Comprehensive Transcriptional Profiling and Mouse Phenotyping Reveals Dispensable Role for Adipose Tissue Selective Long Noncoding RNA Gm15551
Abstract
:1. Introduction
2. Material and Methods
2.1. Animal Experiments
2.2. Generation of Gm15551 Knock out Animals
2.3. Indirect Calorimetry
2.4. IPGTT
2.5. Adipocyte Diameter
2.6. RNA Isolation and Reverse Transcription
2.7. Quantitative Polymerase Chain Reaction (qPCR)
2.8. Total RNA Sequencing
2.9. Poly A RNA Sequencing
2.10. RNA Sequencing Data Analysis
2.11. ChIP Sequencing
2.12. ChIP Sequencing Data Analysis
2.13. Tissue Specificity
2.14. Gene Set Enrichment Analysis
2.15. Assessment of Coding Potential
2.16. Primary Cell Culture
2.17. Cultivation of Brown Adipocyte Cell Lines
2.18. In Vitro Gain and Loss of Function Studies
2.19. Oil Red O Staining
2.20. Statistical Analysis
3. Results
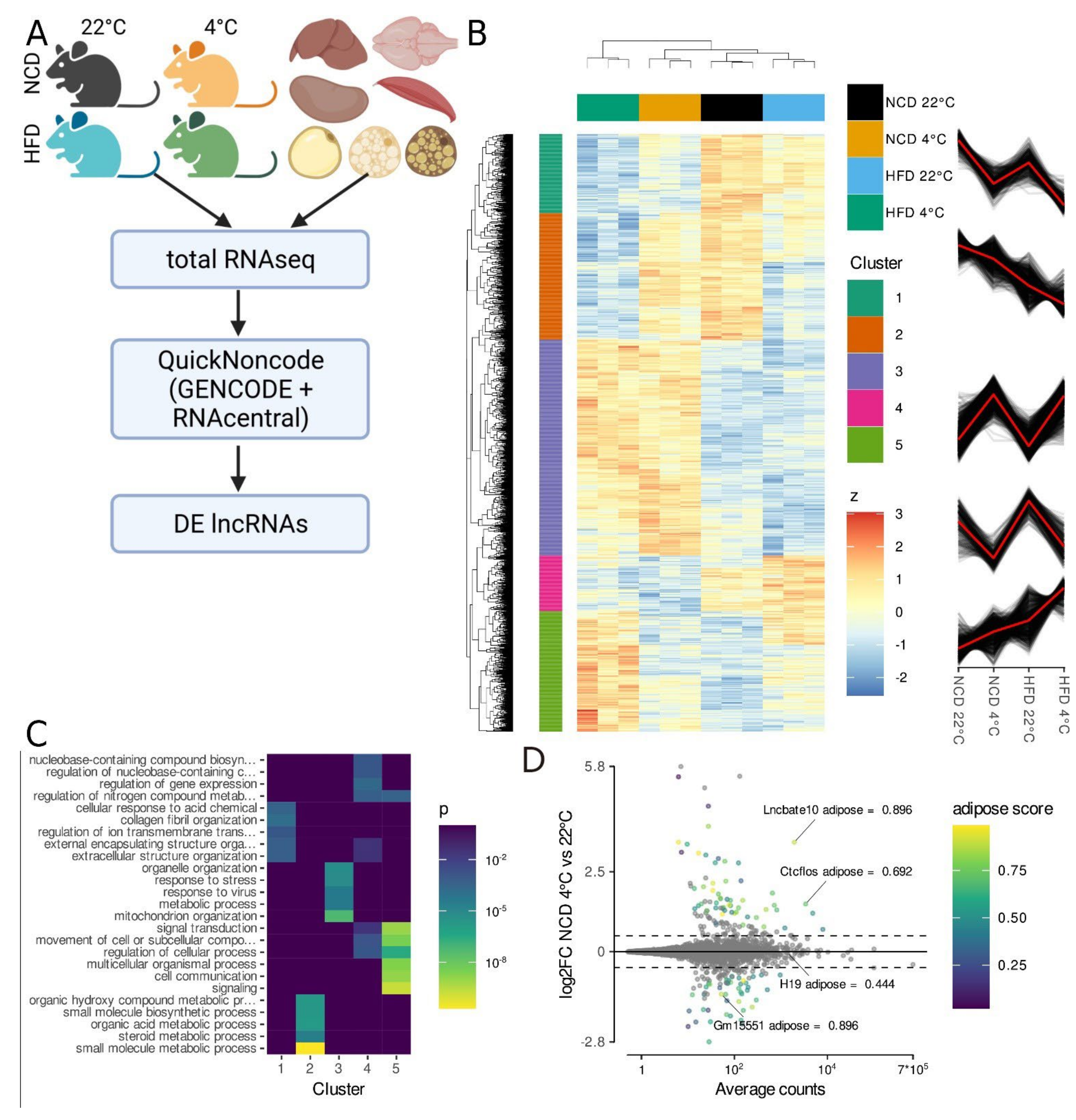
3.1. Total RNA-Seq Identifies lncRNAs Regulated in Activated iBAT
3.2. Gm15551 Is an Adipose Specific, Highly Regulated lncRNA
3.3. Gain- and Loss-of-Function of Gm15551 Does Not Disturb Brown Adipocyte Development and Function In Vitro
3.4. Gm15551 Loss-of-Function Does Not Impair Adipose Tissue Function In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANCOVA | analysis of covariances |
| ANOVA | analysis of variances |
| ATP | adenosine triphosphate |
| BAT | brown adipose tissue |
| cDNA | complementary DNA |
| ChIP | chromatin immunoprecipitation |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DNA | deoxyribonucleic acid |
| eRNA | enhancer RNA |
| eWAT | epididymal white adipose tissue |
| FBS | foetal bovine serum |
| GO | gene ontology |
| HFD | high fat diet |
| iBAT | interscapular brown adipose tissue |
| IBMX | 3-isobutyl-1-methylxanthin |
| IPGTT | intraperitoneal glucose tolerance test |
| iWAT | inguinal white adipose tissue |
| LNA | locked nucleic acid |
| log2FC | log2 fold change |
| lncRNA | long non-coding RNA |
| LRT | likelihood ratio test |
| miRNA | micro-RNA |
| NCD | control diet |
| qPCR | quantitative polymerase chain reaction |
| RNA | ribonucleic acid |
| SDS | sodium dodecyl sulphate |
| sgRNA | single guide RNA |
| siRNA | small interfering RNA |
| T3 | triiodothyronine |
| TRAP | translating ribosome affinity purification |
References
- NCD Risk Factor Collaboration (NCD-RisC). Trends in Adult Body-Mass Index in 200 Countries from 1975 to 2014: A Pooled Analysis of 1698 Population-Based Measurement Studies with 19·2 Million Participants. Lancet Lond. Engl. 2016, 387, 1377–1396. [Google Scholar] [CrossRef] [Green Version]
- Angelantonio, E.D.; Bhupathiraju, S.N.; Wormser, D.; Gao, P.; Kaptoge, S.; de Gonzalez, A.B.; Cairns, B.J.; Huxley, R.; Jackson, C.L.; Joshy, G.; et al. Body-Mass Index and All-Cause Mortality: Individual-Participant-Data Meta-Analysis of 239 Prospective Studies in Four Continents. Lancet 2016, 388, 776–786. [Google Scholar] [CrossRef] [Green Version]
- Prospective Studies Collaboration Body-Mass Index and Cause-Specific Mortality in 900 000 Adults: Collaborative Analyses of 57 Prospective Studies. Lancet 2009, 373, 1083–1096. [CrossRef] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. What We Talk About When We Talk About Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [Green Version]
- Cannon, B.; Nedergaard, J. Brown Adipose Tissue: Function and Physiological Significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef]
- Betz, M.J.; Enerbäck, S. Targeting Thermogenesis in Brown Fat and Muscle to Treat Obesity and Metabolic Disease. Nat. Rev. Endocrinol. 2018, 14, 77–87. [Google Scholar] [CrossRef]
- Klepac, K.; Georgiadi, A.; Tschöp, M.; Herzig, S. The Role of Brown and Beige Adipose Tissue in Glycaemic Control. Mol. Aspects Med. 2019, 68, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Scheele, C.; Wolfrum, C. Brown Adipose Crosstalk in Tissue Plasticity and Human Metabolism. Endocr. Rev. 2020, 41, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Yang, Y.; Xiang, L.; Zhao, Z.; Ye, R. Adipose-Derived Exosomes: A Novel Adipokine in Obesity-Associated Diabetes. J. Cell. Physiol. 2019, 234, 16692–16702. [Google Scholar] [CrossRef]
- Nedergaard, J.; Bengtsson, T.; Cannon, B. Unexpected Evidence for Active Brown Adipose Tissue in Adult Humans. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E444–E452. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Gil, N.; Ulitsky, I. Regulation of Gene Expression by Cis -Acting Long Non-Coding RNAs. Nat. Rev. Genet. 2019, 21, 102–117. [Google Scholar] [CrossRef]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Nguyen, T.C.; Zaleta-Rivera, K.; Huang, X.; Dai, X.; Zhong, S. RNA, Action through Interactions. Trends Genet. TIG 2018, 34, 867–882. [Google Scholar] [CrossRef]
- Yi, W.; Li, J.; Zhu, X.; Wang, X.; Fan, L.; Sun, W.; Liao, L.; Zhang, J.; Li, X.; Ye, J.; et al. CRISPR-Assisted Detection of RNA–Protein Interactions in Living Cells. Nat. Methods 2020, 17, 685–688. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.; Chai, X.; Yoon, M.J.; Kim, H.-J.; Lo, K.A.; Zhang, Z.; Xu, D.; Siang, D.T.C.; Walet, A.C.E.; Xu, S.; et al. Dynamic Transcriptome Changes during Adipose Tissue Energy Expenditure Reveal Critical Roles for Long Noncoding RNA Regulators. PLoS Biol. 2017, 15, e2002176. [Google Scholar] [CrossRef]
- Schmidt, E.; Dhaouadi, I.; Gaziano, I.; Oliverio, M.; Klemm, P.; Awazawa, M.; Mitterer, G.; Fernandez-Rebollo, E.; Pradas-Juni, M.; Wagner, W.; et al. LincRNA H19 Protects from Dietary Obesity by Constraining Expression of Monoallelic Genes in Brown Fat. Nat. Commun. 2018, 9, 3622. [Google Scholar] [CrossRef]
- Bast-Habersbrunner, A.; Kiefer, C.; Weber, P.; Fromme, T.; Schießl, A.; Schwalie, P.C.; Deplancke, B.; Li, Y.; Klingenspor, M. LncRNA Ctcflos Orchestrates Transcription and Alternative Splicing in Thermogenic Adipogenesis. EMBO Rep. 2021, 22, e51289. [Google Scholar] [CrossRef]
- Alcalá, M.; Calderon-Dominguez, M.; Bustos, E.; Ramos, P.; Casals, N.; Serra, D.; Viana, M.; Herrero, L. Increased Inflammation, Oxidative Stress and Mitochondrial Respiration in Brown Adipose Tissue from Obese Mice. Sci. Rep. 2017, 7, 16082. [Google Scholar] [CrossRef] [Green Version]
- Galarraga, M.; Campión, J.; Muñoz-Barrutia, A.; Boqué, N.; Moreno, H.; Martínez, J.A.; Milagro, F.; Ortiz-de-Solórzano, C. Adiposoft: Automated Software for the Analysis of White Adipose Tissue Cellularity in Histological Sections. J. Lipid Res. 2012, 53, 2791–2796. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Dominguez, J.R.; Bai, Z.; Xu, D.; Yuan, B.; Lo, K.A.; Yoon, M.J.; Lim, Y.C.; Knoll, M.; Slavov, N.; Chen, S.; et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Novel Long Non-Coding RNAs That Regulate Brown Adipocyte Development. Cell Metab. 2015, 21, 764–776. [Google Scholar] [CrossRef] [Green Version]
- Pradas-Juni, M.; Hansmeier, N.R.; Link, J.C.; Schmidt, E.; Larsen, B.D.; Klemm, P.; Meola, N.; Topel, H.; Loureiro, R.; Dhaouadi, I.; et al. A MAFG-LncRNA Axis Links Systemic Nutrient Abundance to Hepatic Glucose Metabolism. Nat. Commun. 2020, 11, 644. [Google Scholar] [CrossRef] [Green Version]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved Scoring of Functional Groups from Gene Expression Data by Decorrelating GO Graph Structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor Package for Reactome Pathway Analysis and Visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool Using an Alignment-Free Logistic Regression Model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational Design of Highly Active SgRNAs for CRISPR-Cas9-Mediated Gene Inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-Scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed Prior Distributions for Sequence Count Data: Removing the Noise and Preserving Large Differences. Bioinformatics 2018, 35, 2084–2092. [Google Scholar] [CrossRef]
- Henriques, F.; Bedard, A.H.; Guilherme, A.; Kelly, M.; Chi, J.; Zhang, P.; Lifshitz, L.M.; Bellvé, K.; Rowland, L.A.; Yenilmez, B.; et al. Single-Cell RNA Profiling Reveals Adipocyte to Macrophage Signaling Sufficient to Enhance Thermogenesis. Cell Rep. 2020, 32, 107998. [Google Scholar] [CrossRef]
- Natoli, G.; Andrau, J.-C. Noncoding Transcription at Enhancers: General Principles and Functional Models. Annu. Rev. Genet. 2012, 46, 1–19. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Lundh, M.; Pluciñska, K.; Isidor, M.S.; Petersen, P.S.S.; Emanuelli, B. Bidirectional Manipulation of Gene Expression in Adipocytes Using CRISPRa and SiRNA. Mol. Metab. 2017, 6, 1313–1320. [Google Scholar] [CrossRef]
- Rui, L. Brown and Beige Adipose Tissues in Health and Disease. Compr. Physiol. 2017, 7, 1281–1306. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Perspectives: Noncoding RNAs as Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef] [Green Version]
- Wahlestedt, C. Targeting Long Non-Coding RNA to Therapeutically Upregulate Gene Expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef]
- Sun, L.; Lin, J.D. Function and Mechanism of Long Noncoding RNAs in Adipocyte Biology. Diabetes 2019, 68, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 Determines the Thermogenic Program of Subcutaneous White Adipose Tissue in Mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef] [Green Version]
- Siersbæk, M.S.; Loft, A.; Aagaard, M.M.; Nielsen, R.; Schmidt, S.F.; Petrovic, N.; Nedergaard, J.; Mandrup, S. Genome-Wide Profiling of Peroxisome Proliferator-Activated Receptor γ in Primary Epididymal, Inguinal, and Brown Adipocytes Reveals Depot-Selective Binding Correlated with Gene Expression. Mol. Cell. Biol. 2012, 32, 3452–3463. [Google Scholar] [CrossRef] [Green Version]
- Ji, Z.; Song, R.; Regev, A.; Struhl, K. Many LncRNAs, 5′UTRs, and Pseudogenes Are Translated and Some Are Likely to Express Functional Proteins. eLife 2015, 4, e08890. [Google Scholar] [CrossRef]
- Gil, N.; Ulitsky, I. Production of Spliced Long Noncoding RNAs Specifies Regions with Increased Enhancer Activity. Cell Syst. 2018, 7, 537–547. [Google Scholar] [CrossRef] [Green Version]
- Maffei, M.; Barone, I.; Scabia, G.; Santini, F. The Multifaceted Haptoglobin in the Context of Adipose Tissue and Metabolism. Endocr. Rev. 2016, 37, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Sommer, G.; Weise, S.; Kralisch, S.; Scherer, P.E.; Lössner, U.; Blüher, M.; Stumvoll, M.; Fasshauer, M. The Adipokine SAA3 Is Induced by Interleukin-1β in Mouse Adipocytes. J. Cell. Biochem. 2008, 104, 2241–2247. [Google Scholar] [CrossRef]
- Sommer, G.; Weise, S.; Kralisch, S.; Lossner, U.; Bluher, M.; Stumvoll, M.; Fasshauer, M. Lipocalin-2 Is Induced by Interleukin-1β in Murine Adipocytes in Vitro. J. Cell. Biochem. 2009, 106, 103–108. [Google Scholar] [CrossRef]
- Guerra, C.; Navarro, P.; Valverde, A.M.; Arribas, M.; Brüning, J.; Kozak, L.P.; Kahn, C.R.; Benito, M. Brown Adipose Tissue–Specific Insulin Receptor Knockout Shows Diabetic Phenotype without Insulin Resistance. J. Clin. Investig. 2001, 108, 1205–1213. [Google Scholar] [CrossRef]
- Lowell, B.B.; S-Susulic, V.; Hamann, A.; Lawitts, J.A.; Himms-Hagen, J.; Boyer, B.B.; Kozak, L.P.; Flier, J.S. Development of Obesity in Transgenic Mice after Genetic Ablation of Brown Adipose Tissue. Nature 1993, 366, 740–742. [Google Scholar] [CrossRef]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long Noncoding RNAs Regulate Adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef] [Green Version]
- Hughes, M.R.; Canals Hernaez, D.; Cait, J.; Refaeli, I.; Lo, B.C.; Roskelley, C.D.; McNagny, K.M. A Sticky Wicket: Defining Molecular Functions for CD34 in Hematopoietic Cells. Exp. Hematol. 2020, 86, 1–14. [Google Scholar] [CrossRef]
- Qian, W.; Liao, B.-Y.; Chang, A.Y.-F.; Zhang, J. Maintenance of Duplicate Genes and Their Functional Redundancy by Reduced Expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudarzi, M.; Berg, K.; Pieper, L.M.; Schier, A.F. Individual Long Non-Coding RNAs Have No Overt Functions in Zebrafish Embryogenesis, Viability and Fertility. eLife 2019, 8, e40815. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Luo, S.; Peng, G.; Lu, J.Y.; Cui, G.; Liu, L.; Yan, P.; Yin, Y.; Liu, W.; Wang, R.; et al. Mouse Knockout Models Reveal Largely Dispensable but Context-Dependent Functions of LncRNAs during Development. J. Mol. Cell Biol. 2018, 10, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Gurmaches, J.; Hung, C.-M.; Guertin, D.A. Emerging Complexities in Adipocyte Origins and Identity. Trends Cell Biol. 2016, 26, 313–326. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engelhard, C.A.; Huang, C.; Khani, S.; Kasparek, P.; Prochazka, J.; Rozman, J.; Reguera, D.P.; Sedlacek, R.; Kornfeld, J.-W. Comprehensive Transcriptional Profiling and Mouse Phenotyping Reveals Dispensable Role for Adipose Tissue Selective Long Noncoding RNA Gm15551. Non-Coding RNA 2022, 8, 32. https://doi.org/10.3390/ncrna8030032
Engelhard CA, Huang C, Khani S, Kasparek P, Prochazka J, Rozman J, Reguera DP, Sedlacek R, Kornfeld J-W. Comprehensive Transcriptional Profiling and Mouse Phenotyping Reveals Dispensable Role for Adipose Tissue Selective Long Noncoding RNA Gm15551. Non-Coding RNA. 2022; 8(3):32. https://doi.org/10.3390/ncrna8030032
Chicago/Turabian StyleEngelhard, Christoph Andreas, Chien Huang, Sajjad Khani, Petr Kasparek, Jan Prochazka, Jan Rozman, David Pajuelo Reguera, Radislav Sedlacek, and Jan-Wilhelm Kornfeld. 2022. "Comprehensive Transcriptional Profiling and Mouse Phenotyping Reveals Dispensable Role for Adipose Tissue Selective Long Noncoding RNA Gm15551" Non-Coding RNA 8, no. 3: 32. https://doi.org/10.3390/ncrna8030032
APA StyleEngelhard, C. A., Huang, C., Khani, S., Kasparek, P., Prochazka, J., Rozman, J., Reguera, D. P., Sedlacek, R., & Kornfeld, J. -W. (2022). Comprehensive Transcriptional Profiling and Mouse Phenotyping Reveals Dispensable Role for Adipose Tissue Selective Long Noncoding RNA Gm15551. Non-Coding RNA, 8(3), 32. https://doi.org/10.3390/ncrna8030032






