A Structural View of miRNA Biogenesis and Function
Abstract
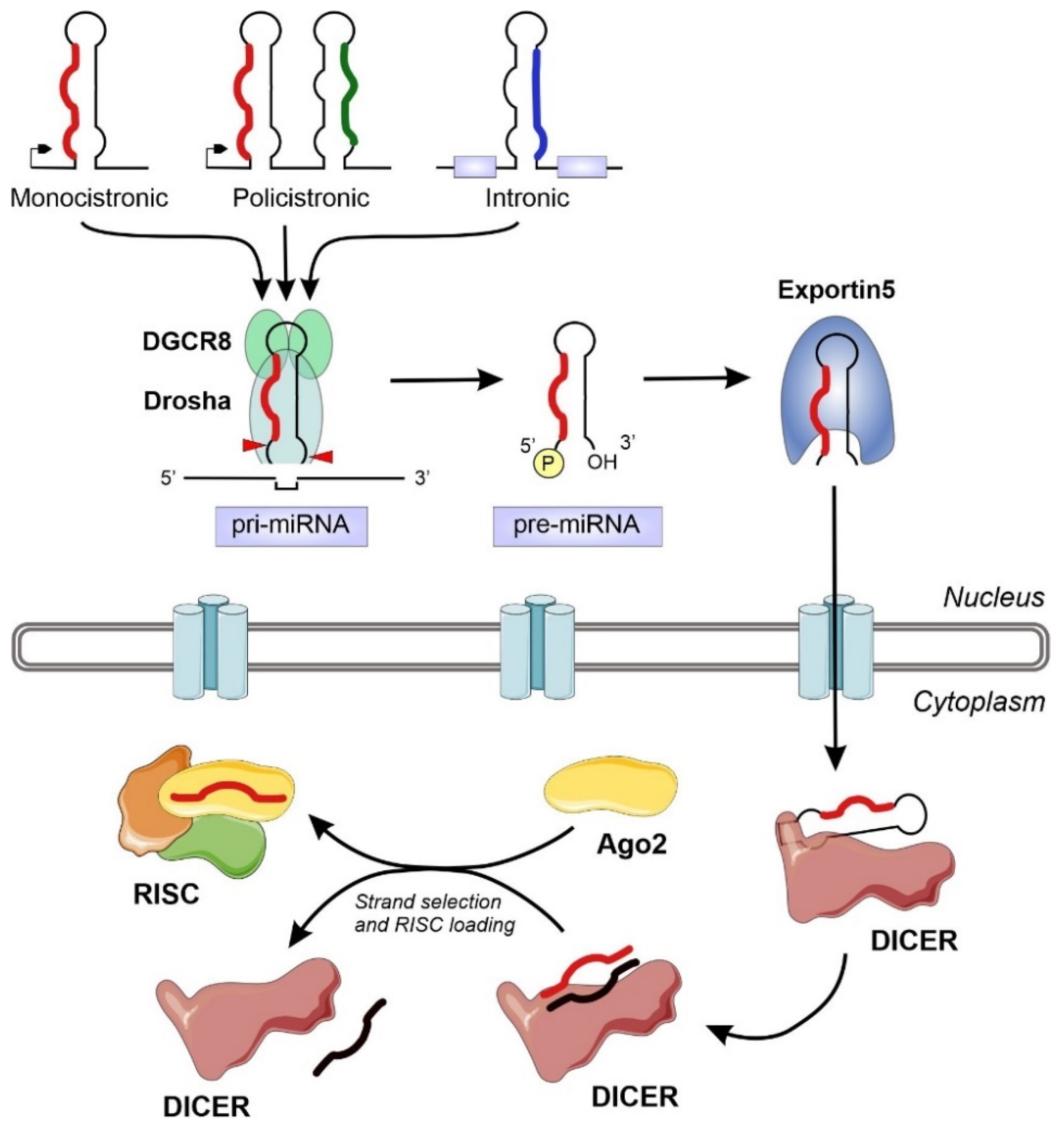
:1. Introduction
2. Nuclear Microprocessor: Drosha and DGCR8
3. Cytoplasmic Processors: Dicer
| PDB Code | Biomolecule | Species | Technique | Resolution | Year | Reference |
|---|---|---|---|---|---|---|
| 2FFL | Apo-Dicer | Giardia intestinalis | X-ray | 3.3 Å | 2006 | [48] |
| 2QVW | Apo-Dicer twinned data | Giardia intestinalis | X-ray | 3.0 Å | 2007 | [61] |
| 2EB1 | Dicer C-terminal domain | Homo sapiens | X-ray | 2.0 Å | 2007 | [62] |
| 3C4B | Dicer RNAseIIIb-dsRNAb domain | Mus musculus | X-ray | 1.6 Å | 2008 | [63] |
| 2KOU | Dicer-like protein | Arabidopsis thaliana | NMR | N/A | 2010 | [64] |
| 3ADL | TRBP2 | Homo sapiens | X-ray | 2.2 Å | 2010 | [65] |
| 3RV0 | Dcr1 without C-terminal domain | Vanderwaltozyma polyspora | X-ray | 2.2 Å | 2011 | [66] |
| 2L6M | Dcr1 C-terminal dsRBD domain | Schizosaccharomyces pombe | NMR | N/A | 2011 | [67] |
| 2LRS | Dicer-like 1 dsRBD domain | Arabidopsis thaliana | NMR | N/A | 2012 | [68] |
| 4NGB | Dicer Platform-PAZ-connector | Homo sapiens | X-ray | 2.2 Å | 2014 | [69] |
| 4WYQ | Dicer-TRBP interface | Homo sapiens | X-ray | 3.2 Å | 2015 | [55] |
| 5F3P | Non-canonical Dicer protein | Entaboeba histolytica | X-ray | 1.9 Å | 2018 | Unpublished |
| 6BU9 | Dicer-2 complexed with dsRNA | Drosophila melanogaster | Cryo-EM | 6.8 Å | 2018 | [56] |
| 5N8L | TRBP dsRBD siRNA complex | Homo sapiens | NMR | N/A | 2018 | [60] |
| 5ZAL | Dicer complex with pre-miRNA | Homo sapiens | Cryo-EM | 4.7 Å | 2018 | [51] |
| 7DEY | Apo-Dicer | Scheffersomyces stipitis | X-ray | 2.8 Å | 2021 | [70] |
| 7ELE | DCL1 in complex with pre-miRNA | Arabidopsis thaliana | Cryo-EM | 4.9 Å | 2021 | [71] |
| 7VG2 | DCL3 in complex with RNA | Arabidopsis thaliana | Cryo-EM | 3.1 Å | 2021 | [72] |

4. Cytoplasmic Effectors: Argonaute Proteins and RISC Complex
| PDB Code | Biomolecule | Species | Technique | Resolution | Year | Reference |
|---|---|---|---|---|---|---|
| 1U04 | Full length Argonaute | Pyrococcus furiosus | X-ray | 2.2 Å | 2004 | [90] |
| 1W9H | Apo form Piwi protein | Archaeoglobus fulgidus | X-ray | 1.9 Å | 2004 | [102] |
| 2BGG | siRNA/Piwi protein complex | Archaeoglobus fulgidus | X-ray | 2.2 Å | 2005 | [103] |
| 1YVU | Full length Argonaute | Aquifex aeolicus | X-ray | 2.9 Å | 2005 | [104] |
| 3DLB | RNA/Ago complex | Thermus thermophilus | X-ray | 2.7 Å | 2008 | [105] |
| 3F73 | dsRNA/Ago complex | Thermus thermophilus | X-ray | 3.0 Å | 2008 | [105] |
| 3HJF | RNA-DNA/Ago complex | Thermus thermophilus | X-ray | 3.0 Å | 2009 | [106] |
| 4F3T | miRNA/Ago2 complex | Homo sapiens | X-ray | 2.2 Å | 2012 | [97] |
| 4KRE | Sf9/Ago1 complex | Homo sapiens | X-ray | 1.7 Å | 2013 | [107] |
| 4N41 | dsDNA/Argonaute complex | Thermus thermophilus | X-ray | 2.2 Å | 2014 | [92] |
| 4OLA | Apo form Argonaute 2 | Homo sapiens | X-ray | 2.3 Å | 2014 | [108] |
| 4W5N | RNA/Argonaute 2 complex | Homo sapiens | X-ray | 2.9 Å | 2014 | [109] |
| 4Z4D | Target RNA/Ago2 complex | Homo sapiens | X-ray | 1.6 Å | 2015 | [110] |
| 5AWH | RNA-DNA/Argonaute complex | Cereibacter sphaeroides | X-ray | 2.0 Å | 2015 | [111] |
| 5JS1 | siRNA/Ago2 complex | Homo sapiens | X-ray | 2.5 Å | 2016 | [112] |
| 5W6V | RNA/Ago1/GW182 complex | Homo sapiens | X-ray | 2.8 Å | 2017 | [113] |
| 5VM9 | Guide RNA/Ago3 complex | Homo sapiens | X-ray | 3.2 Å | 2017 | [114] |
| 6CBD | RNA/Ago2 complex | Homo sapiens | X-ray | 2.2 Å | 2018 | [115] |
| 6D8A | RNA-DNA/Argonaute complex | Cereibacter sphaeroides | X-ray | 2.2 Å | 2018 | [116] |
| 6QZK | DNA/Argonaute complex | Clostridium butyricum | X-ray | 3.5 Å | 2019 | [117] |
| 6N4O | MiR-122/Ago2 complex | Homo sapiens | X-ray | 2.9 Å | 2019 | [118] |
| 6OON | Guide RNA/Ago4 complex | Homo sapiens | X-ray | 1.9 Å | 2019 | [93] |
| 6MFN | MiR-27a/Ago2 complex | Homo sapiens | X-ray | 2.5 Å | 2019 | [119] |
| 7KI3 | MiR-122/target/Ago2 | Homo sapiens | X-ray | 3.0 Å | 2021 | [120] |

5. Conclusions and Further Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Lim, L.P.; Glasner, M.E.; Yekta, S.; Burge, C.B.; Bartel, D.P. Vertebrate microRNA genes. Science 2003, 299, 1540. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Javanmardi, S.; Aghamaali, M.R.; Abolmaali, S.S.; Mohammadi, S.; Tamaddon, A.M. miR-21, An Oncogenic Target miRNA for Cancer Therapy: Molecular Mechanisms and Recent Advancements in Chemo and Radio-resistance. Curr. Gene Ther. 2017, 16, 375–389. [Google Scholar] [CrossRef]
- Nucera, S.; Giustacchini, A.; Boccalatte, F.; Calabria, A.; Fanciullo, C.; Plati, T.; Ranghetti, A.; Garcia-Manteiga, J.; Cittaro, D.; Benedicenti, F.; et al. miRNA-126 Orchestrates an Oncogenic Program in B Cell Precursor Acute Lymphoblastic Leukemia. Cancer Cell 2016, 29, 905–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Yu, X. Cardinal roles of miRNA in cardiac development and disease. Sci. China Life Sci. 2011, 54, 1113–1120. [Google Scholar] [CrossRef] [Green Version]
- Lock, M.C.; Tellam, R.L.; Botting, K.J.; Wang, K.C.W.; Selvanayagam, J.B.; Brooks, D.A.; Seed, M.; Morrison, J.L. The role of miRNA regulation in fetal cardiomyocytes, cardiac maturation and the risk of heart disease in adults. J. Physiol. 2018, 596, 5625–5640. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pastor, A.R.; Infante-Menendez, J.; Escribano, O.; Gomez-Hernandez, A. miRNA Dysregulation in the Development of Non-Alcoholic Fatty Liver Disease and the Related Disorders Type 2 Diabetes Mellitus and Cardiovascular Disease. Front. Med. (Lausanne) 2020, 7, 527059. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, Y.; Shi, Y.; Zhang, Y.; Liu, K.; Liang, R.; Sun, P.; Chang, X.; Tang, W.; Zhang, Y.; et al. Expression of miRNA-29 in Pancreatic beta Cells Promotes Inflammation and Diabetes via TRAF3. Cell Rep. 2021, 34, 108576. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Fejes Toth, K.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Leitao, A.L.; Enguita, F.J. Biogenesis and mechanism of action of small non-coding RNAs: Insights from the point of view of structural biology. Int J. Mol. Sci. 2012, 13, 10268–10295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathore, I.; Mishra, V.; Bhaumik, P. Advancements in macromolecular crystallography: From past to present. Emerg. Top. Life Sci. 2021, 5, 127–149. [Google Scholar] [CrossRef] [PubMed]
- Assaiya, A.; Burada, A.P.; Dhingra, S.; Kumar, J. An overview of the recent advances in cryo-electron microscopy for life sciences. Emerg. Top. Life Sci. 2021, 5, 151–168. [Google Scholar] [CrossRef]
- Yeom, K.H.; Lee, Y.; Han, J.; Suh, M.R.; Kim, V.N. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006, 34, 4622–4629. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.C.; Jang, H.; Shen, S.; Baek, S.C.; Kim, K.; Yang, J.; Kim, J.; Kim, J.S.; Wang, S.; Shi, Y.; et al. ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8. Nucleic Acids Res. 2020, 48, 11097–11112. [Google Scholar] [CrossRef]
- Shiohama, A.; Sasaki, T.; Noda, S.; Minoshima, S.; Shimizu, N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 2003, 304, 184–190. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Faller, M.; Matsunaga, M.; Yin, S.; Loo, J.A.; Guo, F. Heme is involved in microRNA processing. Nat. Struct. Mol. Biol. 2007, 14, 23–29. [Google Scholar] [CrossRef]
- Sohn, S.Y.; Bae, W.J.; Kim, J.J.; Yeom, K.H.; Kim, V.N.; Cho, Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007, 14, 847–853. [Google Scholar] [CrossRef]
- Senturia, R.; Faller, M.; Yin, S.; Loo, J.A.; Cascio, D.; Sawaya, M.R.; Hwang, D.; Clubb, R.T.; Guo, F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 2010, 19, 1354–1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senturia, R.; Laganowsky, A.; Barr, I.; Scheidemantle, B.D.; Guo, F. Dimerization and heme binding are conserved in amphibian and starfish homologues of the microRNA processing protein DGCR8. PLoS ONE 2012, 7, e39688. [Google Scholar] [CrossRef]
- Mueller, G.A.; Miller, M.T.; DeRose, E.F.; Ghosh, M.; London, R.E.; Hall, T.M.T. Solution structure of the Drosha double-stranded RNA-binding domain. Silence 2010, 1, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.C.; Nguyen, T.A.; Choi, Y.G.; Jo, M.H.; Hohng, S.; Kim, V.N.; Woo, J.S. Structure of Human DROSHA. Cell 2016, 164, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Partin, A.C.; Zhang, K.; Jeong, B.C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol. Cell 2020, 78, 411–422 e414. [Google Scholar] [CrossRef]
- Jin, W.; Wang, J.; Liu, C.P.; Wang, H.W.; Xu, R.M. Structural Basis for pri-miRNA Recognition by Drosha. Mol. Cell 2020, 78, 423–433 e425. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, G.; Armenia, I.; Molla, G. The Protein Imager: A full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics 2020, 36, 2909–2911. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Filipek, S.; Vogel, H. PyMOL and Inkscape Bridge the Data and the Data Visualization. Structure 2016, 24, 2041–2042. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002, 21, 5875–5885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeulen, A.; Behlen, L.; Reynolds, A.; Wolfson, A.; Marshall, W.S.; Karpilow, J.; Khvorova, A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 2005, 11, 674–682. [Google Scholar] [CrossRef] [Green Version]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.S.; Yan, S.; Farooq, A.; Han, A.; Zeng, L.; Zhou, M.M. Structure and conserved RNA binding of the PAZ domain. Nature 2003, 426, 468–474. [Google Scholar] [CrossRef]
- Macrae, I.J.; Li, F.; Zhou, K.; Cande, W.Z.; Doudna, J.A. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb Symp. Quant. Biol. 2006, 71, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Wang, J.; Cheng, H.; Ke, X.; Sun, L.; Zhang, Q.C.; Wang, H.W. Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate. Cell 2018, 173, 1549–1550. [Google Scholar] [CrossRef]
- Baldaccini, M.; Pfeffer, S. Untangling the roles of RNA helicases in antiviral innate immunity. PLoS Pathog. 2021, 17, e1010072. [Google Scholar] [CrossRef]
- Cenik, E.S.; Fukunaga, R.; Lu, G.; Dutcher, R.; Wang, Y.; Tanaka Hall, T.M.; Zamore, P.D. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol. Cell 2011, 42, 172–184. [Google Scholar] [CrossRef] [Green Version]
- Lau, P.W.; Potter, C.S.; Carragher, B.; MacRae, I.J. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 2009, 17, 1326–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Sinha, N.K.; Iwasa, J.; Shen, P.S.; Bass, B.L. Dicer uses distinct modules for recognizing dsRNA termini. Science 2018, 359, 329–334. [Google Scholar] [CrossRef] [Green Version]
- Koralewska, N.; Ciechanowska, K.; Pokornowska, M.; Figlerowicz, M.; Kurzynska-Kokorniak, A. Human ribonuclease Dicer—structure and functions. Postepy Biochem. 2019, 65, 173–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Q.J.; Zhang, J.; Li, P.; Wang, Q.; Zhang, Y.; Roy-Chaudhuri, B.; Xu, J.; Kay, M.A.; Zhang, Q.C. RNA structure probing reveals the structural basis of Dicer binding and cleavage. Nat. Commun. 2021, 12, 3397. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wu, Y.; Niu, Q.; Zhang, J.; Jia, G.; Manjunath, N.; Wu, H. A sliding-bulge structure at the Dicer processing site of pre-miRNAs regulates alternative Dicer processing to generate 5’-isomiRs. Heliyon 2016, 2, e00148. [Google Scholar] [CrossRef] [Green Version]
- Masliah, G.; Maris, C.; Konig, S.L.; Yulikov, M.; Aeschimann, F.; Malinowska, A.L.; Mabille, J.; Weiler, J.; Holla, A.; Hunziker, J.; et al. Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains. EMBO J. 2018, 37, e97089. [Google Scholar] [CrossRef] [Green Version]
- MacRae, I.J.; Doudna, J.A. An unusual case of pseudo-merohedral twinning in orthorhombic crystals of Dicer. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 993–999. [Google Scholar] [CrossRef]
- Takeshita, D.; Zenno, S.; Lee, W.C.; Nagata, K.; Saigo, K.; Tanokura, M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J. Mol. Biol. 2007, 374, 106–120. [Google Scholar] [CrossRef]
- Du, Z.; Lee, J.K.; Tjhen, R.; Stroud, R.M.; James, T.L. Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage. Proc. Natl. Acad. Sci. USA 2008, 105, 2391–2396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, H.; Chen, F.; Huan, X.; Machida, S.; Song, J.; Yuan, Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein-protein interaction. RNA 2010, 16, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.W.; Chen, H.Y.; Yang, J.; Machida, S.; Chua, N.H.; Yuan, Y.A. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 2010, 18, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, D.E.; Nakanishi, K.; Patel, D.J.; Bartel, D.P. The inside-out mechanism of Dicers from budding yeasts. Cell 2011, 146, 262–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barraud, P.; Emmerth, S.; Shimada, Y.; Hotz, H.R.; Allain, F.H.; Buhler, M. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J. 2011, 30, 4223–4235. [Google Scholar] [CrossRef] [Green Version]
- Burdisso, P.; Suarez, I.P.; Bologna, N.G.; Palatnik, J.F.; Bersch, B.; Rasia, R.M. Second double-stranded RNA binding domain of dicer-like ribonuclease 1: Structural and biochemical characterization. Biochemistry 2012, 51, 10159–10166. [Google Scholar] [CrossRef]
- Tian, Y.; Simanshu, D.K.; Ma, J.B.; Park, J.E.; Heo, I.; Kim, V.N.; Patel, D.J. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol. Cell 2014, 53, 606–616. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.; Qinqin, F.; Jianwei, L.; Ying, C.; Machida, S.; Wei, C.; Yuan, Y.A.; Jobichen, C. Structural and mechanistic insight into stem-loop RNA processing by yeast Pichia stipitis Dicer. Protein Sci. 2021, 30, 1210–1220. [Google Scholar] [CrossRef]
- Wei, X.; Ke, H.; Wen, A.; Gao, B.; Shi, J.; Feng, Y. Structural basis of microRNA processing by Dicer-like 1. Nat. Plants 2021, 7, 1389–1396. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, Y.; Zhang, L.; Zhong, Z.; Feng, S.; Wang, C.; Xiao, L.; Yang, Z.; Harris, C.J.; Wu, Z.; et al. Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation. Science 2021, 374, 1152–1157. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant. Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z. miRNA regulatory va.a.ariation in human evolution. Trends Genet. 2013, 29, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Cole, C.N.; McPeek, M.A.; Peterson, K.J. The phylogenetic distribution of metazoan microRNAs: Insights into evolutionary complexity and constraint. J. Exp. Zool. B Mol. Dev. Evol. 2006, 306, 575–588. [Google Scholar] [CrossRef]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2017, 130, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhao, H.; Chen, Z.; Feng, L.; Ren, J.; Cai, R.; Xiang, Y. The Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in Populus trichocarpa: Gene structure, gene expression, phylogenetic analysis and evolution. J. Genet. 2015, 94, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Martinez, J.; Schroeder, R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Rivas, F.V.; Tolia, N.H.; Song, J.J.; Aragon, J.P.; Liu, J.; Hannon, G.J.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar] [CrossRef]
- Wang, H.W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chendrimada, T.P.; Finn, K.J.; Ji, X.; Baillat, D.; Gregory, R.I.; Liebhaber, S.A.; Pasquinelli, A.E.; Shiekhattar, R. MicroRNA silencing through RISC recruitment of eIF6. Nature 2007, 447, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Bouasker, S.; Simard, M.J. The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic Acids Res. 2012, 40, 10452–10462. [Google Scholar] [CrossRef] [Green Version]
- Faehnle, C.R.; Joshua-Tor, L. Argonautes confront new small RNAs. Curr. Opin. Chem. Biol. 2007, 11, 569–577. [Google Scholar] [CrossRef] [Green Version]
- van der Oost, J.; Swarts, D.C.; Jore, M.M. Prokaryotic Argonautes—Variations on the RNA interference theme. Microb. Cell 2014, 1, 158–159. [Google Scholar] [CrossRef]
- Swarts, D.C. Prokaryotic Argonautes Function beyond Immunity by Unlinking Replicating Chromosomes. Cell 2020, 182, 1381–1383. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef] [Green Version]
- Rashid, U.J.; Paterok, D.; Koglin, A.; Gohlke, H.; Piehler, J.; Chen, J.C. Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function. J. Biol. Chem. 2007, 282, 13824–13832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, G.; Zhao, H.; Wang, J.; Rao, Y.; Tian, W.; Swarts, D.C.; van der Oost, J.; Patel, D.J.; Wang, Y. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 2014, 111, 652–657. [Google Scholar] [CrossRef] [Green Version]
- Park, M.S.; Araya-Secchi, R.; Brackbill, J.A.; Phan, H.D.; Kehling, A.C.; Abd El-Wahab, E.W.; Dayeh, D.M.; Sotomayor, M.; Nakanishi, K. Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters. Mol. Cell 2019, 75, 725–740 e726. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, C.D.; Joshua-Tor, L. Eukaryotic Argonautes come into focus. Trends Biochem. Sci. 2013, 38, 263–271. [Google Scholar] [CrossRef]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef] [Green Version]
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.M. Structure and function of argonaute proteins. Structure 2005, 13, 1403–1408. [Google Scholar] [CrossRef]
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005, 19, 2837–2848. [Google Scholar] [CrossRef] [Green Version]
- Ruda, V.M.; Chandwani, R.; Sehgal, A.; Bogorad, R.L.; Akinc, A.; Charisse, K.; Tarakhovsky, A.; Novobrantseva, T.I.; Koteliansky, V. The roles of individual mammalian argonautes in RNA interference in vivo. PLoS ONE 2014, 9, e101749. [Google Scholar] [CrossRef] [Green Version]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.S.; Roe, S.M.; Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004, 23, 4727–4737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.S.; Roe, S.M.; Barford, D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 2005, 434, 663–666. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.R.; Pei, Y.; Ma, J.B.; Kuryavyi, V.; Zhadina, M.; Meister, G.; Chen, H.Y.; Dauter, Z.; Tuschl, T.; Patel, D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 2005, 19, 405–419. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Tuschl, T.; Patel, D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 2008, 456, 921–926. [Google Scholar] [CrossRef]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Wardle, G.S.; Tuschl, T.; Patel, D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 2009, 461, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Faehnle, C.R.; Elkayam, E.; Haase, A.D.; Hannon, G.J.; Joshua-Tor, L. The making of a slicer: Activation of human Argonaute-1. Cell Rep. 2013, 3, 1901–1909. [Google Scholar] [CrossRef] [Green Version]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 346, 608–613. [Google Scholar] [CrossRef]
- Schirle, N.T.; Sheu-Gruttadauria, J.; Chandradoss, S.D.; Joo, C.; MacRae, I.J. Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife 2015, 4, e07646. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, T.; Ito, K.; Murakami, R.; Uchiumi, T. Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 2016, 7, 11846. [Google Scholar] [CrossRef] [Green Version]
- Schirle, N.T.; Kinberger, G.A.; Murray, H.F.; Lima, W.F.; Prakash, T.P.; MacRae, I.J. Structural Analysis of Human Argonaute-2 Bound to a Modified siRNA Guide. J. Am. Chem. Soc. 2016, 138, 8694–8697. [Google Scholar] [CrossRef] [Green Version]
- Elkayam, E.; Faehnle, C.R.; Morales, M.; Sun, J.; Li, H.; Joshua-Tor, L. Multivalent Recruitment of Human Argonaute by GW182. Mol. Cell 2017, 67, 646–658 e643. [Google Scholar] [CrossRef]
- Park, M.S.; Phan, H.D.; Busch, F.; Hinckley, S.H.; Brackbill, J.A.; Wysocki, V.H.; Nakanishi, K. Human Argonaute3 has slicer activity. Nucleic Acids Res. 2017, 45, 11867–11877. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; MacRae, I.J. Phase Transitions in the Assembly and Function of Human miRISC. Cell 2018, 173, 946–957 e916. [Google Scholar] [CrossRef]
- Liu, Y.; Esyunina, D.; Olovnikov, I.; Teplova, M.; Kulbachinskiy, A.; Aravin, A.A.; Patel, D.J. Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA. Cell Rep. 2018, 24, 453–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegge, J.W.; Swarts, D.C.; Chandradoss, S.D.; Cui, T.J.; Kneppers, J.; Jinek, M.; Joo, C.; van der Oost, J. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 2019, 47, 5809–5821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheu-Gruttadauria, J.; Xiao, Y.; Gebert, L.F.; MacRae, I.J. Beyond the seed: Structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J. 2019, 38, e101153. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA. Degradation. Mol. Cell 2019, 75, 1243–1255 e1247. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; Law, M.; MacRae, I.J. A structured RNA motif locks Argonaute2:miR-122 onto the 5’ end of the HCV genome. Nat. Commun. 2021, 12, 6836. [Google Scholar] [CrossRef]
- Brown, K.M.; Chu, C.Y.; Rana, T.M. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005, 12, 469–470. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sheng, G.; Cheung, P.P.; Wang, S.; Li, Y.; Gao, X.; Zhang, Y.; Wang, Y.; Huang, X. Two symmetric arginine residues play distinct roles in Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 2019, 116, 845–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.S.; Sim, G.; Kehling, A.C.; Nakanishi, K. Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 28576–28578. [Google Scholar] [CrossRef] [PubMed]
- Dalmadi, A.; Miloro, F.; Balint, J.; Varallyay, E.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef]
- Dueck, A.; Meister, G. Assembly and function of small RNA—argonaute protein complexes. Biol Chem 2014, 395, 611–629. [Google Scholar] [CrossRef]
- Mallick, B.; Sharma, A.R.; Lee, S.S.; Chakraborty, C. Understanding the molecular interaction of human argonaute-2 and miR-20a complex: A molecular dynamics approach. J. Cell Biochem. 2019, 120, 19915–19924. [Google Scholar] [CrossRef]
- Ruijtenberg, S.; Sonneveld, S.; Cui, T.J.; Logister, I.; de Steenwinkel, D.; Xiao, Y.; MacRae, I.J.; Joo, C.; Tanenbaum, M.E. mRNA structural dynamics shape Argonaute-target interactions. Nat. Struct. Mol. Biol. 2020, 27, 790–801. [Google Scholar] [CrossRef]


| PDB Code | Biomolecule | Species | Technique | Resolution | Year | Reference |
|---|---|---|---|---|---|---|
| 1X47 | dsRNA binding domain of DGCR8 | Homo sapiens | NMR | N/A | 2005 | Unpublished |
| 2YT4 | DGCR8 core | Homo sapiens | X-ray | 2.6 Å | 2007 | [32] |
| 2KHX | dsRNA binding domain of Drosha | Homo sapiens | NMR | N/A | 2010 | [35] |
| 3LE4 | DGCR8 dimerization domain | Homo sapiens | X-ray | 1.7 Å | 2010 | [33] |
| 4ER5 | DGCR8 dimerization domain | Xenopus laevis | X-ray | 1.9 Å | 2013 | [34] |
| 5B16 | Drosha–DGCR8 complex | Homo sapiens | X-ray | 3.2 Å | 2016 | [36] |
| 6V5B | Drosha–DGCR8–pri-miRNA complex | Homo sapiens | Cryo-EM | 3.7 Å | 2020 | [37] |
| 6V5C | Drosha–DGCR8–pri-miRNA complex | Homo sapiens | Cryo-EM | 4.4 Å | 2020 | [37] |
| 6XLE | Drosha–DGCR8 complex | Homo sapiens | Cryo-EM | 4.2 Å | 2020 | [38] |
| 6XLD | Pri-miRNA bound to Drosha-DGCR8 | Homo sapiens | Cryo-EM | 3.9 Å | 2020 | [38] |
| 7CNC | ERH in complex with DGCR8 | Homo sapiens | X-ray | 1.6 Å | 2020 | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitão, A.L.; Enguita, F.J. A Structural View of miRNA Biogenesis and Function. Non-Coding RNA 2022, 8, 10. https://doi.org/10.3390/ncrna8010010
Leitão AL, Enguita FJ. A Structural View of miRNA Biogenesis and Function. Non-Coding RNA. 2022; 8(1):10. https://doi.org/10.3390/ncrna8010010
Chicago/Turabian StyleLeitão, Ana Lúcia, and Francisco J. Enguita. 2022. "A Structural View of miRNA Biogenesis and Function" Non-Coding RNA 8, no. 1: 10. https://doi.org/10.3390/ncrna8010010
APA StyleLeitão, A. L., & Enguita, F. J. (2022). A Structural View of miRNA Biogenesis and Function. Non-Coding RNA, 8(1), 10. https://doi.org/10.3390/ncrna8010010








