The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer
Abstract
1. Introduction
2. Results
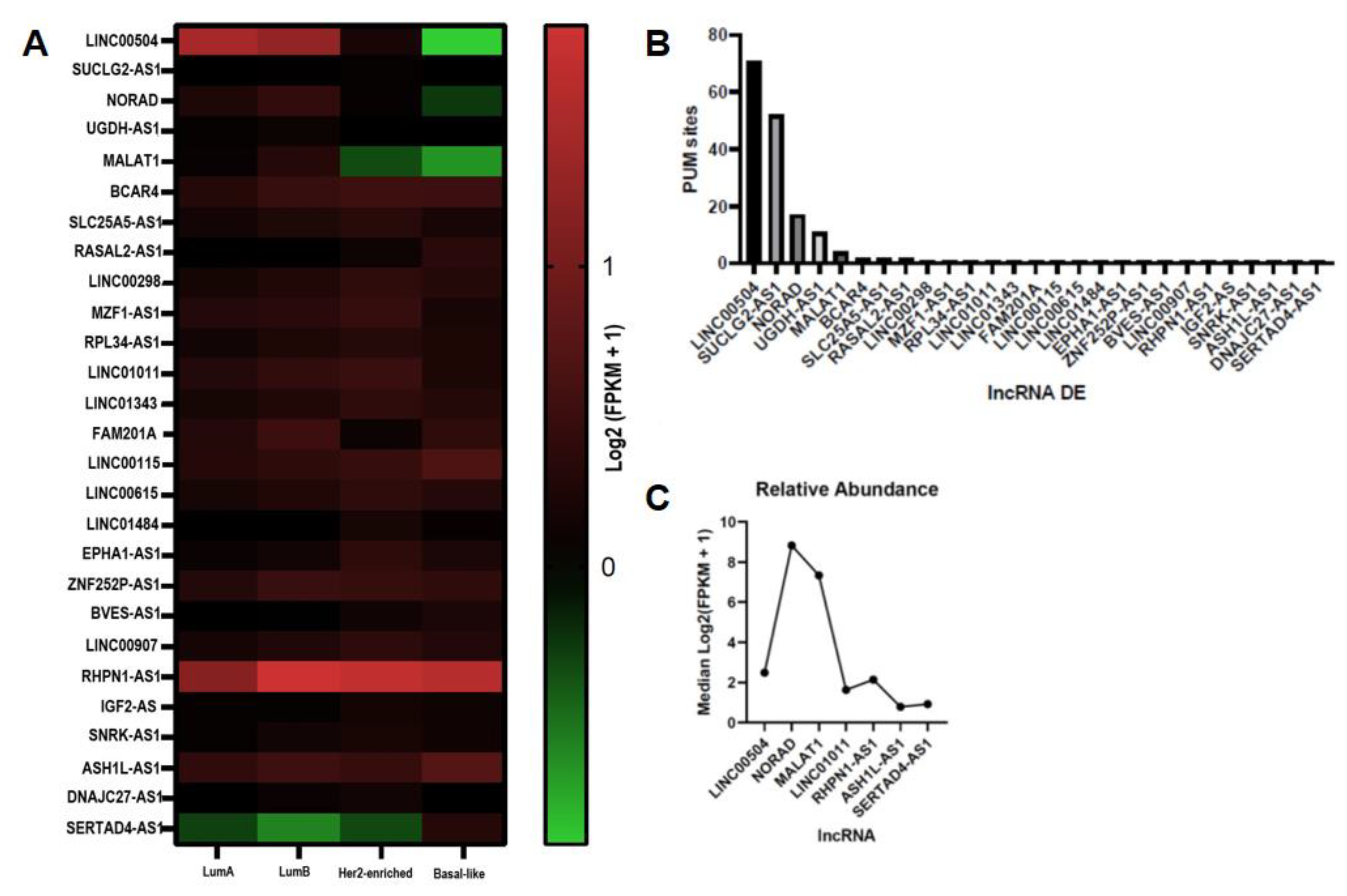
2.1. LncRNA with Sites for PUMILIO Are Differentially Expressed in BRCA
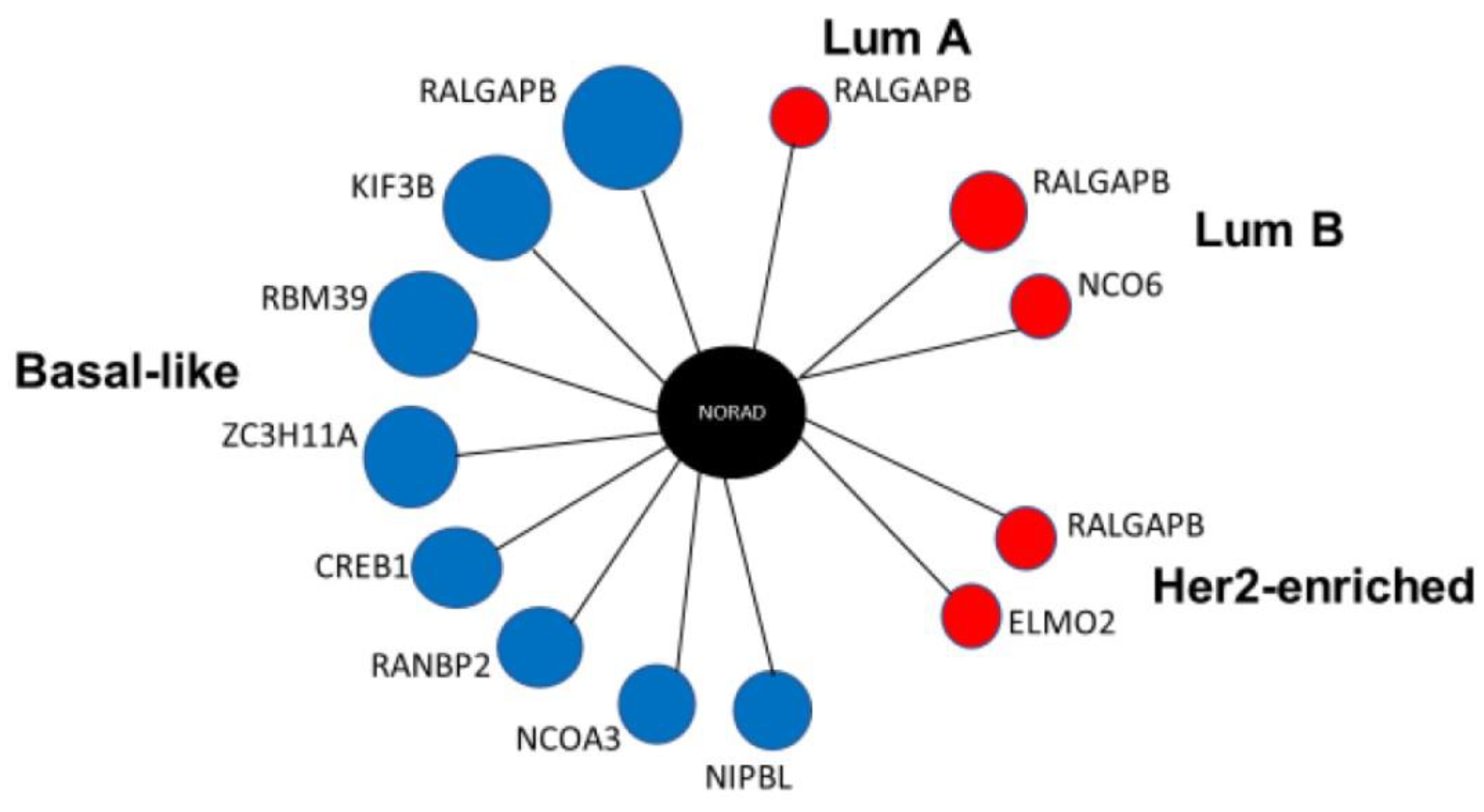
2.2. LncRNA NORAD Shows Co-Expression Networks in All Four BRCA Subtypes
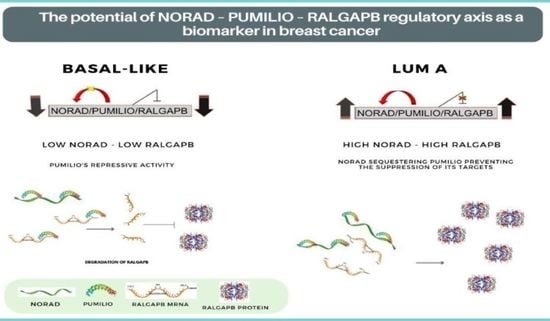
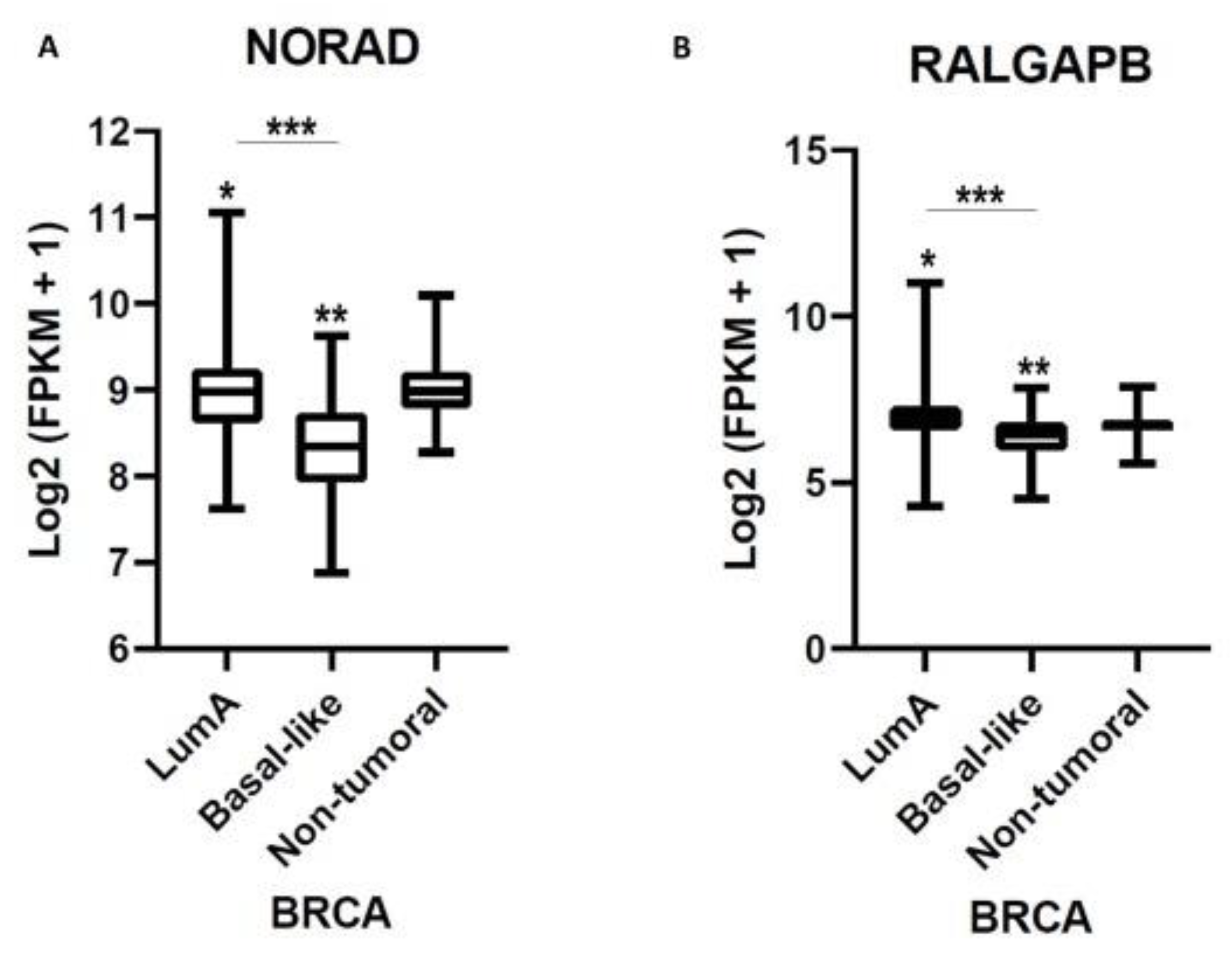
2.3. NORAD and RALGAPB Are Differentially Expressed in Lum A and Basal-Like Subtypes Compared to Non-Tumoral Samples
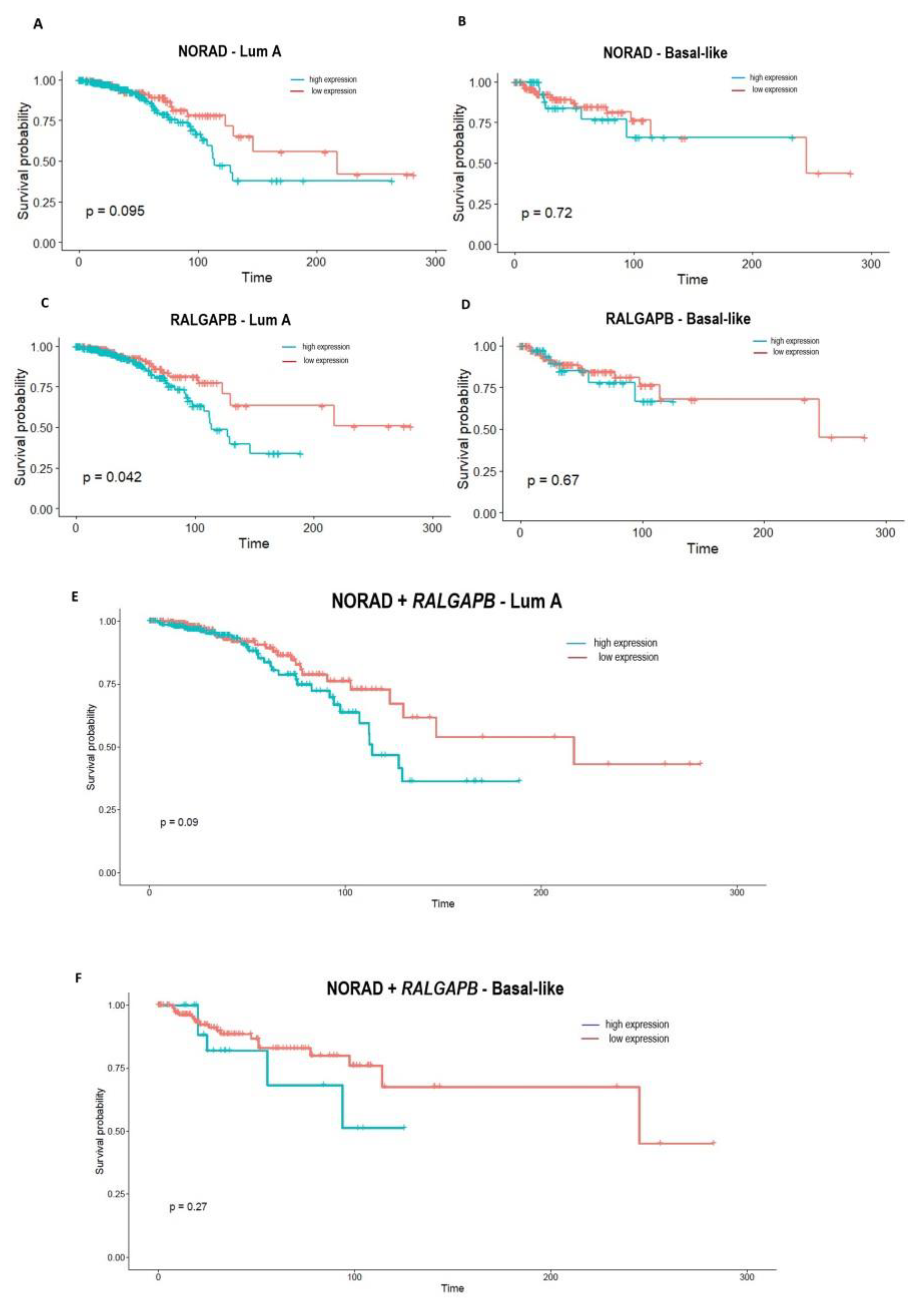
2.4. NORAD and RALGAPB Are up Expressed in Breast Cancer and Related to OS in Patients
2.5. NORAD and RALGAPB Showed a Biomarker Potential, Alone or Combined in Panel
3. Materials and Methods
3.1. TCGA Analysis
3.2. LncRNAs Selection
3.3. Co-Expression Regulatory Network
3.4. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BRCA | breast cancer |
| DFS | disease-free survival |
| FC | fold change |
| FPKM | Fragments Per Kilobase Million |
| HR | Hazard Ratio |
| Lum A | luminal A |
| Lum B | luminal B |
| LncRNA | Long non-coding RNAs |
| NRU | NORAD repeat unit |
| OS | overall survival |
| RBP | RNA-binding protein |
| RNP | ribonucleoprotein particle |
| ROC | receiver operating characteristic |
| PRE | PUM recognition element |
| TCGA | The Cancer Genome Atlas |
| TRN | transcriptional regulatory network |
References
- Eroles, P.; Bosch, A.; Pérez-Fidalgo, J.A.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Urasaki, T. Precision medicine in breast cancer. Chin. Clin. Oncol. 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Fisusi, F.A.; Akala, E.O. Drug Combinations in Breast Cancer Therapy. Pharm Nanotechnol 2019, 7, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.; Braybrooke, J.; Gray, R.; Hills, R.; Liu, Z.; Peto, R.; Davies, L.; Dodwell, D.; McGale, P.; Pan, H. Trastuzumab for early-stage, HER2-positive breast cancer: A meta-analysis of 13,864 women in seven randomised trials. Lancet Oncol. 2021, 22, 1139–1150. [Google Scholar] [CrossRef]
- Gonçalves, A.; Bertucci, A.; Bertucci, F. PARP inhibitors in the treatment of early breast cancer: The step beyond? Cancers 2020, 12, 1378. [Google Scholar] [CrossRef]
- Andre, F.; Ismaila, N.; Henry, N.L.; Somerfield, M.R.; Bast, R.C.; Barlow, W.; Collyar, D.E.; Hammond, M.E.; Kuderer, N.M.; Liu, M.C.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update—Integration of Results From TAILORx. J. Clin. Oncol. 2019, 37, 1956–1964. [Google Scholar] [CrossRef]
- Lavalée, M.; Curdy, N.; Laurent, C.; Fournié, J.-J.; Franchini, D.-M. Cancer cell adaptability: Turning ribonucleoprotein granules into targets. Trends Cancer 2021, 7, 902–915. [Google Scholar] [CrossRef]
- Ferrè, F.; Colantoni, A.; Helmer-Citterich, M. Revealing protein-lncRNA interaction. Brief Bioinform 2016, 17, 106–116. [Google Scholar] [CrossRef]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci.. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Dai, H.; Shen, K.; Yang, Y.; Su, X.; Luo, Y.; Jiang, Y.; Shuai, L.; Zheng, P.; Chen, Z.; Bie, P. PUM1 knockdown prevents tumor progression by activating the PERK/eIF2/ATF4 signaling pathway in pancreatic adenocarcinoma cells. Cell Death Dis. 2019, 10, 595. [Google Scholar] [CrossRef]
- Goldstrohm, A.C.; Hall, T.M.T.; McKenney, K.M. Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins. Trends Genet. 2018, 34, 972–990. [Google Scholar] [CrossRef] [PubMed]
- Galgano, A.; Forrer, M.; Jaskiewicz, L.; Kanitz, A.; Zavolan, M.; Gerber, A.P. Comparative Analysis of mRNA Targets for Human PUF-Family Proteins Suggests Extensive Interaction with the miRNA Regulatory System. PLoS ONE 2008, 3, e3164. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.-C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2015, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Mathias, C.; Groeneveld, C.; Trefflich, S.; Zambalde, E.; Lima, R.; Urban, C.; Prado, K.; Ribeiro, E.; Castro, M.; Gradia, D.; et al. Novel lncRNAs Co-Expression Networks Identifies LINC00504 with Oncogenic Role in Luminal A Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 2420. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Dang, Y.; Li, D.; Lu, G.; Chan, W.Y.; Leung, P.C.K.; Zhao, S.; Qin, Y.; Chen, Z. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020, 48, 4480–4491. [Google Scholar] [CrossRef]
- A Bohn, J.; Van Etten, J.L.; Schagat, T.L.; Bowman, B.M.; McEachin, R.C.; Freddolino, P.L.; Goldstrohm, A.C. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res. 2017, 46, 362–386. [Google Scholar] [CrossRef]
- Smialek, M.J.; Ilaslan, E.; Sajek, M.P.; Swiercz, A.; Janecki, D.M.; Kusz-Zamelczyk, K.; Wozniak, T.; Kotecki, M.; Handschuh, L.; Figlerowicz, M.; et al. Characterization of RNP Networks of PUM1 and PUM2 Post-Transcriptional Regulators in TCam-2 Cells, a Human Male Germ Cell Model. Cells 2020, 9, 984. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, L.; Xu, F. LncRNA NORAD stimulates proliferation and migration of renal cancer via activating the miR-144-3p/MYCN axis. Eur. Rev. Med. Pharm. Sci. 2020, 24, 10426–10432. [Google Scholar]
- Shi, P.; Zhang, J.; Li, X.; Li, W.; Li, H.; Fu, P. Long non-coding RNA NORAD inhibition upregulates microRNA-323a-3p to suppress tumorigenesis and development of breast cancer through the PUM1/eIF2 axis. Cell Cycle 2021, 20, 1295–1307. [Google Scholar] [CrossRef]
- Li, R.; Qu, H.; Wang, S.; Wei, J.; Zhang, L.; Ma, R.; Lu, J.; Zhu, J.; Zhong, W.-D.; Jia, Z. GDCRNATools: An R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018, 34, 2515–2517. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.A.; de Santiago, I.; Campbell, T.M.; Vaughn, C.; Hickey, T.E.; Ross, E.D.; Tilley, W.; Markowetz, F.; Ponder, B.A.; Meyer, K.B. Regulators of genetic risk of breast cancer identified by integrative network analysis. Nat. Genet. 2015, 48, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhou, X.; Li, Y.; Jiang, H.; Chen, A. Long non-coding rna norad inhibits breast cancer cell proliferation and metastasis by regulating mir-155-5p/ socs1 axis. J. Breast Cancer 2021, 24, 330–343. [Google Scholar] [CrossRef]
- Spassov, D.S.; Jurecic, R. The PUF Family of RNA-binding Proteins: Does Evolutionarily Conserved Structure Equal Conserved Function? IUBMB Life 2003, 55, 359–366. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Khan, E.; Ching, S.R.; Khan, M.; Siddiqui, J.K.; Gradia, D.F.; Lin, C.; Bouley, S.J.; Mercadante, D.; Manning, A.L.; et al. PUMILIO competes with AUF1 to control DICER1 RNA levels and miRNA processing. Nucleic Acids Res. 2022, 50, 7048–7066. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, D.; Qian, H.; Tsai, Y.S.; Shao, S.; Liu, Q.; Dominguez, D.; Wang, Z. The Splicing Factor RBM4 Controls Apoptosis, Proliferation, and Migration to Suppress Tumor Progression. Cancer Cell. 2014, 26, 374–389. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Shukla, C.J.; Weiner, C.L.; Rinn, J.L. Function and evolution of local repeats in the Firre locus. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Elguindy, M.M.; Mendell, J.T. NORAD-induced Pumilio phase separation is required for genome stability. Semin. Cell Dev. Biol. 2021, 595, 303–308. [Google Scholar] [CrossRef]
- Kedde, M.; Van Kouwenhove, M.; Zwart, W.; Oude Vrielink, J.A.F.; Elkon, R.; Agami, R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010, 12, 1014–1020. [Google Scholar] [CrossRef]
- Personnic, N.; Lakisic, G.; Gouin, E.; Rousseau, A.; Gautreau, A.; Cossart, P.; Bierne, H. A role for Ral GTPase-activating protein subunit β in mitotic regulation. FEBS J. 2014, 281, 2977–2989. [Google Scholar] [CrossRef] [PubMed]
| Variables Lum A | Univariate Model | Variables Lum A | Multivariate Model | ||||
|---|---|---|---|---|---|---|---|
| HR | CI | p-value | HR | CI | p-value | ||
| NORAD | 2 | 1.2–3.4 | 0.01 | NORAD | 2.73 | 1.07–6.9 | 0.035 |
| RALGAPB | 1.6 | 0.95–2.8 | 0.079 | RALGAPB | 0.72 | 0.29–1.8 | 0.475 |
| AGE | 1.04 | 1.02–1.1 | <0.001 | ||||
| Comparison | RNA | AUC | Sensitivity | Specificity | p-Value |
|---|---|---|---|---|---|
| LA × NT | NORAD | 0.5369 | 86.73 | 29.64 | =0.2149 |
| RALGAPB | 0.6574 | 88.50 | 46.96 | <0.0001 | |
| NORAD + RALGAPB | 0.7242 | 78.79 | 56.79 | <0.0001 | |
| BL × NT | NORAD | 0.8496 | 88.50 | 72.11 | <0.0001 |
| RALGAPB | 0.6659 | 85.84 | 51.58 | <0.0001 | |
| NORAD + RALGAPB | 0.8588 | 70.53 | 91.15 | <0.0001 | |
| LA × BL | NORAD | 0.7953 | 69.47 | 76.07 | <0.0001 |
| RALGAPB | 0.7555 | 81.58 | 57.86 | <0.0001 | |
| NORAD + RALGAPB | 0.7980 | 68.95 | 76.61 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muller, C.S.M.; Giner, I.S.; Zambalde, É.P.; Carvalho, T.M.; Ribeiro, E.M.d.S.F.; Carvalho de Oliveira, J.; Mathias, C.; Gradia, D.F. The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer. Non-Coding RNA 2022, 8, 76. https://doi.org/10.3390/ncrna8060076
Muller CSM, Giner IS, Zambalde ÉP, Carvalho TM, Ribeiro EMdSF, Carvalho de Oliveira J, Mathias C, Gradia DF. The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer. Non-Coding RNA. 2022; 8(6):76. https://doi.org/10.3390/ncrna8060076
Chicago/Turabian StyleMuller, Cristiane Sato Mara, Igor Samesima Giner, Érika Pereira Zambalde, Tamyres Mingorance Carvalho, Enilze Maria de Souza Fonseca Ribeiro, Jaqueline Carvalho de Oliveira, Carolina Mathias, and Daniela Fiori Gradia. 2022. "The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer" Non-Coding RNA 8, no. 6: 76. https://doi.org/10.3390/ncrna8060076
APA StyleMuller, C. S. M., Giner, I. S., Zambalde, É. P., Carvalho, T. M., Ribeiro, E. M. d. S. F., Carvalho de Oliveira, J., Mathias, C., & Gradia, D. F. (2022). The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer. Non-Coding RNA, 8(6), 76. https://doi.org/10.3390/ncrna8060076