 5026. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ahn, C.; Chun, C.H.; Jin, E.J. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J. Orthop. Res. 2014, 32, 1628–1635. [Google Scholar] [CrossRef] [PubMed]

| LncRNA | Translation Factor | Function | Reference |
|---|---|---|---|
| GAS5 | Binds to eIF4E and prevents formation of initiation complex (eIF4F) | Decreases translation of c-Myc | [37] |
| RP1-5O6.5 | Interacts with eIF4E and prevents binding to eIF4G | Promotes breast cancer metastasis by inhibiting translation of p27Kip1 | [59] |
| SNHG1 and SNGH4 | Bind to eIF4E and dysregulate it | Enhance translation and contribute aggressiveness of lymphoma cells | [60] |
| treRNA | Promotes the formation of a treRNA-associated protein (treRNP) complex and suppresses translation by binding to eEIF4G1 | treRNP complex reduces translation efficiency of E-cadherin and decreases tumor metastasis | [61] |
| BC1 | Interacts with eIF4A and poly(A)-binding protein (PABP) | Represses translation | [62,63] |
| GAPLINC | Positively regulates eEF2K expression by sponging miR-661 | Promotes tumorigenesis of non-small cell lung cancer cells | [64] |
| SRA | Binds and increases the expression of eIF4E-binding protein 1 (eIF4E-BP1) | Increases the activity of Wnt/ β-catenin signaling and promotes aggressive characteristics of endometrial cancer | [66] |
| MCM3AP-AS1 | Positively regulates the expression of eIF4E by using miR15a as a sponge | Promotes translation and contributes doxorubicin resistance | [67] |
| SNGH12 | Binds to miR-766-5p, which is a negative regulator of eIF5A | Targets miR-766-5p/eIF5A axis and enhances invasion of vascular smooth muscle cells | [68] |
| LNC00278 | Decreases eEF2K expression | Micropeptide of lncRNA, YY1BM, represses the eEF2K/eEF2 axis | [69] |
| LncRNA | Target | Function | Reference |
|---|---|---|---|
| MALAT1 | mTOR signaling | Improves glucose metabolism to contribute aggressiveness in hepatocellular carcinoma cells | [87] |
| HOXB-AS3 | PI3K/AKT signaling | Increases proliferation, migration, and invasion of lung cancer cells | [88] |
| AK023391 | PI3K/AKT signaling | Promotes tumorigenesis and invasion of gastric cancer | [89] |
| LOC101928316 | PI3K/AKT/mTOR signaling | Inhibits cell proliferation, invasion and tumorigenesis of gastric cancer cells | [90] |
| UCA1 | PI3K/AKT signaling | Promotes cell proliferation and inhibits apoptosis in retinoblastoma cells | [91] |
| OECC | PI3K/AKT/mTOR signaling | Increases proliferation, migration and invasion of lung cancer cells | [92] |
| GAS5 | PTEN/PI3K/AKT signaling | Suppresses proliferation and invasion of osteosarcoma cells and promotes PTEN expression by sponging miR-23a-3p | [93] |
| LINC01503 | MAPK/ERK signaling | Increases proliferation and tumor forming-ability of hepatocellular carcinoma cells | [94] |
| ST8SIA6-AS1 | p38 MAPK signaling | Promotes proliferation, migration and invasion of breast cancer cells | [95] |
| FENDRR | p38 MAPK signaling | Inhibits cell proliferation and induces apoptosis in hepatocellular carcinoma cells | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karakas, D.; Ozpolat, B. The Role of LncRNAs in Translation. Non-Coding RNA 2021, 7, 16. https://doi.org/10.3390/ncrna7010016
Karakas D, Ozpolat B. The Role of LncRNAs in Translation. Non-Coding RNA. 2021; 7(1):16. https://doi.org/10.3390/ncrna7010016
Chicago/Turabian StyleKarakas, Didem, and Bulent Ozpolat. 2021. "The Role of LncRNAs in Translation" Non-Coding RNA 7, no. 1: 16. https://doi.org/10.3390/ncrna7010016
APA StyleKarakas, D., & Ozpolat, B. (2021). The Role of LncRNAs in Translation. Non-Coding RNA, 7(1), 16. https://doi.org/10.3390/ncrna7010016






