miR-218: A Stress-Responsive Epigenetic Modifier
(This article belongs to the Section Small Non-Coding RNA)
Abstract
:1. Introduction
2. miRNA Biogenesis following Canonical Pathways
3. miRNAs as Epigenetic Regulators of Brain Functions and Dysfunctions
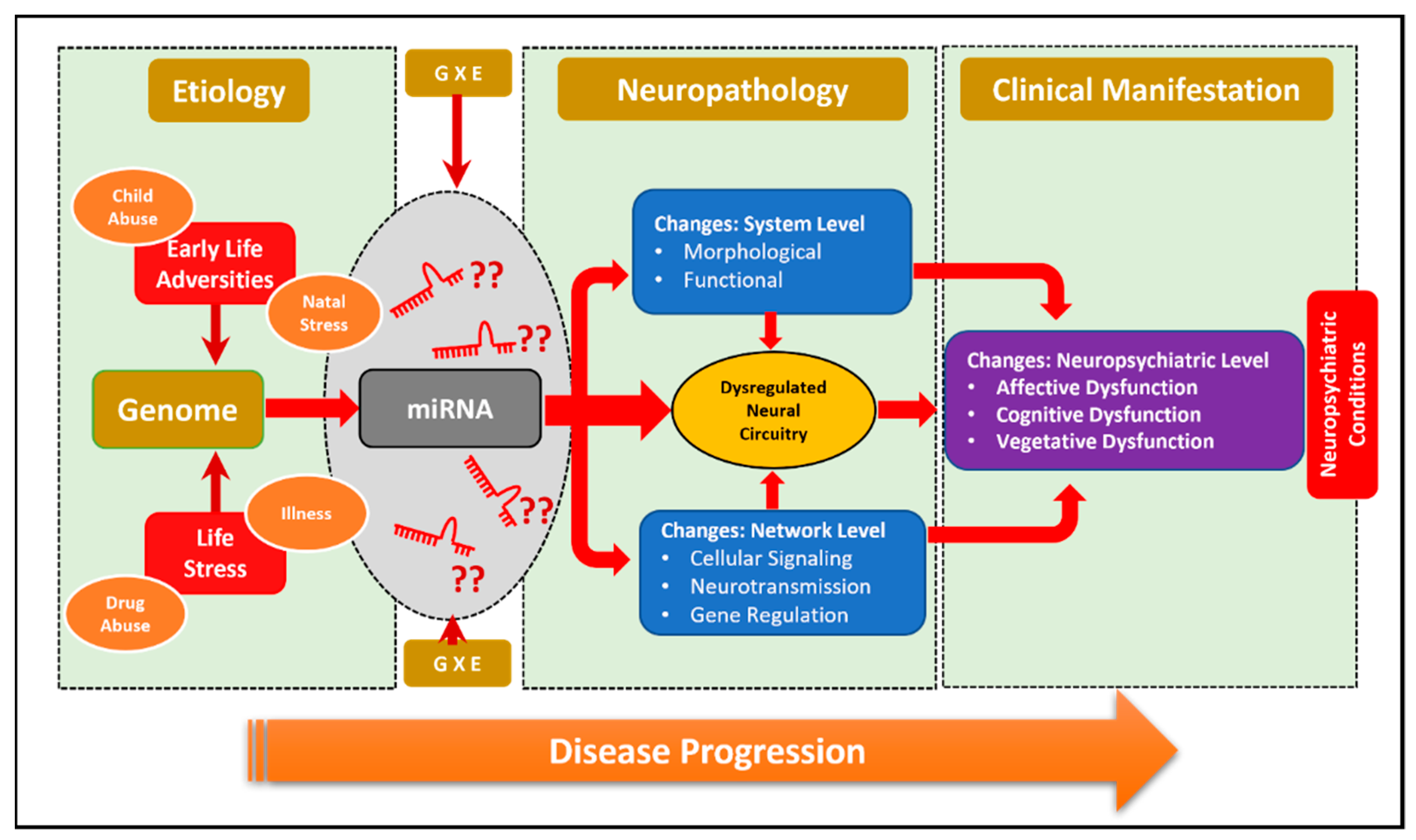
4. Role of miRNAs in Neuropsychiatric Disorders
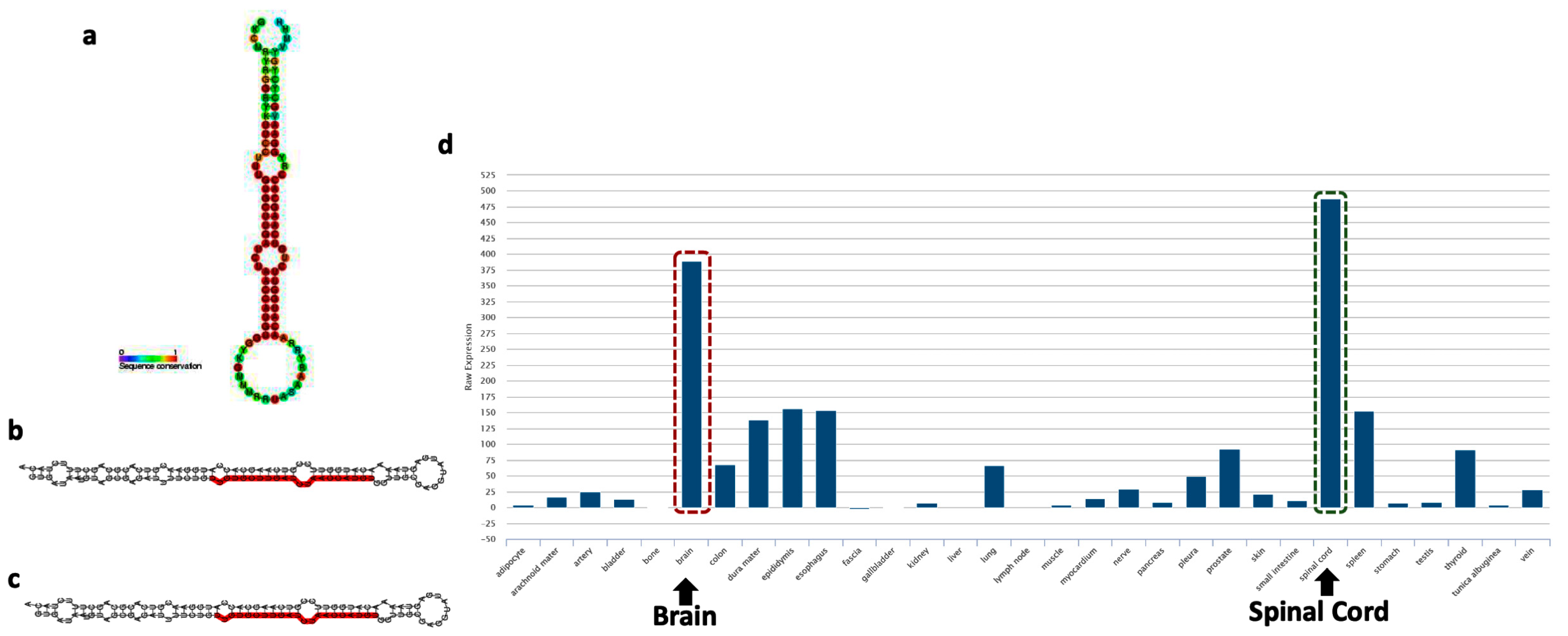
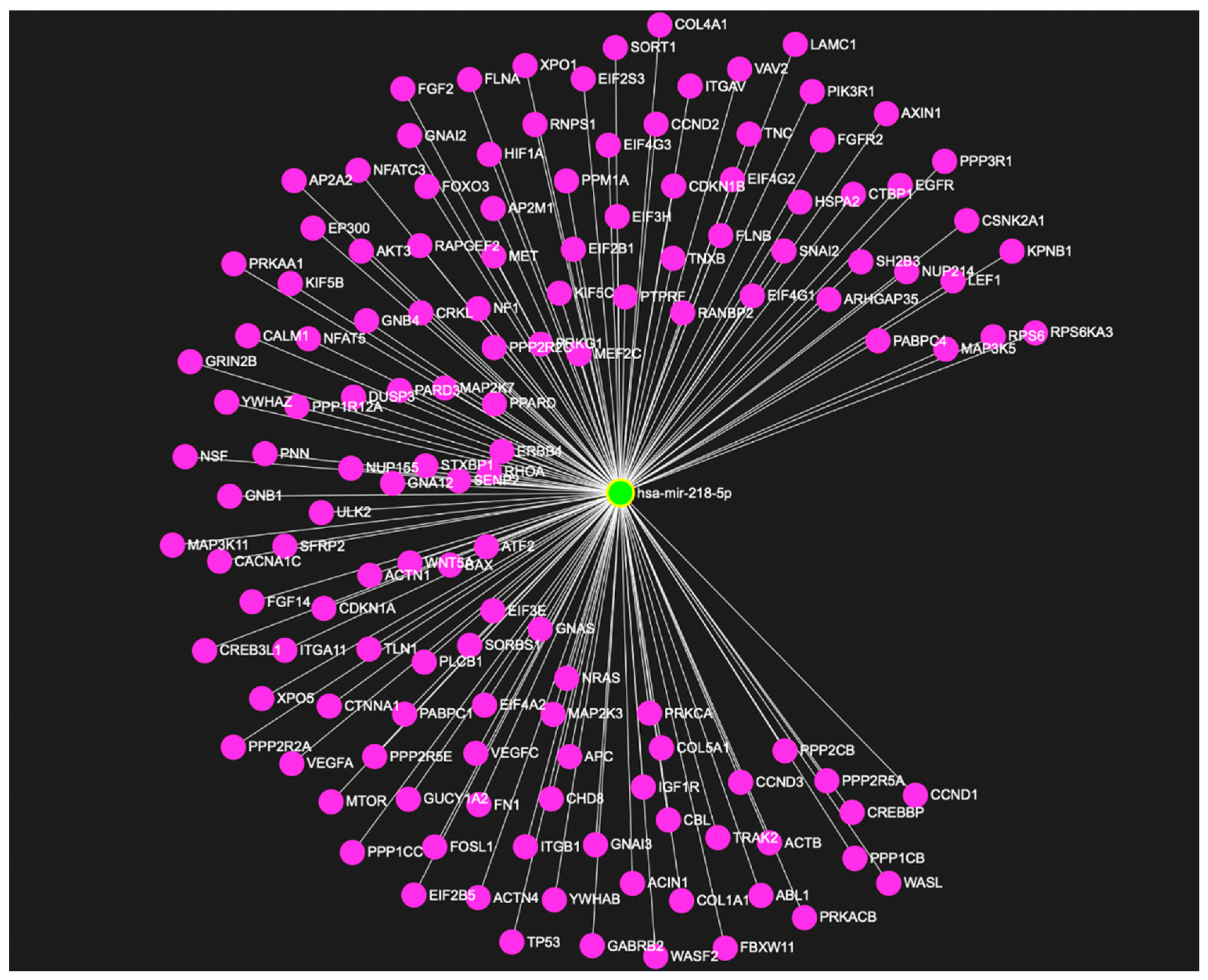
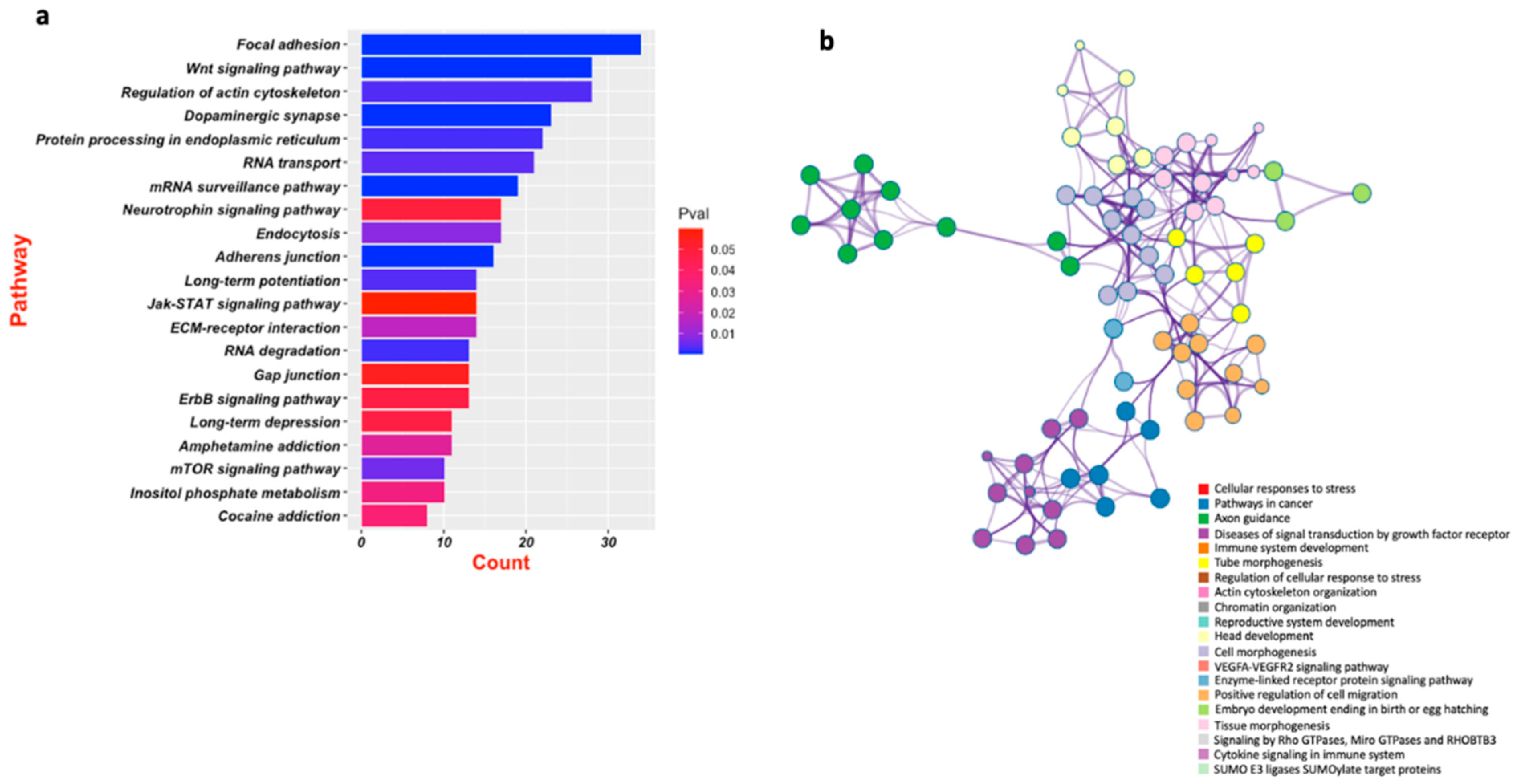
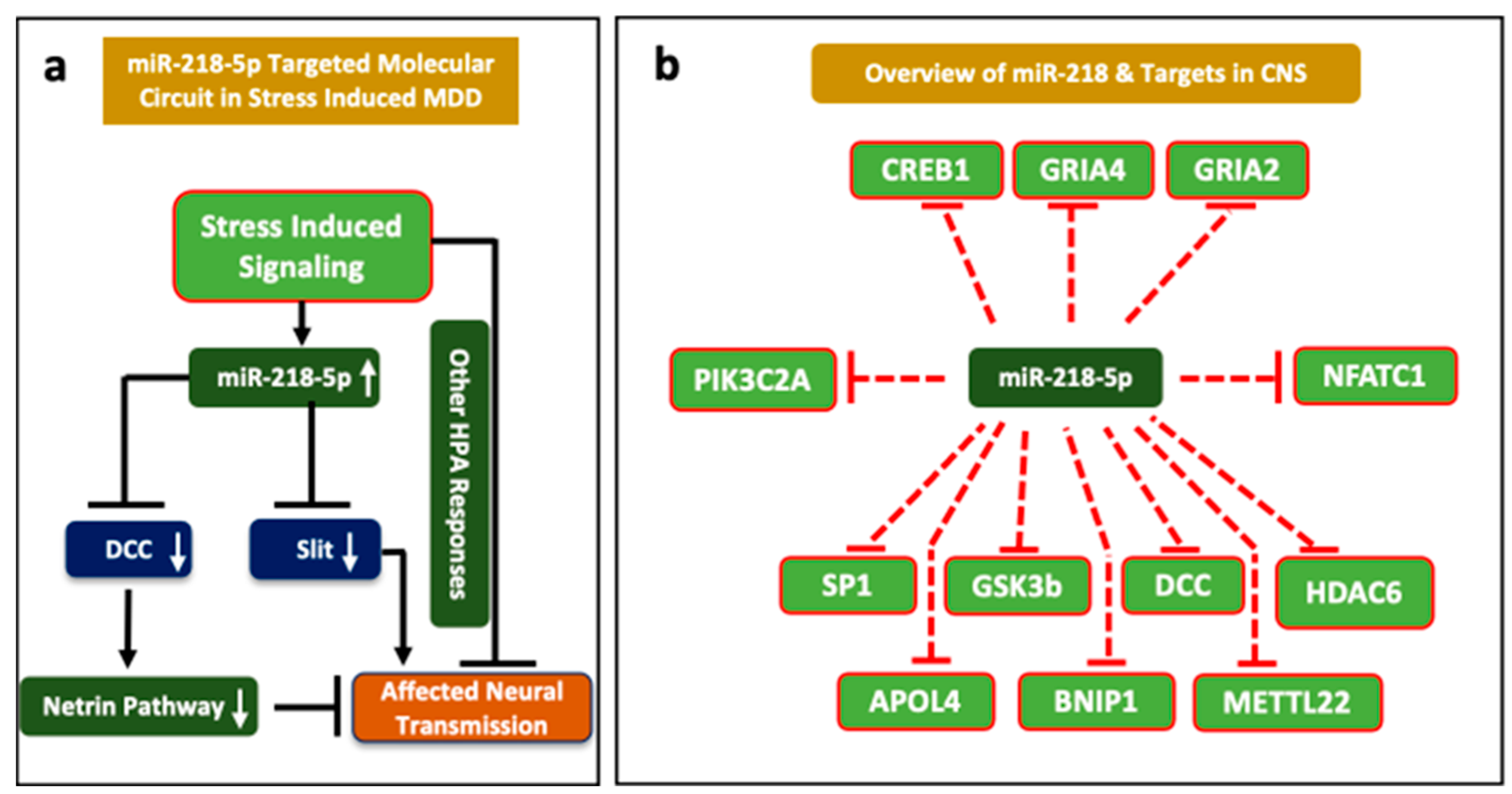
5. Emerging Role of miR-218 in Disease Pathogenesis
6. Role of miRNA-218 in Stress Susceptibility
7. Conclusions
8. Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Miyoshi, K.; Morimura, Y. Clinical Manifestations of Neuropsychiatric Disorders. In Neuropsychiatric Disorders; Miyoshi, K., Morimura, Y., Maeda, K., Eds.; Springer: Japan, Tokyo, 2010; pp. 1–14. [Google Scholar] [CrossRef]
- Ardila, A. Psychiatric disorders associated with acquired brain pathology. Appl. Neuropsychol. Adult 2019, 26, 591–597. [Google Scholar] [CrossRef] [PubMed]
- McGuire, J.L.; Depasquale, E.A.; Funk, A.J.; O’Donnovan, S.M.; Hasselfeld, K.; Marwaha, S.; Hammond, J.H.; Hartounian, V.; Meador-Woodruff, J.H.; Meller, J.; et al. Abnormalities of signal transduction networks in chronic schizophrenia. NPJ Schizophr. 2017, 3, 30. [Google Scholar] [CrossRef]
- Ramocki, M.B.; Zoghbi, H.Y. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature 2008, 455, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Geng, R.; Huang, X. Identification of major depressive disorder disease-related genes and functional pathways based on system dynamic changes of network connectivity. BMC Med. Genom. 2021, 14, 55. [Google Scholar] [CrossRef]
- Kuehner, J.N.; Bruggeman, E.C.; Wen, Z.; Yao, B. Epigenetic Regulations in Neuropsychiatric Disorders. Front. Genet. 2019, 10, 268. [Google Scholar] [CrossRef] [Green Version]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. In Non-Coding RNAs in Colorectal Cancer; Slaby, O., Calin, G.A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–17. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [Green Version]
- Mehler, M.F.; Mattick, J.S. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 2007, 87, 799–823. [Google Scholar] [CrossRef] [Green Version]
- Taft, R.J.; Simons, C.; Nahkuri, S.; Oey, H.; Korbie, D.J.; Mercer, T.R.; Holst, J.; Ritchie, W.; Wong, J.J.; Rasko, J.E.; et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010, 17, 1030–1034. [Google Scholar] [CrossRef]
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwivedi, Y. Emerging role of microRNAs in major depressive disorder: Diagnosis and therapeutic implications. Dialogues Clin. Neurosci. 2014, 16, 43–61. [Google Scholar] [CrossRef]
- Roy, B.; Dwivedi, Y. miRNAs as Critical Epigenetic Players in Determining Neurobiological Correlates of Major Depressive Disorder. In Understanding Depression: Volume 1. Biomedical and Neurobiological Background; Kim, Y.-K., Ed.; Springer: Singapore, 2018; pp. 51–69. [Google Scholar] [CrossRef]
- Schiele, M.A.; Gottschalk, M.G.; Domschke, K. The applied implications of epigenetics in anxiety, affective and stress-related disorders—A review and synthesis on psychosocial stress, psychotherapy and prevention. Clin. Psychol. Rev. 2020, 77, 101830. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Malchow, B.; Hasan, A.; Falkai, P. The impact of environmental factors in severe psychiatric disorders. Front. Neurosci. 2014, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Schulze-Hentrich, J.M.; Jakovcevski, M. Editorial: Neuroepigenetics of Neuropsychiatric Disease-Hope, Success and Obstacles for Translational Findings and Applications. Front. Neurosci. 2022, 16, 886695. [Google Scholar] [CrossRef]
- Issler, O.; Chen, A. Determining the role of microRNAs in psychiatric disorders. Nat. Rev. Neurosci. 2015, 16, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Yoshino, Y.; Allen, L.; Prall, K.; Schell, G.; Dwivedi, Y. Exploiting Circulating MicroRNAs as Biomarkers in Psychiatric Disorders. Mol. Diagn. Ther. 2020, 24, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Dwivedi, Y. Non-Coding RNAs in Psychiatric Disorders and Suicidal Behavior. Front. Psychiatry 2020, 11, 543893. [Google Scholar] [CrossRef] [PubMed]
- Camkurt, M.A.; Gunes, S.; Coskun, S.; Findikli, E. Peripheral Signatures of Psychiatric Disorders: MicroRNAs. Clin. Psychopharmacol. Neurosci. Off. Sci. J. Korean Coll. Neuropsychopharmacol. 2017, 15, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geaghan, M.; Cairns, M.J. MicroRNA and Posttranscriptional Dysregulation in Psychiatry. Biol. Psychiatry 2015, 78, 231–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollins, S.L.; Cairns, M.J. MicroRNA: Small RNA mediators of the brains genomic response to environmental stress. Prog. Neurobiol. 2016, 143, 61–81. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, C.; Savelsbergh, A.; Heusser, P. MicroRNAs in Psychological Stress Reactions and Their Use as Stress-Associated Biomarkers, Especially in Human Saliva. Biomed. Hub. 2017, 2, 481126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Berrio, A.; Nouel, D.; Cuesta, S.; Parise, E.M.; Restrepo-Lozano, J.M.; Larochelle, P.; Nestler, E.J.; Flores, C. MiR-218: A molecular switch and potential biomarker of susceptibility to stress. Mol. Psychiatry 2020, 25, 951–964. [Google Scholar] [CrossRef]
- Rocchi, A.; Moretti, D.; Lignani, G.; Colombo, E.; Scholz-Starke, J.; Baldelli, P.; Tkatch, T.; Benfenati, F. Neurite-Enriched MicroRNA-218 Stimulates Translation of the GluA2 Subunit and Increases Excitatory Synaptic Strength. Mol. Neurobiol 2019, 56, 5701–5714. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.Y.; Fu, C.L.; Liang, L.; Yang, B.; Shen, W.; Wang, Q.W.; Chen, Y.; Chen, Y.F.; Liu, Y.N.; Zhu, L.; et al. miR-218-2 regulates cognitive functions in the hippocampus through complement component 3-dependent modulation of synaptic vesicle release. Proc. Natl. Acad. Sci. USA 2021, 118, e2021770118. [Google Scholar] [CrossRef] [PubMed]
- Torres-Berrio, A.; Hernandez, G.; Nestler, E.J.; Flores, C. The Netrin-1/DCC Guidance Cue Pathway as a Molecular Target in Depression: Translational Evidence. Biol. Psychiatry 2020, 88, 611–624. [Google Scholar] [CrossRef]
- Dwivedi, Y.; Roy, B.; Lugli, G.; Rizavi, H.; Zhang, H.; Smalheiser, N.R. Chronic corticosterone-mediated dysregulation of microRNA network in prefrontal cortex of rats: Relevance to depression pathophysiology. Transl. Psychiatry 2015, 5, e682. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Corticosterone-mediated regulation and functions of miR-218-5p in rat brain. Sci. Rep. 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Roy, B.; Wang, Q.; Palkovits, M.; Faludi, G.; Dwivedi, Y. Altered miRNA expression network in locus coeruleus of depressed suicide subjects. Sci. Rep. 2017, 7, 4387. [Google Scholar] [CrossRef] [Green Version]
- Melamed, Z.E.; Levy, A.; Ashwal-Fluss, R.; Lev-Maor, G.; Mekahel, K.; Atias, N.; Gilad, S.; Sharan, R.; Levy, C.; Kadener, S.; et al. Alternative Splicing Regulates Biogenesis of miRNAs Located across Exon-Intron Junctions. Mol. Cell 2013, 50, 869–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Paturi, S.; Deshmukh, M.V. A Glimpse of “Dicer Biology” Through the Structural and Functional Perspective. Front. Mol. Biosci. 2021, 8, 643657. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurtan, A.M.; Lu, V.; Bhutkar, A.; Sharp, P.A. In Vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 2012, 18, 1116–1122. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef]
- Westholm, J.O.; Lai, E.C. Mirtrons: microRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvy-Liivrand, M.; de Sande, A.H.; Polonen, P.; Mehtonen, J.; Vuorenmaa, T.; Niskanen, H.; Sinkkonen, L.; Kaikkonen, M.U.; Heinaniemi, M. Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture. Nucleic Acids Res. 2017, 45, 9837–9849. [Google Scholar] [CrossRef] [Green Version]
- McEwen, B.S. Allostasis and Allostatic Load: Implications for Neuropsychopharmacology. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2000, 22, 108–124. [Google Scholar] [CrossRef]
- Lopizzo, N.; Zonca, V.; Cattane, N.; Pariante, C.M.; Cattaneo, A. miRNAs in depression vulnerability and resilience: Novel targets for preventive strategies. J. Neural Transm. 2019, 126, 1241–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, Y.; Roy, B.; Kumar, N.; Shahid Mukhtar, M.; Dwivedi, Y. Molecular pathology associated with altered synaptic transcriptome in the dorsolateral prefrontal cortex of depressed subjects. Transl. Psychiatry 2021, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Differential and unique patterns of synaptic miRNA expression in dorsolateral prefrontal cortex of depressed subjects. Neuropsychopharmacology 2021, 46, 900–910. [Google Scholar] [CrossRef]
- Colameo, D.; Rajman, M.; Soutschek, M.; Bicker, S.; von Ziegler, L.; Bohacek, J.; Winterer, J.; Germain, P.-L.; Dieterich, C.; Schratt, G. Pervasive compartment-specific regulation of gene expression during homeostatic synaptic scaling. EMBO Rep. 2021, 22, e52094. [Google Scholar] [CrossRef]
- O’Carroll, D.; Schaefer, A. General Principals of miRNA Biogenesis and Regulation in the Brain. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2013, 38, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Stappert, L.; Klaus, F.; Brüstle, O. MicroRNAs Engage in Complex Circuits Regulating Adult Neurogenesis. Front. Neurosci. 2018, 12, 707. [Google Scholar] [CrossRef]
- Follert, P.; Cremer, H.; Beclin, C. MicroRNAs in brain development and function: A matter of flexibility and stability. Front. Mol. Neurosci. 2014, 7, 24570654. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, M.M.J.; Krauskopf, J.; Ramaekers, J.G.; Kleinjans, J.C.S.; Prickaerts, J.; Briedé, J.J. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog. Neurobiol. 2020, 185, 101732. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Hsu, P.K.; Karayiorgou, M.; Gogos, J.A. MicroRNA dysregulation in neuropsychiatric disorders and cognitive dysfunction. Neurobiol. Dis. 2012, 46, 291–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, H.C.; Schratt, G. MicroRNA-dependent control of neuroplasticity in affective disorders. Transl. Psychiatry 2021, 11, 263. [Google Scholar] [CrossRef]
- Smalheiser, N.R.; Lugli, G.; Rizavi, H.S.; Torvik, V.I.; Turecki, G.; Dwivedi, Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE 2012, 7, e33201. [Google Scholar] [CrossRef]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Żurawek, D.; Turecki, G. The miRNome of Depression. Int. J. Mol. Sci. 2021, 22, 11312. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.P.; Lim, R.; Cruceanu, C.; Crapper, L.; Fasano, C.; Labonte, B.; Maussion, G.; Yang, J.P.; Yerko, V.; Vigneault, E.; et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat. Med. 2014, 20, 764–768. [Google Scholar] [CrossRef]
- Dadkhah, T.; Rahimi-Aliabadi, S.; Jamshidi, J.; Ghaedi, H.; Taghavi, S.; Shokraeian, P.; Akhavan-Niaki, H.; Tafakhori, A.; Ohadi, M.; Darvish, H. A genetic variant in miRNA binding site of glutamate receptor 4, metabotropic (GRM4) is associated with increased risk of major depressive disorder. J. Affect. Disord. 2017, 208, 218–222. [Google Scholar] [CrossRef]
- Roy, B.; Dunbar, M.; Shelton, R.C.; Dwivedi, Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2017, 42, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, J.A.; Carter, B.S.; Meng, F.; Turner, D.L.; Dai, M.; Schatzberg, A.F.; Barchas, J.D.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; et al. The microRNA network is altered in anterior cingulate cortex of patients with unipolar and bipolar depression. J. Psychiatr. Res. 2016, 82, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Bavamian, S.; Mellios, N.; Lalonde, J.; Fass, D.M.; Wang, J.; Sheridan, S.D.; Madison, J.M.; Zhou, F.; Rueckert, E.H.; Barker, D.; et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol. Psychiatry 2015, 20, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.P.; Bruse, S.E.; David-Rus, R.; Buyske, S.; Brzustowicz, L.M. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 2011, 69, 188–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camkurt, M.A.; Karababa, I.F.; Erdal, M.E.; Kandemir, S.B.; Fries, G.R.; Bayazit, H.; Ay, M.E.; Kandemir, H.; Ay, O.I.; Coskun, S.; et al. MicroRNA dysregulation in manic and euthymic patients with bipolar disorder. J. Affect. Disord. 2020, 261, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.T.; Zakharenko, S.S. MicroRNAs in the Onset of Schizophrenia. Cells 2021, 10, 2679. [Google Scholar] [CrossRef] [PubMed]
- Punzi, G.; Bharadwaj, R.; Ursini, G. Neuroepigenetics of Schizophrenia. Prog. Mol. Biol. Transl. Sci. 2018, 158, 195–226. [Google Scholar] [CrossRef]
- Khavari, B.; Cairns, M.J. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells 2020, 9, 1837. [Google Scholar] [CrossRef]
- Cao, T.; Zhen, X.C. Dysregulation of miRNA and its potential therapeutic application in schizophrenia. CNS Neurosci. Ther. 2018, 24, 586–597. [Google Scholar] [CrossRef]
- Hauberg, M.E.; Roussos, P.; Grove, J.; Borglum, A.D.; Mattheisen, M.; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Analyzing the Role of MicroRNAs in Schizophrenia in the Context of Common Genetic Risk Variants. JAMA Psychiatry 2016, 73, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Cattaneo, A.; Riva, M.A. Stress-induced mechanisms in mental illness: A role for glucocorticoid signalling. J. Steroid Biochem. Mol. Biol. 2016, 160, 169–174. [Google Scholar] [CrossRef]
- Gjerstad, J.K.; Lightman, S.L.; Spiga, F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 2018, 21, 403–416. [Google Scholar] [CrossRef] [Green Version]
- McEwen, B.S. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol. Rev. 2007, 87, 873–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smalheiser, N.R.; Lugli, G.; Rizavi, H.S.; Zhang, H.; Torvik, V.I.; Pandey, G.N.; Davis, J.M.; Dwivedi, Y. MicroRNA expression in rat brain exposed to repeated inescapable shock: Differential alterations in learned helplessness vs. non-learned helplessness. Int. J. Neuropsychopharmacol./Off. Sci. J. Coll. Int. Neuropsychopharmacol. CINP 2011, 14, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Nishida, A.; Hara, K.; Kamemoto, T.; Suetsugi, M.; Fujimoto, M.; Watanuki, T.; Wakabayashi, Y.; Otsuki, K.; McEwen, B.S.; et al. Characterization of the vulnerability to repeated stress in Fischer 344 rats: Possible involvement of microRNA-mediated down-regulation of the glucocorticoid receptor. Eur. J. Neurosci. 2008, 27, 2250–2261. [Google Scholar] [CrossRef]
- Kawashima, H.; Numakawa, T.; Kumamaru, E.; Adachi, N.; Mizuno, H.; Ninomiya, M.; Kunugi, H.; Hashido, K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 2010, 165, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Zhu, X.-Z.; Zhang, Y.; Zhang, S.; Zhang, L.; Xue, L.; Zhong, M.; Zhang, X. Anhedonia was associated with the dysregulation of hippocampal HTR4 and microRNA Let-7a in rats. Physiol. Behav. 2014, 129, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, R.; Liu, Y.; Liu, D.; Jiang, H.; Pan, F. FKBP5 and specific microRNAs via glucocorticoid receptor in the basolateral amygdala involved in the susceptibility to depressive disorder in early adolescent stressed rats. J. Psychiatr. Res. 2017, 95, 102–113. [Google Scholar] [CrossRef]
- Beveridge, N.J.; Tooney, P.A.; Carroll, A.P.; Tran, N.; Cairns, M.J. Down-regulation of miR-17 family expression in response to retinoic acid induced neuronal differentiation. Cell. Signal. 2009, 21, 1837–1845. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef]
- Yu, J.Y.; Chung, K.H.; Deo, M.; Thompson, R.C.; Turner, D.L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008, 314, 2618–2633. [Google Scholar] [CrossRef] [Green Version]
- Haramati, S.; Navon, I.; Issler, O.; Ezra-Nevo, G.; Gil, S.; Zwang, R.; Hornstein, E.; Chen, A. MicroRNA as repressors of stress-induced anxiety: The case of amygdalar miR-34. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 14191–14203. [Google Scholar] [CrossRef]
- Alural, B.; Genc, S.; Haggarty, S.J. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: Past, present, and future. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2017, 73, 87–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, F.; Uchida, S.; Yamagata, H.; Abe-Higuchi, N.; Hobara, T.; Hara, K.; Kobayashi, A.; Shintaku, T.; Itoh, Y.; Suzuki, T.; et al. Hippocampal MicroRNA-124 Enhances Chronic Stress Resilience in Mice. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 7253–7267. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.G.; Domschke, K.; Schiele, M.A. Epigenetics Underlying Susceptibility and Resilience Relating to Daily Life Stress, Work Stress, and Socioeconomic Status. Front. Psychiatry 2020, 11, 163. [Google Scholar] [CrossRef] [Green Version]
- Nowakowski, T.J.; Rani, N.; Golkaram, M.; Zhou, H.R.; Alvarado, B.; Huch, K.; West, J.A.; Leyrat, A.; Pollen, A.A.; Kriegstein, A.R.; et al. Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nat. Neurosci. 2018, 21, 1784–1792. [Google Scholar] [CrossRef]
- McKibben, L.A.; Dwivedi, Y. Early-life stress induces genome-wide sex-dependent miRNA expression and correlation across limbic brain areas in rats. Epigenomics 2021, 13, 1031–1056. [Google Scholar] [CrossRef] [PubMed]
- Torres-Berrio, A.; Morgunova, A.; Giroux, M.; Cuesta, S.; Nestler, E.J.; Flores, C. miR-218 in Adolescence Predicts and Mediates Vulnerability to Stress. Biol. Psychiatry 2021, 89, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Tran, N. MicroRNAs Regulating MicroRNAs in Cancer. Trends Cancer 2018, 4, 465–468. [Google Scholar] [CrossRef]
- Rheinheimer, B.A.; Vrba, L.; Futscher, B.W.; Heimark, R.L. Identification and transcriptional regulation of the mir-218-1 alternative promoter. bioRxiv 2020. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2018, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Gonda, Y.; Namba, T.; Hanashima, C. Beyond Axon Guidance: Roles of Slit-Robo Signaling in Neocortical Formation. Front. Cell Dev. Biol. 2020, 8, 607415. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Sutherland, L.B.; Rajagopalan, K.N.; Wang, S.; Olson, E.N. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ. Res. 2010, 107, 1336–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, H.; Yan, Y.; Hu, M.; Wang, Y.; Wang, Y.; Dai, Y.; Chen, J.; Di, G.; Chen, X.; Jiang, X. MiR-218 sensitizes glioma cells to apoptosis and inhibits tumorigenicity by regulating ECOP-mediated suppression of NF-kappaB activity. Neuro. Oncol. 2013, 15, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Amin, N.D.; Bai, G.; Klug, J.R.; Bonanomi, D.; Pankratz, M.T.; Gifford, W.D.; Hinckley, C.A.; Sternfeld, M.J.; Driscoll, S.P.; Dominguez, B.; et al. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 2015, 350, 1525–1529. [Google Scholar] [CrossRef] [Green Version]
- Thiebes, K.P.; Nam, H.; Cambronne, X.A.; Shen, R.; Glasgow, S.M.; Cho, H.-H.; Kwon, J.-S.; Goodman, R.H.; Lee, J.W.; Lee, S.; et al. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1–Lhx3. Nat. Commun. 2015, 6, 7718. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.; Choi, H.; Kim, J. Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 2014, 1587, 23–32. [Google Scholar] [CrossRef]
- Kaalund, S.S.; Venø, M.T.; Bak, M.; Møller, R.S.; Laursen, H.; Madsen, F.; Broholm, H.; Quistorff, B.; Uldall, P.; Tommerup, N.; et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia 2014, 55, 2017–2027. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Geng, X.; Ban, J.; Liu, Y. MicroRNA-218 inhibits type I interferon production and facilitates virus immune evasion via targeting RIG-I. Biotechnol. Appl. Biochem. 2020, 67, 396–403. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Cui, J.; Sun, M.; Du, W.; Chen, T.; Ming, X.; Zhang, L.; Tian, J.; Li, J.; et al. MiR218 Modulates Wnt Signaling in Mouse Cardiac Stem Cells by Promoting Proliferation and Inhibiting Differentiation through a Positive Feedback Loop. Sci. Rep. 2016, 6, 20968. [Google Scholar] [CrossRef] [Green Version]
- Mariani, N.; Cattane, N.; Pariante, C.; Cattaneo, A. Gene expression studies in Depression development and treatment: An overview of the underlying molecular mechanisms and biological processes to identify biomarkers. Transl. Psychiatry 2021, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, S.; Restrepo-Lozano, J.M.; Silvestrin, S.; Nouel, D.; Torres-Berrio, A.; Reynolds, L.M.; Arvanitogiannis, A.; Flores, C. Non-Contingent Exposure to Amphetamine in Adolescence Recruits miR-218 to Regulate Dcc Expression in the VTA. Neuropsychopharmacology 2018, 43, 900–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Berrio, A.; Lopez, J.P.; Bagot, R.C.; Nouel, D.; Dal Bo, G.; Cuesta, S.; Zhu, L.; Manitt, C.; Eng, C.; Cooper, H.M.; et al. DCC Confers Susceptibility to Depression-like Behaviors in Humans and Mice and Is Regulated by miR-218. Biol. Psychiatry 2017, 81, 306–315. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.Y.; Choi, J.E.; Kim, M.; Hong, J.; Park, Y. N-3 PUFA Have Antidepressant-like Effects via Improvement of the HPA-Axis and Neurotransmission in Rats Exposed to Combined Stress. Mol. Neurobiol. 2020, 57, 3860–3874. [Google Scholar] [CrossRef]
- Choi, J.E.; Kim, E.Y.; Park, Y. N-3 PUFA improved pup separation-induced postpartum depression via serotonergic pathway regulated by miRNA. J. Nutr. Biochem. 2020, 84, 108417. [Google Scholar] [CrossRef] [PubMed]
- Patchev, V.K.; Patchev, A.V. Experimental models of stress. Dialogues Clin. Neurosci. 2006, 8, 417–432. [Google Scholar] [CrossRef]
- Vosberg, D.E.; Leyton, M.; Flores, C. The Netrin-1/DCC guidance system: Dopamine pathway maturation and psychiatric disorders emerging in adolescence. Mol. Psychiatry 2020, 25, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Sapolsky, R.M.; Romero, L.M.; Munck, A.U. How Do Glucocorticoids Influence Stress Responses? Integrating Permissive, Suppressive, Stimulatory, and Preparative Actions*. Endocr. Rev. 2000, 21, 55–89. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Sun, B.; Xu, P.; Zhu, Y.; Meng, X.-H.; Teng, G.-J.; Xiao, Z.-D. MiR-218 Induces Neuronal Differentiation of ASCs in a Temporally Sequential Manner with Fibroblast Growth Factor by Regulation of the Wnt Signaling Pathway. Sci. Rep. 2017, 7, 39427. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhao, G. Downregulation of microRNA-218 relieves neuropathic pain by regulating suppressor of cytokine signaling 3. Int. J. Mol. Med. 2016, 37, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Bak, M.; Silahtaroglu, A.; Moller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA expression in the adult mouse central nervous system. RNA 2008, 14, 432–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, L.; Klausen, M.; Helboe, L.; Nielsen, F.C.; Werge, T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS ONE 2009, 4, e7225. [Google Scholar] [CrossRef] [PubMed]
- Jurkiewicz, M.; Moser, D.; Koller, A.; Yu, L.; Chen, E.I.; Bennett, D.A.; Canli, T. Integration of postmortem amygdala expression profiling, GWAS, and functional cell culture assays: Neuroticism-associated synaptic vesicle glycoprotein 2A (SV2A) gene is regulated by miR-133a and miR-218. Transl. Psychiatry 2020, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, S.; Nagy, C.; Théroux, J.-F.; Wakid, M.; Fiori, L.M.; Ibrahim, P.; Yang, J.; Rotzinger, S.; Foster, J.A.; Mechawar, N.; et al. Neuron-derived extracellular vesicles extracted from plasma show altered size and miRNA cargo as a function of antidepressant drug response. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, W.; Yang, L.; Zhang, D.; Gao, C.; Wu, J.; Zhu, Y.; Zhang, H. MicroRNA-218 Negatively Regulates Osteoclastogenic Differentiation by Repressing the Nuclear Factor-kappaB Signaling Pathway and Targeting Tumor Necrosis Factor Receptor 1. Cell Physiol. Biochem. 2018, 48, 339–347. [Google Scholar] [CrossRef]
- Salk, R.H.; Hyde, J.S.; Abramson, L.Y. Gender differences in depression in representative national samples: Meta-analyses of diagnoses and symptoms. Psychol. Bull. 2017, 143, 783–822. [Google Scholar] [CrossRef]
- Piccinelli, M.; Wilkinson, G. Gender differences in depression: Critical review. Br. J. Psychiatry 2000, 177, 486–492. [Google Scholar] [CrossRef] [Green Version]
- Strand, N.; Fang, L.; Carlson, J.M. Sex Differences in Anxiety: An Investigation of the Moderating Role of Sex in Performance Monitoring and Attentional Bias to Threat in High Trait Anxious Individuals. Front. Hum. Neurosci. 2021, 15. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schell, G.; Roy, B.; Prall, K.; Dwivedi, Y. miR-218: A Stress-Responsive Epigenetic Modifier. Non-Coding RNA 2022, 8, 55. https://doi.org/10.3390/ncrna8040055
Schell G, Roy B, Prall K, Dwivedi Y. miR-218: A Stress-Responsive Epigenetic Modifier. Non-Coding RNA. 2022; 8(4):55. https://doi.org/10.3390/ncrna8040055
Chicago/Turabian StyleSchell, Grant, Bhaskar Roy, Kevin Prall, and Yogesh Dwivedi. 2022. "miR-218: A Stress-Responsive Epigenetic Modifier" Non-Coding RNA 8, no. 4: 55. https://doi.org/10.3390/ncrna8040055
APA StyleSchell, G., Roy, B., Prall, K., & Dwivedi, Y. (2022). miR-218: A Stress-Responsive Epigenetic Modifier. Non-Coding RNA, 8(4), 55. https://doi.org/10.3390/ncrna8040055





