Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far
Abstract
1. Introduction
2. lncRNA Are Critical Regulators of Gene Expression
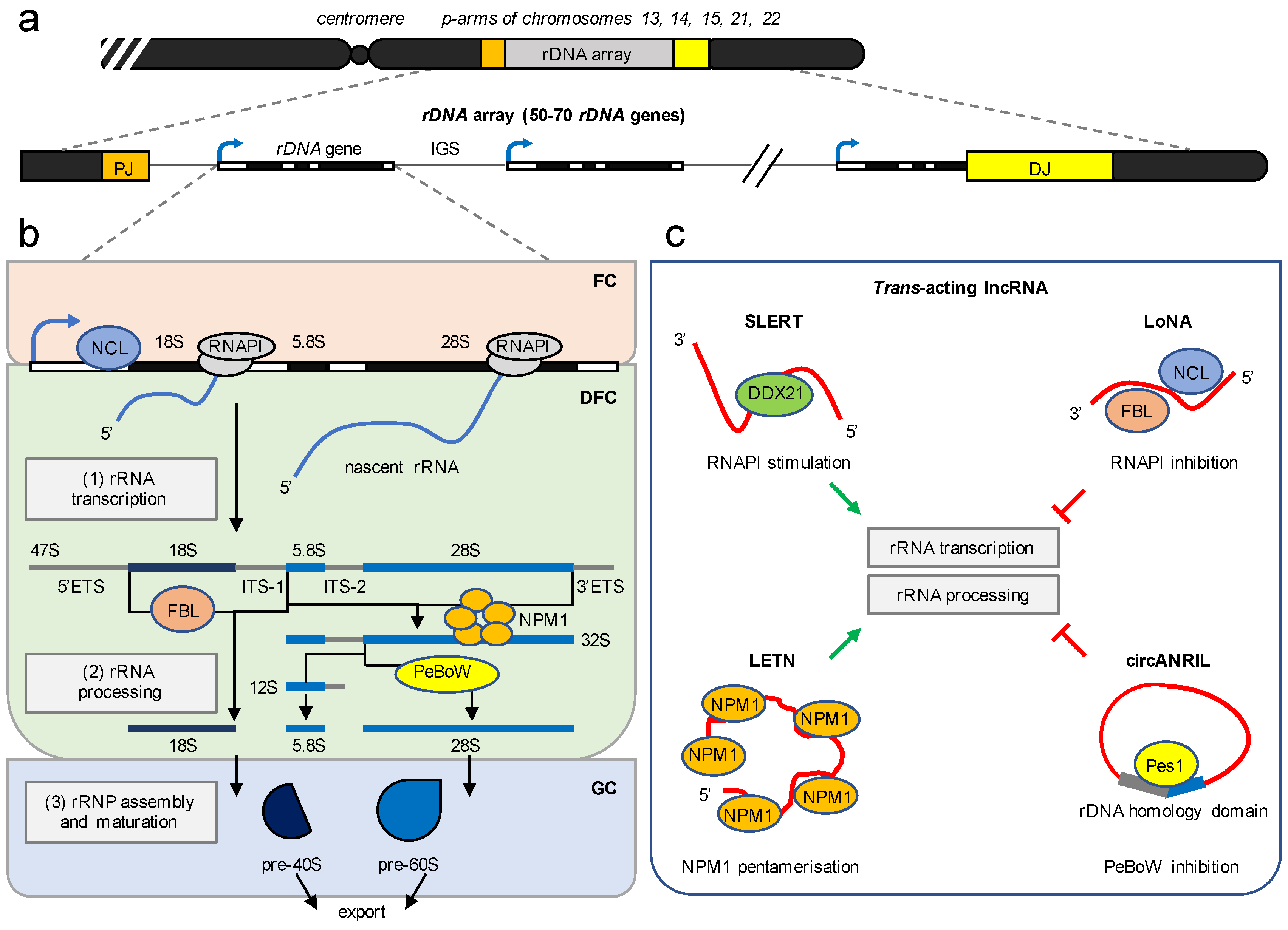
3. The Nucleolus Is a Multifunctional RNA Metabolic Hub
4. lncRNA Modulate Ribosome Biogenesis in Trans
5. lncRNA Modulate Ribosome Biogenesis in Cis
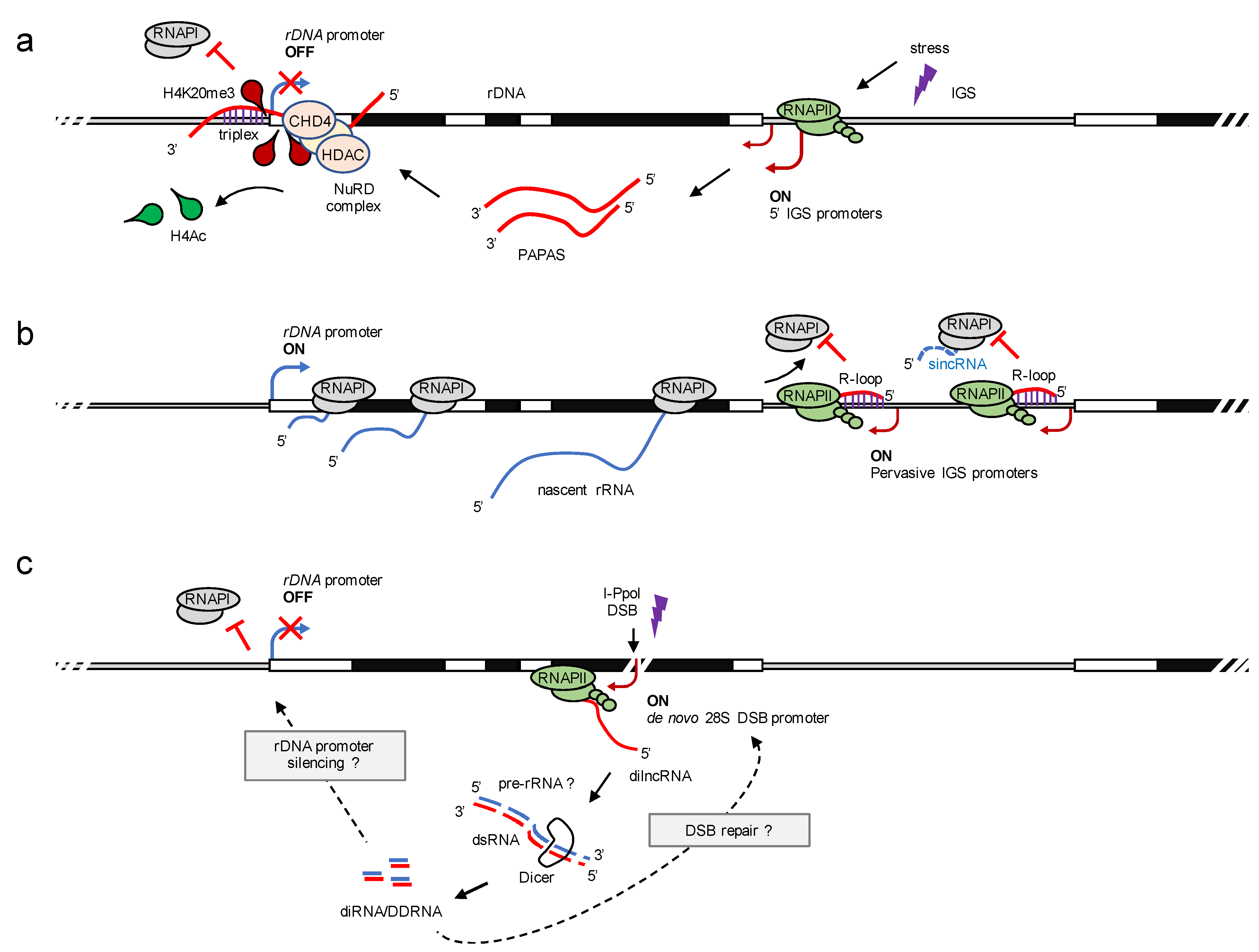
5.1. RNAPI-Dependent Nucleolar lncRNA
5.2. RNAPII-Dependent Nucleolar lncRNA
6. DNA Damage Stimulates Nucleolar RNAPII Transcription
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ezkurdia, I.; Juan, D.; Rodriguez, J.M.; Frankish, A.; Diekhans, M.; Harrow, J.; Vazquez, J.; Valencia, A.; Tress, M.L. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 2014, 23, 5866–5878. [Google Scholar] [CrossRef] [PubMed]
- Hillier, L.W.; Coulson, A.; Murray, J.I.; Bao, Z.; Sulston, J.E.; Waterston, R.H. Genomics in C. elegans: So many genes, such a little worm. Genome Res. 2005, 15, 1651–1660. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563, Erratum in 2006, 311, 1713. [Google Scholar] [CrossRef]
- Kapranov, P.; Cawley, S.E.; Drenkow, J.; Bekiranov, S.; Strausberg, R.L.; Fodor, S.P.; Gingeras, T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science 2002, 296, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef]
- ENCODE Project Consortium; Birney, E.; Stamatoyannopoulos, J.A.; Dutta, A.; Guigó, R.; Gingeras, T.R.; Margulies, E.H.; Weng, Z.; Snyder, M.; Dermitzakis, E.T.; et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118, Erratum in 2021, 22, 159. [Google Scholar] [CrossRef]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 2011, 3, a000638. [Google Scholar] [CrossRef]
- Shaw, P.J.; Jordan, E.G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995, 11, 93–121. [Google Scholar] [CrossRef]
- Van Sluis, M.; McStay, B. Nucleolar reorganization in response to rDNA damage. Curr. Opin. Cell Biol. 2017, 46, 81–86. [Google Scholar] [CrossRef]
- Fatica, A.; Tollervey, D. Making ribosomes. Curr. Opin. Cell Biol. 2002, 14, 313–318. [Google Scholar] [CrossRef]
- Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, R.; Roeder, R.G. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl. Acad. Sci. USA 1974, 71, 1790–1794. [Google Scholar] [CrossRef] [PubMed]
- Kass, S.; Tyc, K.; Steitz, J.A.; Sollner-Webb, B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 1990, 60, 897–908. [Google Scholar] [CrossRef]
- Peculis, B.A.; Steitz, J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 1993, 73, 1233–1245. [Google Scholar] [CrossRef]
- Ginisty, H.; Amalric, F.; Bouvet, P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998, 17, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Mongelard, F.; Bouvet, P. Nucleolin: A multiFACeTed protein. Trends Cell Biol. 2007, 17, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, D.; Lehtonen, H.; Jansen, R.; Kern, H.; Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993, 72, 443–457. [Google Scholar] [CrossRef]
- Guillen-Chable, F.; Corona, U.R.; Pereira-Santana, A.; Bayona, A.; Rodríguez-Zapata, L.C.; Aquino, C.; Šebestová, L.; Vitale, N.; Hozak, P.; Castano, E. Fibrillarin Ribonuclease Activity is Dependent on the GAR Domain and Modulated by Phospholipids. Cells 2020, 9, 1143. [Google Scholar] [CrossRef] [PubMed]
- Sztacho, M.; Šalovská, B.; Červenka, J.; Balaban, C.; Hoboth, P.; Hozák, P. Limited Proteolysis-Coupled Mass Spectrometry Identifies Phosphatidylinositol 4,5-Bisphosphate Effectors in Human Nuclear Proteome. Cells 2021, 10, 68. [Google Scholar] [CrossRef]
- Farley, K.I.; Surovtseva, Y.; Merkel, J.; Baserga, S.J. Determinants of mammalian nucleolar architecture. Chromosoma 2015, 124, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Frankowski, K.J.; Wang, C.; Patnaik, S.; Schoenen, F.J.; Southall, N.; Li, D.; Teper, Y.; Sun, W.; Kandela, I.; Hu, D.; et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci. Transl. Med. 2018, 10, eaap8307. [Google Scholar] [CrossRef] [PubMed]
- Stenström, L.; Mahdessian, D.; Gnann, C.; Cesnik, A.J.; Ouyang, W.; Leonetti, M.D.; Uhlén, M.; Cuylen-Haering, S.; Thul, P.J.; Lundberg, E. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 2020, 16, e9469. [Google Scholar] [CrossRef]
- Lewis, J.D.; Tollervey, D. Like attracts like: Getting RNA processing together in the nucleus. Science 2000, 288, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell. 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Dundr, M.; Misteli, T.; Olson, M.O. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 2000, 150, 433–446. [Google Scholar] [CrossRef]
- Leung, A.; Gerlich, D.W.; Miller, G.; Lyon, C.; Lam, Y.W.; Lleres, D.; Daigle, N.; Zomerdijk, J.; Ellenberg, J.; Lamond, A.I. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004, 166, 787–800. [Google Scholar] [CrossRef]
- Sirri, V.; Hernandez-Verdun, D.; Roussel, P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 2002, 156, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA Polymerase I with an Oral Small Molecule CX-5461 Inhibits Ribosomal RNA Synthesis and Solid Tumor Growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Mars, J.-C.; Tremblay, M.G.; Valere, M.; Sibai, D.S.; Sabourin-Felix, M.; Lessard, F.; Moss, T. The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. NAR Cancer 2020, 2, zcaa032. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic Sorting of Nuclear Components into Distinct Nucleolar Caps during Transcriptional Inhibition. Mol. Biol. Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef]
- Fumagalli, S.; Di Cara, A.; Neb-Gulati, A.; Natt, F.; Schwemberger, S.; Hall, J.; Babcock, G.F.; Bernardi, R.; Pandolfi, P.P.; Thomas, G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009, 11, 501–508. [Google Scholar] [CrossRef]
- Nicolas, E.; Parisot, P.; Pinto-Monteiro, C.; De Walque, R.; De Vleeschouwer, P.P.C.; Lafontaine, D.L. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016, 7, 11390. [Google Scholar] [CrossRef]
- Burger, K.; Eick, D. Functional ribosome biogenesis is a prerequisite for p53 destabilization: Impact of chemotherapy on nucleolar functions and RNA metabolism. Biol. Chem. 2013, 394, 1133–1143. [Google Scholar] [CrossRef]
- Gentilella, A.; Morón-Duran, F.; Fuentes, P.; Zweig-Rocha, G.; Riaño-Canalias, F.; Pelletier, J.; Ruiz, M.; Turón, G.; Castaño, J.; Tauler, A.; et al. Autogenous Control of 5′TOP mRNA Stability by 40S Ribosomes. Mol. Cell 2017, 67, 55–70.e4. [Google Scholar] [CrossRef]
- Weinmann, R.; Raskas, H.J.; Roeder, R.G. Role of DNA-Dependent RNA Polymerases II and III in Transcription of the Adenovirus Genome Late in Productive Infection. Proc. Natl. Acad. Sci. USA 1974, 71, 3426–3430. [Google Scholar] [CrossRef]
- Burger, K.; Mühl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Hölzel, M.; et al. Chemotherapeutic Drugs Inhibit Ribosome Biogenesis at Various Levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef]
- Burger, K.; Mühl, B.; Rohrmoser, M.; Coordes, B.; Heidemann, M.; Kellner, M.; Gruber-Eber, A.; Heissmeyer, V.; Sträßer, K.; Eick, D. Cyclin-dependent Kinase 9 Links RNA Polymerase II Transcription to Processing of Ribosomal RNA. J. Biol. Chem. 2013, 288, 21173–21183. [Google Scholar] [CrossRef]
- Xing, Y.-H.; Yao, R.-W.; Zhang, Y.; Guo, C.-J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.-L. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 2017, 169, 664–678.e16. [Google Scholar] [CrossRef]
- Calo, E.; Flynn, R.; Martin, L.; Spitale, R.C.; Chang, H.Y.; Wysocka, J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nat. Cell Biol. 2015, 518, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Leisegang, M.S.; Doebele, C.; Ramirez, A.S.; Simm, S.; Safferthal, C.; Kretschmer, J.; Schorge, T.; Markoutsa, S.; Haag, S.; et al. The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX. Nucleic Acids Res. 2015, 43, 553–564. [Google Scholar] [CrossRef]
- Wang, X.; Hu, X.; Song, W.; Xu, H.; Xiao, Z.; Huang, R.; Bai, Q.; Zhang, F.; Chen, Y.; Liu, Y.; et al. Mutual dependency between lncRNA LETN and protein NPM1 in controlling the nucleolar structure and functions sustaining cell proliferation. Cell Res. 2021, 31, 664–683. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, e13571. [Google Scholar] [CrossRef] [PubMed]
- Floutsakou, I.; Agrawal, S.; Nguyen, T.; Seoighe, C.; Ganley, A.; McStay, B. The shared genomic architecture of human nucleolar organizer regions. Genome Res. 2013, 23, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Van Sluis, M.; Gailín, M.Ó.; McCarter, J.G.; Mangan, H.; Grob, A.; McStay, B. Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions. Genes Dev. 2019, 33, 1688–1701. [Google Scholar] [CrossRef]
- Häsler, J.; Samuelsson, T.; Strub, K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell. Mol. Life Sci. 2007, 64, 1793–1800. [Google Scholar] [CrossRef]
- Conti, A.; Carnevali, D.; Bollati, V.; Fustinoni, S.; Pellegrini, M.; Dieci, G. Identification of RNA polymerase III-transcribed Alu loci by computational screening of RNA-Seq data. Nucleic Acids Res. 2015, 43, 817–835. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. ALU ternative Regulation for Gene Expression. Trends Cell Biol. 2017, 27, 480–490. [Google Scholar] [CrossRef]
- Caudron-Herger, M.; Pankert, T.; Seiler, J.; Németh, A.; Voit, R.; Grummt, I.; Rippe, K. Alu element-containing RNA s maintain nucleolar structure and function. EMBO J. 2015, 34, 2758–2774. [Google Scholar] [CrossRef]
- Manning, K.S.; Cooper, K.S.M.T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017, 18, 102–114. [Google Scholar] [CrossRef]
- Li, D.; Zhang, J.; Wang, M.; Li, X.; Gong, H.; Tang, H.; Chen, L.; Wan, L.; Liu, Q. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 2018, 9, 1726. [Google Scholar] [CrossRef]
- Pietrzak, M.; Rempala, G.; Nelson, P.T.; Zheng, J.-J.; Hetman, M. Epigenetic Silencing of Nucleolar rRNA Genes in Alzheimer’s Disease. PLoS ONE 2011, 6, e22585. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Sánchez, J.S.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef]
- McRae, E.K.; Booy, E.P.; Moya-Torres, A.; Ezzati, P.; Stetefeld, J.; McKenna, S.A. Human DDX21 binds and unwinds RNA guanine quadruplexes. Nucleic Acids Res. 2017, 45, 6656–6668. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.; Shaw, P.G.; Kim, M.-S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nat. Cell Biol. 2014, 507, 195–200. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Lapik, Y.R.; Fernandes, C.J.; Lau, L.; Pestov, D.G. Physical and Functional Interaction between Pes1 and Bop1 in Mammalian Ribosome Biogenesis. Mol. Cell 2004, 15, 17–29. [Google Scholar] [CrossRef]
- Hölzel, M.; Rohrmoser, M.; Schlee, M.; Grimm, T.; Harasim, T.; Malamoussi, A.; Gruber-Eber, A.; Kremmer, E.; Hiddemann, W.; Bornkamm, G.W.; et al. Mammalian WDR12 is a novel member of the Pes1–Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005, 170, 367–378. [Google Scholar] [CrossRef]
- Rohrmoser, M.; HölzelM.; Grimm, T.; Malamoussi, A.; Harasim, T.; Orban, M.; Pfisterer, I.; Gruber-Eber, A.; Kremmer, E.; Eick, D. Interdependence of Pes1, Bop1, and WDR12 Controls Nucleolar Localization and Assembly of the PeBoW Complex Required for Maturation of the 60S Ribosomal Subunit. Mol. Cell. Biol. 2007, 27, 3682–3694. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.-M.; Van Koningsbruggen, S.; Navascués, J.; Lamond, A. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Körner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef]
- Audas, T.; Jacob, M.D.; Lee, S. Immobilization of Proteins in the Nucleolus by Ribosomal Intergenic Spacer Noncoding RNA. Mol. Cell 2012, 45, 147–157. [Google Scholar] [CrossRef]
- Mekhail, K.; Gunaratnam, L.; Bonicalzi, M.-E.; Lee, S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004, 6, 642–647. [Google Scholar] [CrossRef]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540.e13. [Google Scholar] [CrossRef] [PubMed]
- McStay, B.; Grummt, I. The Epigenetics of rRNA Genes: From Molecular to Chromosome Biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef]
- Mayer, C.; Schmitz, K.-M.; Li, J.; Grummt, I.; Santoro, R. Intergenic Transcripts Regulate the Epigenetic State of rRNA Genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef]
- Schmitz, K.-M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef]
- Bierhoff, H.; Schmitz, K.; Maass, F.; Ye, J.; Grummt, I. Noncoding Transcripts in Sense and Antisense Orientation Regulate the Epigenetic State of Ribosomal RNA Genes. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Grummt, I.; Pikaard, C. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.S.; Boeke, J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997, 11, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Fritze, C.E.; Verschueren, K.; Strich, R.; Esposito, R.E. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997, 16, 6495–6509. [Google Scholar] [CrossRef] [PubMed]
- Bryk, M.; Briggs, S.D.; Strahl, B.D.; Curcio, M.J.; Allis, C.; Winston, F. Evidence that Set1, a Factor Required for Methylation of Histone H3, Regulates rDNA Silencing in S. cerevisiae by a Sir2-Independent Mechanism. Curr. Biol. 2002, 12, 165–170. [Google Scholar] [CrossRef]
- Kobayashi, T.; Horiuchi, T.; Tongaonkar, P.; Vu, L.; Nomura, M. SIR2 Regulates Recombination between Different rDNA Repeats, but Not Recombination within Individual rRNA Genes in Yeast. Cell 2004, 117, 441–453. [Google Scholar] [CrossRef]
- Moazed, D. Common Themes in Mechanisms of Gene Silencing. Mol. Cell 2001, 8, 489–498. [Google Scholar] [CrossRef]
- Rusche, L.N.; Kirchmaier, A.; Rine, J. The Establishment, Inheritance, and Function of Silenced Chromatin inSaccharomyces cerevisiae. Annu. Rev. Biochem. 2003, 72, 481–516. [Google Scholar] [CrossRef]
- Buck, S.W.; Sandmeier, J.J.; Smith, J.S. RNA Polymerase I Propagates Unidirectional Spreading of rDNA Silent Chromatin. Cell 2002, 111, 1003–1014. [Google Scholar] [CrossRef]
- Cioci, F.; Vu, L.; Eliason, K.; Oakes, M.; Siddiqi, I.; Nomura, M. Silencing in Yeast rDNA Chromatin: Reciprocal Relationship in Gene Expression between RNA Polymerase I and II. Mol. Cell 2003, 12, 135–145. [Google Scholar] [CrossRef]
- Conrad-Webb, H.; Butow, R.A. A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.; Siddiqi, I.; Lee, B.-S.; Josaitis, C.A.; Nomura, M. RNA polymerase switch in transcription of yeast rDNA: Role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1999, 96, 4390–4395. [Google Scholar] [CrossRef]
- Li, C.; Mueller, J.E.; Bryk, M. Sir2 Represses Endogenous Polymerase II Transcription Units in the Ribosomal DNA Nontranscribed Spacer. Mol. Biol. Cell 2006, 17, 3848–3859. [Google Scholar] [CrossRef][Green Version]
- Houseley, J.; Kotovic, K.; el Hage, A.; Tollervey, D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007, 26, 4996–5006. [Google Scholar] [CrossRef]
- Gagnon-Kugler, T.; Langlois, F.; Stefanovsky, V.; Lessard, F.; Moss, T. Loss of Human Ribosomal Gene CpG Methylation Enhances Cryptic RNA Polymerase II Transcription and Disrupts Ribosomal RNA Processing. Mol. Cell 2009, 35, 414–425. [Google Scholar] [CrossRef]
- Zhao, Z.; Dammert, M.A.; Hoppe, S.; Bierhoff, H.; Grummt, I. Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 2016, 44, 8144–8152. [Google Scholar] [CrossRef]
- Bierhoff, H.; Dammert, M.A.; Brocks, D.; Dambacher, S.; Schotta, G.; Grummt, I. Quiescence-Induced lncRNAs Trigger H4K20 Trimethylation and Transcriptional Silencing. Mol. Cell 2014, 54, 675–682. [Google Scholar] [CrossRef]
- Zhao, Z.; Sentürk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef]
- Abraham, K.J.; Khosraviani, N.; Chan, J.N.Y.; Gorthi, A.; Samman, A.; Zhao, D.Y.; Wang, M.; Bokros, M.; Vidya, E.; Ostrowski, L.A.; et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nat. Cell Biol. 2020, 585, 298–302. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.-G.; Qi, Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; Hoon, D.; Anelli, V.V.; Mione, M.C.; Carninci, P.; Di Fagagna, F.D. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nat. Cell Biol. 2012, 488, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.; Schlackow, M.; Potts, M.; Hester, S.; Mohammed, S.; Gullerova, M. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J. Cell Biol. 2017, 216, 2373–2389. [Google Scholar] [CrossRef]
- Burger, K.; Schlackow, M.; Gullerova, M. Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks. Nucleic Acids Res. 2019, 47, 3467–3484. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.; Gullerova, M. Nuclear re-localization of Dicer in primary mouse embryonic fibroblast nuclei following DNA damage. PLoS Genet. 2018, 14, e1007151. [Google Scholar] [CrossRef]
- Francia, S.; Cabrini, M.; Matti, V.; Oldani, A.; di Fagagna, F.D. DICER, DROSHA and DNA damage-response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016, 129, 1468–1476. [Google Scholar] [CrossRef]
- Bonath, F.; Prim, J.D.; Tarbier, M.; Friedländer, M.R.; Visa, N. Next-generation sequencing reveals two populations of damage-induced small RNAs at endogenous DNA double-strand breaks. Nucleic Acids Res. 2018, 46, 11869–11882. [Google Scholar] [CrossRef]
- Lee, H.-C.; Chang, S.-S.; Choudhary, S.; Aalto, A.P.; Maiti, M.; Bamford, D.; Liu, Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nat. Cell Biol. 2009, 459, 274–277. [Google Scholar] [CrossRef]
- Montanaro, L.; Treré, D.; Derenzini, M. Nucleolus, Ribosomes, and Cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.S.; Jurada, D.; Bursac, S.; Oršolić, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef]
- Schlosser, I.; Hölzel, M.; Mürnseer, M.; Burtscher, H.; Weidle, U.H.; Eick, D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003, 31, 6148–6156. [Google Scholar] [CrossRef] [PubMed]
- Arabi, A.; Wu, S.; Ridderstråle, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlén, S.; Hydbring, P.; Söderberg, O.; Grummt, I.; et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Pirogov, S.A.; Gvozdev, V.A.; Klenov, M.S. Long Noncoding RNAs and Stress Response in the Nucleolus. Cells 2019, 8, 668. [Google Scholar] [CrossRef]
- Kaliatsi, E.G.; Giarimoglou, N.; Stathopoulos, C.; Stamatopoulou, V. Non-Coding RNA-Driven Regulation of rRNA Biogenesis. Int. J. Mol. Sci. 2020, 21, 9738. [Google Scholar] [CrossRef] [PubMed]
- McCool, M.A.; Bryant, C.; Baserga, S.J. MicroRNAs and long non-coding RNAs as novel regulators of ribosome biogenesis. Biochem. Soc. Trans. 2020, 48, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Khot, A.; Brajanovski, N.; Cameron, D.P.; Hein, N.; Maclachlan, K.H.; Sanij, E.; Lim, J.; Soong, J.; Link, E.; Blombery, P.; et al. First-in-Human RNA Polymerase I Transcription Inhibitor CX-5461 in Patients with Advanced Hematologic Cancers: Results of a Phase I Dose-Escalation Study. Cancer Discov. 2019, 9, 1036–1049. [Google Scholar] [CrossRef]
- Pessina, F.; Giavazzi, F.; Yin, Y.; Gioia, U.; Vitelli, V.; Galbiati, A.; Barozzi, S.; Garre’, M.; Oldani, A.; Flaus, A.; et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat. Cell Biol. 2019, 21, 1286–1299. [Google Scholar] [CrossRef]
- Shore, S.M.; Byers, S.A.; Maury, W.; Price, D.H. Identification of a novel isoform of Cdk. Gene 2003, 307, 175–182. [Google Scholar] [CrossRef]
- Shore, S.M.; Byers, S.A.; Dent, P.; Price, D. Characterization of Cdk955 and differential regulation of two Cdk9 isoforms. Gene 2005, 350, 51–58. [Google Scholar] [CrossRef]
- Liu, H.; Herrmann, C.H.; Chiang, K.; Sung, T.-L.; Moon, S.-H.; Donehower, L.A.; Rice, A.P. 55K isoform of CDK9 associates with Ku70 and is involved in DNA repair. Biochem. Biophys. Res. Commun. 2010, 397, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Flynn, R.A.; Crowe, J.L.; Zhu, Y.; Liang, J.; Jiang, W.; Aryan, F.; Aoude, P.; Bertozzi, C.R.; Estes, V.M.; et al. DNA-PKcs has KU-dependent function in rRNA processing and haematopoiesis. Nat. Cell Biol. 2020, 579, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.; Ketley, R.F.; Gullerova, M. Beyond the Trinity of ATM, ATR, and DNA-PK: Multiple Kinases Shape the DNA Damage Response in Concert With RNA Metabolism. Front. Mol. Biosci. 2019, 6, 61. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamontova, V.; Trifault, B.; Boten, L.; Burger, K. Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far. Non-Coding RNA 2021, 7, 42. https://doi.org/10.3390/ncrna7030042
Mamontova V, Trifault B, Boten L, Burger K. Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far. Non-Coding RNA. 2021; 7(3):42. https://doi.org/10.3390/ncrna7030042
Chicago/Turabian StyleMamontova, Victoria, Barbara Trifault, Lea Boten, and Kaspar Burger. 2021. "Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far" Non-Coding RNA 7, no. 3: 42. https://doi.org/10.3390/ncrna7030042
APA StyleMamontova, V., Trifault, B., Boten, L., & Burger, K. (2021). Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far. Non-Coding RNA, 7(3), 42. https://doi.org/10.3390/ncrna7030042





