Abstract
During alcoholic fermentation, ethanol accumulation significantly impacts yeast cells by disrupting membrane integrity, increasing permeability, and reducing cell viability. This study evaluated the effects of ethanol stress on the growth, membrane fluidity, and cell surface morphology of Saccharomyces cerevisiae and non-Saccharomyces yeast strains, specifically Torulaspora delbrueckii and Metschnikowia pulcherrima. These strains, commercialized by AEB SpA and preserved at the Unimore Microbial Culture Collection (UMCC), were tested for fermentative performance in grape must and grown under varying ethanol concentrations. Membrane fluidity was measured using Laurdan generalized polarization (GP), while Atomic Force Microscopy (AFM) assessed cell surface morphology. Results indicated that at 10% ethanol, membrane fluidity increased, particularly in strains able to tolerate up to 16% ethanol, which also demonstrated superior fermentative performance. Less tolerant strains, such as T. delbrueckii UMCC 5 and M. pulcherrima UMCC 15, showed smaller increases in fluidity. At 18% ethanol, these strains exhibited severely altered surface morphology and larger surface roughness values, suggesting increased instability under high ethanol stress, while more tolerant strains displayed better-preserved surface morphology and lower roughness values, reflecting enhanced adaptability. These findings offer insights into yeast responses to ethanol stress, supporting the development of more resilient strains for improved fermentation.
1. Introduction
Yeasts play a crucial role in various biotechnological processes, particularly in producing alcoholic beverages. During alcoholic fermentation, yeast cells encounter multiple stress factors, including osmotic stress due to high sugar concentrations in the initial stages, temperature fluctuations, nutrient depletion, and the accumulation of metabolites, especially ethanol [1,2,3,4]. Yeasts of the Saccharomyces cerevisiae species have a native capability to withstand high levels of ethanol that would be lethal to or severely impair the physiology of other organisms. However, ethanol is one of the most significant stressors because of its ability to penetrate cell membranes due to its small size and amphiphilic nature—possessing both a hydroxyl group and a methyl group. This dual solubility allows ethanol to integrate into both the aqueous and lipid phases of the cell, facilitating its penetration of the cell membrane, which is primarily composed of phospholipids, sterols, and proteins. Once ethanol permeates the membrane, it disrupts membrane integrity by altering its fluidity and permeability, leading to the leakage of intracellular components such as amino acids and ions, thereby compromising cellular homeostasis [5,6,7]. Furthermore, ethanol impacts mitochondrial structure, reducing ATP levels and respiration rates while promoting the production of acetaldehyde and reactive oxygen species. These events can result in lipid peroxidation, DNA damage, and oxidative stress, ultimately decreasing cell viability [8,9]. The alteration of membrane properties is central to the mechanism underlying ethanol toxicity. The composition of membrane fatty acids and sterols, such as ergosterol, plays a key role in determining membrane fluidity, which is essential for maintaining the proper structure and function of the membrane [10]. Membrane fluidity is influenced by the chain length and degree of saturation of fatty acids: a higher proportion of saturated fatty acids (SFAs) results in increased membrane rigidity, which can lead to lipid bilayer stress and trigger the unfolded protein response (UPR) [11,12]. On the other hand, the failure to adequately regulate unsaturated fatty acids (UFAs) can disrupt organelle organization and, in severe cases, lead to cell death. In S. cerevisiae, the production of unsaturated fatty acids largely depends on the Δ9 acyl-CoA desaturase, Ole1p. This enzyme’s activity is tightly controlled through the OLE pathway, which plays a critical role in preserving membrane fluidity, especially under ethanol-induced stress [13]. The OLE pathway is responsible for the regulated activation of the transcription factors Mga2 and Spt23 from the endoplasmic reticulum (ER) [14,15]. These factors then migrate to the nucleus to promote the expression of the OLE1 gene, which encodes the essential Δ9-fatty acid desaturase. This enzyme is responsible for the de novo biosynthesis of UFAs, which serve as fundamental lipid building blocks for maintaining membrane integrity and cellular function [16]. Although S. cerevisiae yeasts have traditionally been regarded as the primary agents in alcoholic fermentation, the role of non-Saccharomyces yeasts should not be overlooked. A diverse range of yeast species is known to inhabit freshly crushed grape juice, primarily from the genera Hanseniaspora (anamorph Kloeckera), Pichia, Candida, Metschnikowia, Kluyveromyces, and Saccharomyces. Additionally, species from other genera, such as Zygosaccharomyces, Saccharomycodes, Torulaspora, Brettanomyces, and Schizosaccharomyces, may also be present [17]. While many non-Saccharomyces species, particularly Hanseniaspora, Candida, Pichia, and Metschnikowia, initiate the early stages of indigenous alcoholic fermentation, they are soon overtaken by S. cerevisiae, which dominates the mid to final stages of the process, often becoming the only species present in the fermenting juice [18]. Historically, non-Saccharomyces yeasts were regarded as spoilage organisms due to their association with stuck fermentations, low ethanol tolerance, reduced acidity production, high residual sugar levels, and off-flavors [19,20,21]. However, the role of non-Saccharomyces yeasts in winemaking has been re-evaluated. Nowadays, winemakers demand yeasts as starter cultures with a whole range of specific properties that largely differ according to the type and style of wine to be made, as well as the technical requirements of the winery [22]. Controlled mixed fermentations involving both Saccharomyces and non-Saccharomyces species have shown numerous benefits [23]. This approach enhances both the complexity and specificity of wine, as non-Saccharomyces species contribute by providing specific compounds during the early stages of fermentation before S. cerevisiae takes over, ensuring the optimal progression of alcoholic fermentation and ultimately improving wine quality [19,24,25]. Mixed fermentations have been shown to increase the production of desirable metabolites, such as acetate esters [26] and glycerol [27,28], while certain non-Saccharomyces yeasts release more polysaccharides than S. cerevisiae, improving mouthfeel and improving wine stability [29]. Among these, Torulaspora delbrueckii and Metschnikowia pulcherrima have attracted particular attention for their unique contributions. T. delbrueckii has been demonstrated to boost the presence of volatile compounds [30,31], produce higher polysaccharide levels compared to S. cerevisiae, exhibit strong β-glucosidase activity that releases aromatic compounds [32], and reduce volatile acidity. Similarly, M. pulcherrima produces β-glucosidase, which not only lowers volatile acidity but also increases the production of higher alcohols, esters, terpenols, and glycerol [27,33]. Furthermore, M. pulcherrima has been reported to reduce titratable acidity, enhancing the sensory balance of the final wine product [27,34].
In this study, we investigated the effect of ethanol stress on the growth, membrane fluidity, and surface morphology of industrial oenological yeast strains, including both S. cerevisiae and non-Saccharomyces strains with distinct fermentative profiles. Specifically, we assessed the growth of selected strains in both solid and liquid media with varying ethanol concentrations, monitored membrane fluidity changes using Laurdan generalized polarization (GP), and examined morphological alterations on the cell surface through atomic force microscopy (AFM). The results of this study provide novel insights into the effects of ethanol in Saccharomyces and non-Saccharomyces yeasts that can contribute to optimizing oenological industrial fermentation processes.
2. Materials and Methods
2.1. Yeast Strains and Culture Conditions
In the present study, eleven strains of commercial active dry yeast (ADY) were used, which were produced and supplied by the AEB Group (Brescia, BS, Italy). Specifically, nine strains belonged to the Saccharomyces cerevisiae species, while the other two were non-Saccharomyces. All the strains used in this study were deposited at the Unimore Microbial Culture Collection (UMCC) of the University of Modena and Reggio Emilia, Italy. The yeast strains, their commercial names, and corresponding UMCC codes are listed in Table 1. ADY strains were revitalized in YPD broth (1% yeast extract, 2% peptone, and 2% glucose) (Biolife, Milan, Italy) at the temperature specified in the technical data sheet for each strain. Cultures were stored for long-term preservation at −80 °C in cryovials containing YPD broth supplemented with 25% glycerol (v/v) as a cryopreservative. A working copy of the culture strains was kept on YPDA medium (1% yeast extract, 1% peptone, 2% glucose, 2% agar) (Biolife, Milan, Italy) and stored at +4 °C.

Table 1.
Yeast strains used in this study.
2.2. Microvinification Assay in Grape Juice
For the microvinification experiments, 1 g of ADY culture was rehydrated in 10 mL of sucrose solution (20 g/L) at a controlled temperature according to the manufacturer-recommended range. Specifically, S. cerevisiae strains were rehydrated at 37 °C, while non-Saccharomyces strains were rehydrated at 30 °C, as these temperatures were optimized for their physiological characteristics. Microvinifications were performed in triplicate using 100 mL glass bottles filled with 90 mL of commercial filtered grape juice (pH 3.36, Titratable Acidity 5.7 g/L, and sugars 170.83 g/L). Subsequently, 5 mL of the rehydrated ADY was inoculated, and the flasks were sealed with 5 mL of paraffin oil (Carlo Erba, Milan, Italy) to ensure anaerobic conditions. Filtered grape juice without the addition of ADY was used as a control. The incubation was conducted at 25 °C under static conditions, and the weight loss of the flasks was measured daily to monitor the progress of fermentation over 12 days. The released carbon dioxide (CO2) was calculated based on the observed weight loss, which directly corresponds to the amount of CO2 produced during the fermentation process. At the end of the fermentation, the samples were collected and immediately analyzed for pH and titratable acidity. Samples for high-performance liquid chromatography (HPLC) analysis were stored at −20 °C until use. The fermentative fitness, defined as the ability to ferment in relation to the CO2 produced, was evaluated using the method described by Bonciani et al. [35]. This was achieved through the interpolation of the fermentation curves, performed with a fifth-degree polynomial function using the software GraphPad Prism v.10.1.1 (GraphPad Software Inc., San Diego, CA, USA). Two kinetic parameters were then defined as follows:
- -
- tR1/2: the time (in days) required to release half of the total CO2 produced by the best fermenter;
- -
- tF1/2: the time (in days) required to release half of the total CO2 at the end of fermentation for each strain.
Both these terms were calculated for each strain, considering the average values of the three replicates. The ratio (fermentative ratio, FR) between these variables was termed FR = tR1/2/tF1/2. An arbitrary cutoff value, FR ≥ 0.90, was used to designate high-performance strains. Additionally, another value was obtained for each fermentation replicate by measuring the amount of CO2 developed (g) after 2 days of fermentation, referred to as fermentative vigor (FV).
Analytical Methods
The pH and titratable acidity of the samples were determined with an XSPH 80 PRO STIRRER (Securlab, Roma, Italy). Titratable acidity was measured by titration with a 0.1 M NaOH solution. After filtration through 0.45 μm nitrocellulose membranes, 20 µL of each filtered sample collected at the end of the fermentation trial were injected into a Jasco LC-Net II/ADC HPLC system (Jasco Inc., Hachioji, Japan), equipped with a Jasco PU-2080 Plus pump, for the analysis of sugars and organic acids. Isocratic elution was carried out using a 300 × 7.8 mm Aminex® HPX-87H column (Bio-Rad Laboratories, Segrate, Italy) maintained at 40 °C with an Eldex CH-150 oven (Eldex Corp., Napa, CA, USA). The mobile phase consisted of 0.005 N H2SO4 and 5% v/v acetonitrile, with a flow rate of 0.6 mL/min. Calibration curves for the standards were generated using Jasco ChromNav software v. 1.18.03 (Tokyo, Japan), which was also used for peak integration and adjustment.
2.3. Phenotypic Growth Test
2.3.1. Assay in Culture Media at Different Ethanol Concentrations
The preliminary screening of the yeast strains involved evaluating their growth ability in a selective medium supplemented with different ethanol concentrations. The culture strains were rehydrated from −80 °C storage and subsequently cultured in YPD broth at 27 °C for 24 h. Cell counts were performed using a Bürker chamber (Brand, Wertheim, Germany) to standardize the inoculum concentration for plating. Growth tests were carried out on YPDA medium supplemented with various ethanol concentrations (0, 10, 12, 13, 14, 16, and 18% v/v). For each strain, 5 µL of culture at a standardized concentration of 106 cells/mL were spotted onto the agar plates, which were then incubated at 27 °C for 96 h. All tests were performed in triplicate.
2.3.2. Assay in Broth at Different Ethanol Concentrations
Pre-cultures of each strain were prepared in YPD broth and incubated at 27 °C for 16 h. A concentration of 0.5 OD600 for all strains was inoculated into 20 mL YPD broth with different ethanol percentages (0, 10, and 14% v/v). Measurements were taken at 600 nm with a spectrophotometer (Jasko V-550, Tokyo, Japan) after 0, 4, 6, 24, 32, 48, 56, and 72 h. The experiment was carried out in triplicate.
2.4. Laurdan Membrane Fluidity Assay
Yeast cultures preserved at −80 °C were rehydrated in YPD broth and incubated at 27 °C for 16 h. Subsequently, 0.5 OD600 of the cultured strains was inoculated into 5 mL of YPD broth containing ethanol at concentrations of 10%, 14%, 16%, and 18% (v/v). YPD broth without ethanol was also inoculated as a control. After 24 h of fermentation, a cell aliquot was standardized by diluting to 0.4 OD600. The samples were centrifuged (8800× g for 5 min), and the cells were washed with phosphate-buffered saline (PBS) (Sigma-Aldrich St. Louis, MO, USA) at pH 7.4 and resuspended in fresh PBS. To incorporate the fluorescent probe, the standardized samples were incubated with 5 μM Laurdan (Cayman Chemical, Ann Arbor, MI, USA) for 1 h at 30 °C [3]. Membrane fluidity measurements were performed using a Jasco FP-6200 spectrofluorometer (Jasco Inc., Hachioji, Japan). A cuvette containing an unlabeled cell suspension at the same cell density was used to measure background fluorescence, which was subtracted from the fluorescence readings obtained from the labeled cell suspension. The experiment was carried out in triplicate. The results were expressed as Generalized Polarization (GP), calculated using the following equation:
where
GP = (I440 − I490)/(I440 + I490)
- -
- I440: Emission intensity at 440 nm;
- -
- I490: Emission intensity at 490 nm.
To better compare the data, GP values are expressed as:
GP (relative value) = GP sample treated with ethanol/GP control
2.5. Atomic Force Microscopy
The local morphology and surface topography of yeasts subjected to different concentrations of ethanol for 24 h was assessed by AFM measurements. Samples were processed according to Canetta et al. [36] with minor modifications. Briefly, after inoculating 0.5 OD600 of cultured strains in YPD broth medium supplemented with 0%, 10%, and 18% (v/v) ethanol, the cells were centrifuged at 8800× g for 5 min, then washed and resuspended in filtered distilled water (Arium® Pro system, Sartorius AG, Göttingen, Germany) (0.22 μm). Then, 250 μL of cell suspensions were spread on the surfaces of empty Petri dishes. The cells were then left to dry overnight at room temperature. AFM was performed using a Bioscope I microscope equipped with a Nanoscope IIIA controller (Veeco Metrology, Plainview, NY, USA), operating in a noncontact mode in air and at room temperature. A silicon cantilever with a nominal spring constant of 0.8 Nm−1 and a nominal resonance frequency of 40 kHz (MikroMaschHQ: CSC37/NoAl, MikroMasch, Germany) was used. For each sample, different sets of height and error mode images of decreasing size (from 25 × 25 µm2 to 5 × 5 µm2) were acquired on different areas of the surface at relatively low scanning speeds. The images were analyzed using Gwiddion 2.61 free software (http://gwyddion.net/, accessed on 6 May 2024). The Sq root mean square (RMS) roughness was extracted from 1 × 1 µm2 cropped topographic images (obtained from different 5 × 5 µm2 images) after data leveling by mean plane subtraction and removal of a third-order x-y polynomial background.
2.6. Statistical Analyses
Statistical analysis was carried out by analysis of variance (ANOVA) using SPSS software v. 20.0.0 (IBM, Chicago, IL, USA). Tukey’s multiple comparison test was used to identify significant differences between strains (p < 0.05). All experiments were performed in triplicate.
3. Results and Discussion
This study examined the effects of ethanol stress on the physiological and morphological characteristics of oenological yeast strains. The use of well-documented, commercially available strains provided a solid basis for investigating tolerance mechanisms. We evaluated growth in media supplemented with different ethanol concentrations and assessed membrane fluidity and cell surface changes to optimize industrial fermentation processes.
3.1. Evaluation of the Fermentative Performance of Yeast Strains
In this study, the yeast strains were subjected to fermentation trials in grape juice to assess their winemaking potential on a laboratory scale. Microvinification tests revealed that, among tested strains, four S. cerevisiae strains (UMCC 3065, 3064, 20, and 19) demonstrated strong fermentative performance (FR > 0.90), as indicated by the CO2 production plot (Figure 1). In contrast, the non-Saccharomyces strains showed lower FR and FV values compared to the S. cerevisiae strains, reflecting a reduced fermentative capacity. Within the S. cerevisiae group, UMCC 3066 reached an FR value of 0.90 but exhibited lower FV than the other S. cerevisiae strains. Fermentative vigor at the start of fermentation is essential for technological performance, as it enhances the starter’s ability to dominate the fermentation process and outcompete other microbial populations. Among the non-Saccharomyces strains, T. delbrueckii UMCC 5 and M. pulcherrima UMCC 15 showed the lowest fermentative performance. However, UMCC 5 had higher FR and FV values than UMCC 15, indicating a stronger fermentative capacity compared to the M. pulcherrima strain (Table 2).
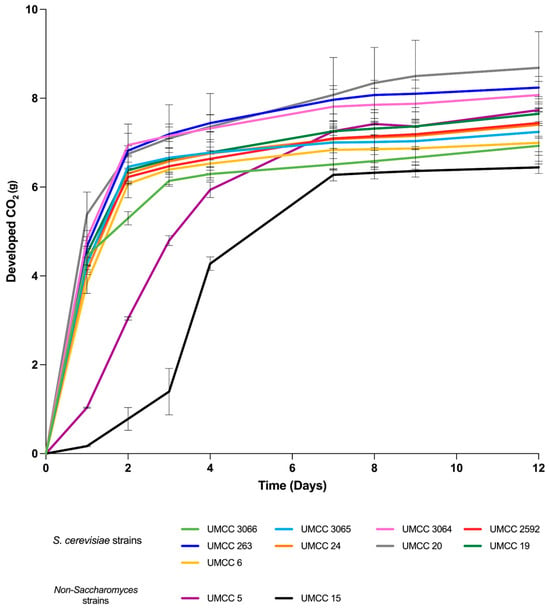
Figure 1.
Developed CO2 over 12 days of microfermentative trials in grape juice. Data are expressed as the mean of triplicate measurements ± standard deviation.

Table 2.
Fermentative ratio (FR) and fermentative vigor (FV) for the eleven yeast strains. ANOVA with Tukey post hoc test (p < 0.05) divided strains into homogeneous subgroups indicated with lowercase alphabetical letters.
At the end of fermentation (12 days), the resulting wines were analyzed for key oenological parameters, including total acidity (TA), residual sugar, ethanol, glycerol, and organic acids (Table 3). In terms of residual sugars, there were no substantial differences among the strains, with even the non-Saccharomyces strains showing low levels of residual sugars. Similarly, no significant differences in ethanol production were observed. Regarding total acidity, the lowest value was attributed to M. pulcherrima (UMCC 15). Some authors have also reported that M. pulcherrima can decrease the titratable acidity of the final wines [27,34]. Notably, UMCC 15 showed the highest glycerol production among the strains, supporting the idea that Metschnikowia tends to increase glycerol levels in wines. Glycerol is an important compound for wine quality, enhancing attributes such as flavor persistence and mouthfeel [37]. During fermentation, glycolysis produces energy without affecting the redox balance, but biomass formation and organic acid synthesis lead to excess NADH [38,39,40,41]. Maintaining redox balance is essential, yet wine yeasts, including S. cerevisiae, lack the enzymes to interconvert NADH and NADPH [42,43,44]. To manage this, S. cerevisiae reduces dihydroxyacetone-3-phosphate to glycerol-3-phosphate, while M. pulcherrima, less adapted to anaerobic fermentation conditions, relies heavily on glycerol production for NAD+ regeneration. This shift to glycerol synthesis helps balance excess NADH, increasing glycerol yield in the final wine [45].

Table 3.
Values of residual sugars (glucose and fructose, expressed in g/L), ethanol and glycerol yields (both expressed as g/100 g of consumed sugar), tartaric acid (expressed in g/L), and succinic acid, acetic acid, citric acid (each expressed as g/100 g of consumed sugar), along with titratable acidity (TA) (expressed in g/L) for 11 strains. The data are the average of three replicates. ANOVA with Tukey post hoc test (p < 0.05) divided strains into homogeneous subgroups indicated with lowercase alphabetical letters.
3.2. Growth at Different Ethanol Concentrations in Solid Media
Ethanol tolerance differences among yeast strains are crucial in selecting strains for winemaking. In particular, ethanol tolerance is especially important due to its strong correlation with fermentative aptitude. After assessing the fermentative aptitude of the strains, phenotypic tests were conducted to evaluate the ethanol resistance in solid media. The screening of the eleven strains on YPDA medium supplemented with varying ethanol concentrations revealed differences in growth, both within strains of the same species and between different species (Figure 2). All S. cerevisiae strains grew up to ethanol concentrations of 14% (v/v), while UMCC 3066, UMCC 3065, UMCC 3064, UMCC 2592, and UMCC 19 grew up to 16% (v/v). All of these strains, except for UMCC 2592 (FR = 0.89), exhibited an FR ≥ 0.90. In contrast, non-Saccharomyces strains exhibited impaired growth when exposed to ethanol concentrations exceeding 10% (v/v) and showed the lowest FR values. The strong correlation observed between ethanol tolerance and fermentative aptitude suggests that strains capable of withstanding higher ethanol concentrations are more likely to maintain robust fermentative aptitude under stressful conditions typical of the later stages of alcoholic fermentation.

Figure 2.
Growth on YPDA medium supplemented with 0, 10, 12, 13, 14, 16, and 18% (v/v) ethanol. The yeast cultures were spotted on plates at a concentration of 106 cells/mL.
3.3. Growth at Different Ethanol Concentrations in Liquid Media
The ethanol tolerance of these eleven strains was further evaluated in YPD broth at 27 °C over 72 h of fermentation. All S. cerevisiae strains demonstrated a moderate decrease in relative optical density (OD600) at a 10% (v/v) ethanol concentration, while a drastic reduction was observed at higher concentrations of 14%, 16%, and 18% (v/v) (Figure 3). This contrasts with the results from ethanol tolerance tests conducted on solid media, where the inhibitory effects were less pronounced. The reduced inhibition observed in solid media is likely due to the immobilizing effect of agar, which provides a more protective environment for yeast cells, as well as the possible evaporation of ethanol from the solid media. Indeed, immobilized yeast cells have been shown to outperform free cells, exhibiting higher ethanol tolerance and reduced substrate inhibition [46]. Moreover, multiple studies have confirmed that immobilized S. cerevisiae produces more ethanol compared to free cells [46,47,48,49]. Except for M. pulcherrima UMCC 15, which showed compromised growth, the other yeast strains, including T. delbrueckii UMCC 5, did not exhibit significantly inhibited growth at 10% (v/v) ethanol concentration. This test allowed for a distinction between the two non-Saccharomyces strains. Specifically, UMCC 15 was strongly inhibited at 10% (v/v), while UMCC 5 exhibited a response similar to the S. cerevisiae strains, indicating better ethanol tolerance.
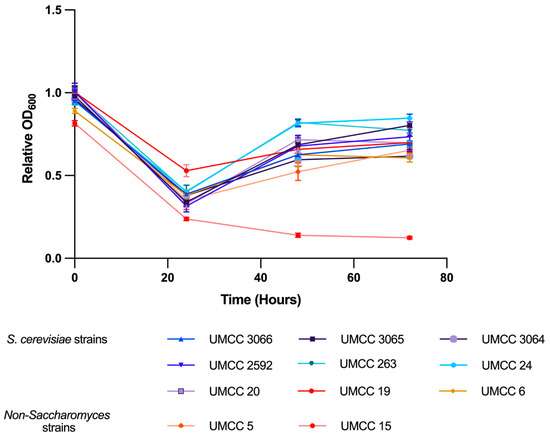
Figure 3.
Growth curves of yeast strains in YPD broth supplemented with 10% (v/v) ethanol. Data are presented as relative OD600 values, normalized to the OD600 of the control (yeast grown without ethanol).
For subsequent analyses, measurements were conducted after 24 h of fermentation, as this time point was identified as the stage where ethanol exerted the most significant inhibitory effect on growth. This time frame led to the greatest growth inhibition across all strains except for M. pulcherrima UMCC 15, where growth remained highly compromised for up to 72 h.
3.4. Evaluation of Membrane Fluidity
Membrane fluidity is a critical factor for maintaining membrane integrity and enabling yeast cells to adapt to ethanol stress. In this study, six yeast strains with varying fermentative aptitudes and ethanol tolerance were selected for membrane fluidity assays. Strain UMCC 3064 was chosen due to its ethanol tolerance up to 16% (v/v), with an FR > 0.90 and the highest FV value. Moreover, strain UMCC 2592 was included because, despite having an FR < 0.90, it exhibited moderately high fermentative vigor, while strain UMCC 3066, with an FR = 0.90, displayed lower fermentative vigor. Among the S. cerevisiae strains, UMCC 24 was also tested, which showed ethanol tolerance up to 14% (v/v) and an FR < 0.90. Additionally, the two non-Saccharomyces strains, UMCC 5 and UMCC 15, were also tested.
The six yeast strains exhibited varying changes in membrane fluidity at 10% (v/v) ethanol, with all showing an increase, though to different extents. These variations corresponded to their levels of ethanol resistance and fermentative capacity. Among the S. cerevisiae strains, UMCC 3066 displayed the highest membrane fluidity, while the non-Saccharomyces strains, T. delbrueckii UMCC 5 and, particularly, M. pulcherrima UMCC 15, had the lowest values. At higher ethanol concentrations (14%, 16%, and 18% v/v), UMCC 15 continued to exhibit the lowest membrane fluidity among the strains, indicating a pronounced stiffening of the membrane (Figure 4).
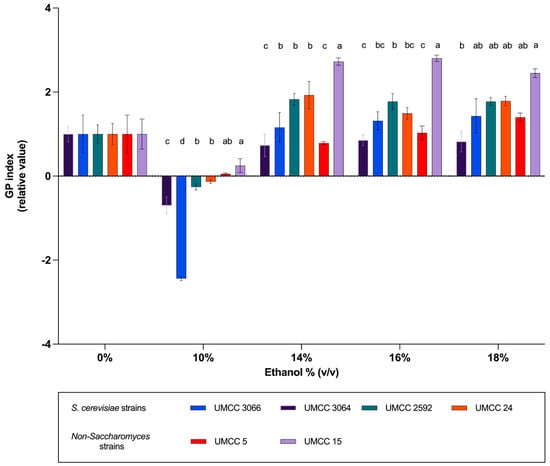
Figure 4.
Membrane fluidity expressed as relative GP index values for strains UMCC 3066, UMCC 3064, UMCC 2592, UMCC 5, and UMCC 15. Significant differences among strains at the same ethanol concentration are indicated by different letters (p < 0.05).
The correlation between membrane fluidity and ethanol tolerance has been extensively documented, although some studies yielded inconsistent results. Ethanol-induced stress primarily targeted the cell membrane, and several authors proposed a link between the fatty acid composition of phospholipid membranes and ethanol tolerance. A well-established relationship existed between ethanol tolerance and an increased degree of fatty acid unsaturation in the membrane lipids of S. cerevisiae [50,51,52,53,54,55,56,57]. In contrast, other researchers, such as Ishmayana et al. [58], suggest an inverse correlation, proposing that lower membrane fluidity might contribute to higher ethanol tolerance in some strains. Our findings indicated that membrane fluidity increased under low ethanol stress, consistent with the observations of Yang et al. [7], who reported that yeast cells adjusted membrane fluidity to counteract ethanol’s disruptive effects. In particular, a correlation between ethanol tolerance and the increase in membrane fluidity at 10% (v/v) ethanol was observed. Strains with higher ethanol tolerance and superior fermentative performance exhibited a greater increase in membrane fluidity compared to less tolerant strains. This pattern was consistent with the findings of Lairón-Peris et al. [59], who noted that the most ethanol-tolerant strain underwent the greatest changes in fluidity, with membranes becoming significantly more fluid at 10% (v/v) ethanol compared to the control without ethanol. This aligned with the findings of Jones and Greenfield [60], who demonstrated that elevated membrane fluidity led to increased cell membrane permeability. As a result, cells with higher membrane fluidity could more effectively expel ethanol, maintaining lower intracellular ethanol concentrations and mitigating ethanol-induced stress. In our study, this explanation was further supported by the behavior of M. pulcherrima UMCC 15, identified as the least ethanol-tolerant strain. This strain exhibited slower fermentative kinetics, showed the smallest increase in membrane fluidity at 10% (v/v) ethanol, and experienced a significant reduction in fluidity at higher ethanol concentrations. Its limited ability to adjust membrane fluidity under ethanol stress likely accounted for its reduced ethanol tolerance and impaired fermentation performance.
3.5. Evaluation of Cell Surface Morphology Under Ethanol Stress
Atomic force microscopy is a powerful tool for directly visualizing and quantifying physical, morphological, and structural changes occurring on the yeast cell surface in response to different stress conditions [61]. In this study, two non-Saccharomyces strains, UMCC 5 (T. delbrueckii) and UMCC 15 (M. pulcherrima), alongside two S. cerevisiae strains, UMCC 3066 and UMCC 24, have been selected for the AFM investigation as they exhibited different fermentative capacities and membrane fluidity changes under ethanol stress. Figure 5 displays representative AFM error mode images of the surface of the selected yeasts. It should be noted that, in this study, both height and error mode signals have been acquired. However, to better visualize small topographical details, we report AFM images obtained by error mode signal.
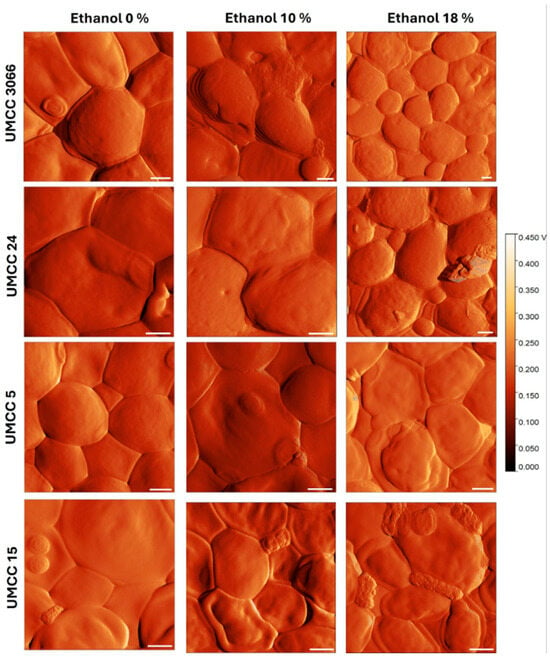
Figure 5.
AFM error mode images of UMCC 3066, UMCC 24, UMCC 5, and UMCC 15 after exposure to 0%, 10%, and 18% (v/v) of ethanol. Scale bars: 1 µm.
As can be observed, the degree of compactness of cell aggregates, as well as the morphology of the cell surface, were barely altered for UMCC 3066 and UMCC 24 upon exposure to ethanol, even at the highest tested concentration. In contrast, deformation of cell morphology and loss of smoothness were found for UMCC 5 and UMCC 15, especially when exposed to 18% (v/v) ethanol. These findings corroborate the RMS data. In fact, the two non-Saccharomyces strains exhibited larger RMS values when exposed to 18% (v/v) ethanol (Figure 6). Conversely, the lowest RMS values at the various ethanol concentrations were found for UMCC 24 and UMCC 3066, which are the most tolerant strains tested in this study (Figure 6). Notably, UMCC 3066 consistently had the lowest RMS values across ethanol concentrations. Consistent with the findings of Canetta et al. [36], the more ethanol-tolerant strains showed minimal morphological changes compared to less tolerant strains. The significant changes in cell surface morphology observed in the non-Saccharomyces strains suggest a greater loss of membrane integrity under ethanol-induced stress than that observed for S. cerevisiae strains. Similar responses have been observed under other stressors, such as osmotic and thermal stress [62]. In summary, results from AFM assays confirm that S. cerevisiae may possess greater resilience to ethanol toxicity. This pattern further indicates that non-Saccharomyces strains may experience increased variability or instability under high ethanol stress. In contrast, the superior stability and tolerance to ethanol of the S. cerevisiae strain UMCC 3066 were further confirmed by AFM analysis.
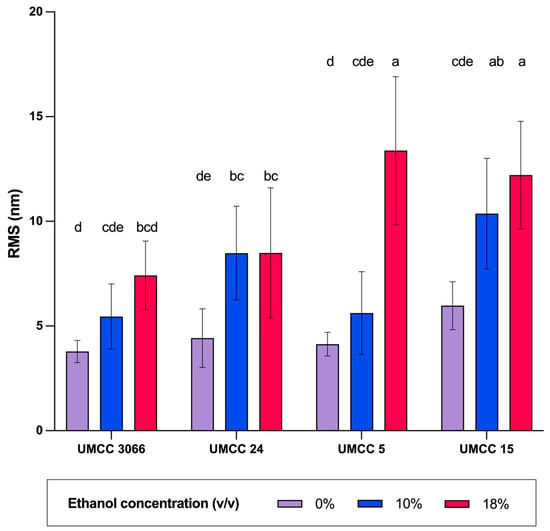
Figure 6.
RMS values of UMCC 3066, UMCC 24, UMCC 5, and UMCC 15 after exposure to 0%, 10%, and 18% (v/v) of ethanol for 24 h. Data are expressed as mean ± standard deviation, obtained from 10 images for each sample. Significant differences are indicated by different letters (p < 0.05).
4. Conclusions
In this study, Saccharomyces and non-Saccharomyces wine yeasts, each with unique fermentative characteristics and profiles in secondary compound production, displayed varied responses in growth, membrane fluidity, and cell morphology under ethanol stress. Notably, strains capable of tolerating ethanol concentrations up to 16% (v/v) and demonstrating superior fermentative performance exhibited increased membrane fluidity at the lowest ethanol concentration tested. In contrast, less tolerant strains with lower fermentative capacity but with significant contributions to final product complexity—such as T. delbrueckii UMCC 5 and M. pulcherrima UMCC 15—showed smaller increases in fluidity. At the highest ethanol concentration tested (18% v/v), these less tolerant strains displayed higher surface roughness, suggesting greater instability, whereas more tolerant strains exhibited lower RMS values, indicative of enhanced adaptability. These findings contribute to the development of multiple selected starters for oenological applications, optimizing S. cerevisiae strains for essential fermentative functions while harnessing the distinct attributes of non-Saccharomyces yeasts to enrich product complexity, thereby expanding the range of commercial options available. Based on the outcomes of this work, these strains can be further explored to understand their genetic background related to the pathway involved in preserving membrane fluidity, especially under ethanol-induced stress.
Author Contributions
Methodology, investigation, writing—original draft preparation, formal analysis, E.A.; writing—review and editing, data curation, M.P.A.; methodology, writing—review and editing, L.D.V.; writing—review and editing, C.M.; AFM methodology and experiment, writing—review and editing, M.B.; AFM methodology—AFM experiment setup, writing—review and editing, A.M.; resources, AFM experiments supervision, writing—review and editing, A.A.; resources, supervision, writing—review and editing, A.P.; conceptualization, resources, funding acquisition, supervision and editing, M.G. All authors have read and agreed to the published version of the manuscript.
Funding
Part of this work was granted by the European Commission—NextGenerationEU, Project “Strengthening the MIRRI Italian Research Infrastructure for Sustainable Bioscience and Bioeconomy”, code n. IR0000005 and by the European Union-NextGenerationEUGrant, CN_00000033, Project “National Biodiversity Future Center-NBFC”, CUP E93C22001090001. AEB S.p.a (Brescia) group supported part of the research; Project CUP: E83C24002050007.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to thank Maicol Ronchi for his contribution to conducting the microvinification experiments.
Conflicts of Interest
Author Carlo Montanini was employed by the company AEB S.p.a. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Dinh, T.N.; Nagahisa, K.; Hirasawa, T.; Furusawa, C.; Shimizu, H. Adaptation of Saccharomyces cerevisiae cells to high ethanol concentration and changes in fatty acid composition of membrane and cell size. PLoS ONE 2008, 3, e2623. [Google Scholar] [CrossRef] [PubMed]
- Learmonth, R.P.; Gratton, E. Assessment of membrane fluidity in individual yeast cells by Laurdan generalised polarisation and multi-photon scanning fluorescence microscopy. In Fluorescence Spectroscopy, Imaging and Probes: New Tools in Chemical, Physical and Life Sciences; Kraayenhof, R., Visser, A.J.W.G., Gerritsen, H.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 241–252. ISBN 978-3-642-56067-5. [Google Scholar]
- Learmonth, R.P. Membrane fluidity in yeast adaptation: Insights from fluorescence spectroscopy and microscopy. In Reviews in Fluorescence 2010; Geddes, C.D., Ed.; Springer: New York, NY, USA, 2012; pp. 67–93. ISBN 978-1-4419-9828-6. [Google Scholar]
- Sunyer-Figueres, M.; Mas, A.; Beltran, G.; Torija, M.-J. Protective effects of melatonin on Saccharomyces cerevisiae under ethanol stress. Antioxidants 2021, 10, 1735. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cen, N.; Liu, L.; Chen, Y.; Yang, X.; Yu, K.; Guo, J.; Liao, X.; Shi, B. Collagen peptide provides Saccharomyces cerevisiae with robust stress tolerance for enhanced bioethanol production. ACS Appl. Mater. Interfaces 2020, 12, 53879–53890. [Google Scholar] [CrossRef] [PubMed]
- Charoenbhakdi, S.; Dokpikul, T.; Burphan, T.; Techo, T.; Auesukaree, C. Vacuolar H+-ATPase protects Saccharomyces cerevisiae cells against ethanol-induced oxidative and cell wall stresses. Appl. Environ. Microbiol. 2016, 82, 3121–3130. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xia, Y.; Hu, W.; Tao, L.; Ni, L.; Yu, J.; Ai, L. Membrane fluidity of Saccharomyces cerevisiae from Huangjiu (Chinese rice wine) is variably regulated by OLE1 to offset the disruptive effect of ethanol. Appl. Environ. Microbiol. 2019, 85, e01620-19. [Google Scholar] [CrossRef]
- Alexandre, H.; Ansanay-Galeote, V.; Dequin, S.; Blondin, B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 2001, 498, 98–103. [Google Scholar] [CrossRef]
- Yang, K.-M.; Lee, N.-R.; Woo, J.-M.; Choi, W.; Zimmermann, M.; Blank, L.M.; Park, J.-B. Ethanol reduces mitochondrial membrane integrity and thereby impacts carbon metabolism of Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 675–684. [Google Scholar] [CrossRef]
- Bagnat, M.; Keränen, S.; Shevchenko, A.; Shevchenko, A.; Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 3254–3259. [Google Scholar] [CrossRef]
- Deguil, J.; Pineau, L.; Rowland Snyder, E.C.; Dupont, S.; Beney, L.; Gil, A.; Frapper, G.; Ferreira, T. Modulation of lipid-induced ER stress by fatty acid shape. Traffic 2011, 12, 349–362. [Google Scholar] [CrossRef]
- Surma, M.A.; Klose, C.; Peng, D.; Shales, M.; Mrejen, C.; Stefanko, A.; Braberg, H.; Gordon, D.E.; Vorkel, D.; Ejsing, C.S.; et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol. Cell 2013, 51, 519–530. [Google Scholar] [CrossRef]
- Hoppe, T.; Rape, M.; Jentsch, S. Membrane-bound transcription factors: Regulated release by RIP or RUP. Curr. Opin. Cell Biol. 2001, 13, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Burkett, T.J.; Yamashita, I.; Garfinkel, D.J. Genetic redundancy between SPT23 and MGA2: Regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 4718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Skalsky, Y.; Garfinkel, D.J. MGA2 or SPT23 is required for transcription of the Δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 1999, 151, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Ballweg, S.; Ernst, R. Control of membrane fluidity: The OLE pathway in focus. Biol. Chem. 2017, 398, 215–228. [Google Scholar] [CrossRef]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and future of non-Saccharomyces yeasts: From spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 2016, 7, 411. [Google Scholar] [CrossRef]
- Varela, C. The impact of non-Saccharomyces yeasts in the production of alcoholic beverages. Appl. Microbiol. Biotechnol. 2016, 100, 9861–9874. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Benito, S. The influence of non-Saccharomyces species on wine fermentation quality parameters. Fermentation 2019, 5, 54. [Google Scholar] [CrossRef]
- Pulvirenti, A.; De Vero, L.; Blaiotta, G.; Sidari, R.; Iosca, G.; Gullo, M.; Caridi, A. Selection of wine Saccharomyces cerevisiae strains and their screening for the adsorption activity of pigments, phenolics and ochratoxin A. Fermentation 2020, 6, 80. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef]
- Amerine, M.A. The Technology of Wine Making, 4th ed.; Avi Publishing Company: Westport, CT, USA, 1980; ISBN 978-0-87055-333-2. [Google Scholar]
- Rojas, V.; Gil, J.V.; Piñaga, F.; Manzanares, P. Acetate ester formation in wine by mixed cultures in laboratory fermentations. Int. J. Food Microbiol. 2003, 86, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Sadineni, V.; Kondapalli, N.; Obulam, V.S.R. Effect of co-fermentation with Saccharomyces cerevisiae and Torulaspora delbrueckii or Metschnikowia pulcherrima on the aroma and sensory properties of mango wine. Ann. Microbiol. 2012, 62, 1353–1360. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 2014, 80, 1670. [Google Scholar] [CrossRef] [PubMed]
- Giovani, G.; Rosi, I.; Bertuccioli, M. Quantification and characterization of cell wall polysaccharides released by non-Saccharomyces yeast strains during alcoholic fermentation. Int. J. Food Microbiol. 2012, 160, 113–118. [Google Scholar] [CrossRef]
- Azzolini, M.; Fedrizzi, B.; Tosi, E.; Finato, F.; Vagnoli, P.; Scrinzi, C.; Zapparoli, G. Effects of Torulaspora delbrueckii and Saccharomyces cerevisiae mixed cultures on fermentation and aroma of Amarone wine. Eur. Food Res. Technol. 2012, 235, 303–313. [Google Scholar] [CrossRef]
- Roca-Mesa, H.; Delgado-Yuste, E.; Mas, A.; Torija, M.-J.; Beltran, G. Importance of micronutrients and organic nitrogen in fermentations with Torulaspora delbrueckii and Saccharomyces cerevisiae. Int. J. Food Microbiol. 2022, 381, 109915. [Google Scholar] [CrossRef]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii—Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef]
- Rodríguez, M.E.; Lopes, C.A.; Barbagelata, R.J.; Barda, N.B.; Caballero, A.C. Influence of Candida pulcherrima Patagonian strain on alcoholic fermentation behaviour and wine aroma. Int. J. Food Microbiol. 2010, 138, 19–25. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Bonciani, T.; De Vero, L.; Mezzetti, F.; Fay, J.C.; Giudici, P. A multi-phase approach to select new wine yeast strains with enhanced fermentative fitness and glutathione production. Appl. Microbiol. Biotechnol. 2018, 102, 2269–2278. [Google Scholar] [CrossRef] [PubMed]
- Canetta, E.; Adya, A.K.; Walker, G.M. Atomic force microscopic study of the effects of ethanol on yeast cell surface morphology. FEMS Microbiol. Lett. 2006, 255, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Laguna, L.; Bartolomé, B.; Moreno-Arribas, M.V. Mouthfeel perception of wine: Oral physiology, components and instrumental characterization. Trends Food Sci. Technol. 2017, 59, 49–59. [Google Scholar] [CrossRef]
- Oura, E. Reaction products of yeast fermentations. Process Biochem. 1977, 12, 19–21. [Google Scholar]
- Bruinenberg, P.; Dijken, J.; Scheffers, W. A theoretical analysis of NADPH production and consumption in yeasts. J. Gen. Microbiol. 1983, 129, 953–964. [Google Scholar] [CrossRef][Green Version]
- Cortassa, S.; Aon, J.C.; Aon, M.A. Fluxes of carbon, phosphorylation, and redox intermediates during growth of Saccharomyces cerevisiae on different carbon sources. Biotechnol. Bioeng. 1995, 47, 193–208. [Google Scholar] [CrossRef]
- van Dijken, J.P.; Scheffers, W.A. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Rev. 1986, 1, 199–224. [Google Scholar] [CrossRef]
- Gonzalez, R.; Quirós, M.; Morales, P. Yeast respiration of sugars by non-Saccharomyces yeast species: A promising and barely explored approach to lowering alcohol content of wines. Trends Food Sci. Technol. 2013, 29, 55–61. [Google Scholar] [CrossRef]
- Pettersen, J.P.; Castillo, S.; Jouhten, P.; Almaas, E. Genome-scale metabolic models reveal determinants of phenotypic differences in non-Saccharomyces yeasts. BMC Bioinform. 2023, 24, 438. [Google Scholar] [CrossRef]
- Flores, C.L.; Rodríguez, C.; Petit, T.; Gancedo, C. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol. Rev. 2000, 24, 507–529. [Google Scholar] [CrossRef] [PubMed]
- Tyibilika, V.; Setati, M.E.; Bloem, A.; Divol, B.; Camarasa, C. Differences in the management of intracellular redox state between wine yeast species dictate their fermentation performances and metabolite production. Int. J. Food Microbiol. 2024, 411, 110537. [Google Scholar] [CrossRef] [PubMed]
- Nikolić, S.; Mojović, L.; Pejin, D.; Rakin, M.; Vukašinović, M. Production of bioethanol from corn meal hydrolyzates by free and immobilized cells of Saccharomyces cerevisiae var. Ellipsoideus. Biomass Bioenergy 2010, 34, 1449–1456. [Google Scholar] [CrossRef]
- Quevedo-Hidalgo, B.; Monsalve-Marín, F.; Narváez-Rincón, P.C.; Pedroza-Rodríguez, A.M.; Velásquez-Lozano, M.E. Ethanol production by Saccharomyces cerevisiae using lignocellulosic hydrolysate from Chrysanthemum waste degradation. World J. Microbiol. Biotechnol. 2013, 29, 459–466. [Google Scholar] [CrossRef]
- Behera, S.; Kar, S.; Mohanty, R.C.; Ray, R.C. Comparative study of bio-ethanol production from Mahula (Madhuca latifolia L.) flowers by Saccharomyces cerevisiae cells immobilized in agar agar and Ca-alginate matrices. Appl. Energy 2010, 87, 96–100. [Google Scholar] [CrossRef]
- Razmovski, R.; Vučurović, V. Ethanol production from sugar beet molasses by S. cerevisiae entrapped in an alginate-maize stem ground tissue matrix. Enzyme Microb. Technol. 2011, 48, 378–385. [Google Scholar] [CrossRef]
- Beaven, M.J.; Charpentier, C.; Rose, A.H. Production and tolerance of ethanol in relation to phospholipid fatty-acyl composition in Saccharomyces cerevisiae NCYC 431. Microbiology 1982, 128, 1447–1455. [Google Scholar] [CrossRef]
- Ghareib, M.; Youssef, K.A.; Khalil, A.A. Ethanol tolerance of Saccharomyces cerevisiae and its relationship to lipid content and composition. Folia Microbiol. 1988, 33, 447–452. [Google Scholar] [CrossRef]
- Mishra, P.; Prasad, R. Relationship between ethanol tolerance and fatty acyl composition of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1989, 30, 294–298. [Google Scholar] [CrossRef]
- Mishra, P.; Kaur, S. Lipids as modulators of ethanol tolerance in yeast. Appl. Microbiol. Biotechnol. 1991, 34, 697–702. [Google Scholar] [CrossRef]
- Šajbidor, J.; Grego, J. Fatty acid alterations in Saccharomyces cerevisiae exposed to ethanol stress. FEMS Microbiol. Lett. 1992, 93, 13–16. [Google Scholar] [CrossRef]
- Alexandre, H.; Rousseaux, I.; Charpentier, C. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol. Lett. 1994, 124, 17–22. [Google Scholar] [CrossRef] [PubMed]
- You, K.M.; Rosenfield, C.-L.; Knipple, D.C. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl. Environ. Microbiol. 2003, 69, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, F.; Peinado, R.A.; Millán, C.; Ortega, J.M.; Mauricio, J.C. Relationship between ethanol tolerance, H+-ATPase activity and the lipid composition of the plasma membrane in different wine yeast strains. Int. J. Food Microbiol. 2006, 110, 34–42. [Google Scholar] [CrossRef]
- Ishmayana, S.; Kennedy, U.J.; Learmonth, R.P. Further investigation of relationships between membrane fluidity and ethanol tolerance in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 218. [Google Scholar] [CrossRef]
- Lairón-Peris, M.; Routledge, S.J.; Linney, J.A.; Alonso-del-Real, J.; Spickett, C.M.; Pitt, A.R.; Guillamón, J.M.; Barrio, E.; Goddard, A.D.; Querol, A. Lipid composition analysis reveals mechanisms of ethanol tolerance in the model yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2021, 87, e00440-21. [Google Scholar] [CrossRef]
- Jones, R.P.; Greenfield, P.F. Ethanol and the fluidity of the yeast plasma membrane. Yeast 1987, 3, 223–232. [Google Scholar] [CrossRef]
- Francois, J.M.; Formosa, C.; Schiavone, M.; Pillet, F.; Martin-Yken, H.; Dague, E. Use of Atomic Force Microscopy (AFM) to explore cell wall properties and response to stress in the yeast Saccharomyces cerevisiae. Curr. Genet. 2013, 59, 187–196. [Google Scholar] [CrossRef]
- Adya, A.K.; Canetta, E.; Walker, G.M. Atomic force microscopic study of the influence of physical stresses on Saccharomyces cerevisiae and Schizosaccharomyces pombe. FEMS Yeast Res. 2006, 6, 120–128. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).