Using Omics Techniques to Analyze the Effects of Gene Mutations and Culture Conditions on the Synthesis of β-Carotene in Pantoea dispersa
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Measurement of β-Carotene
2.3. UV Mutagenesis
2.4. RT-qPCR
2.5. Whole-Genome Resequencing
2.6. Transcriptomics Analysis
2.7. Structure and Activity Analysis of Phosphogluconate Dehydrogenase
2.8. Overexpression Strains
2.9. Statistical Analysis
3. Results
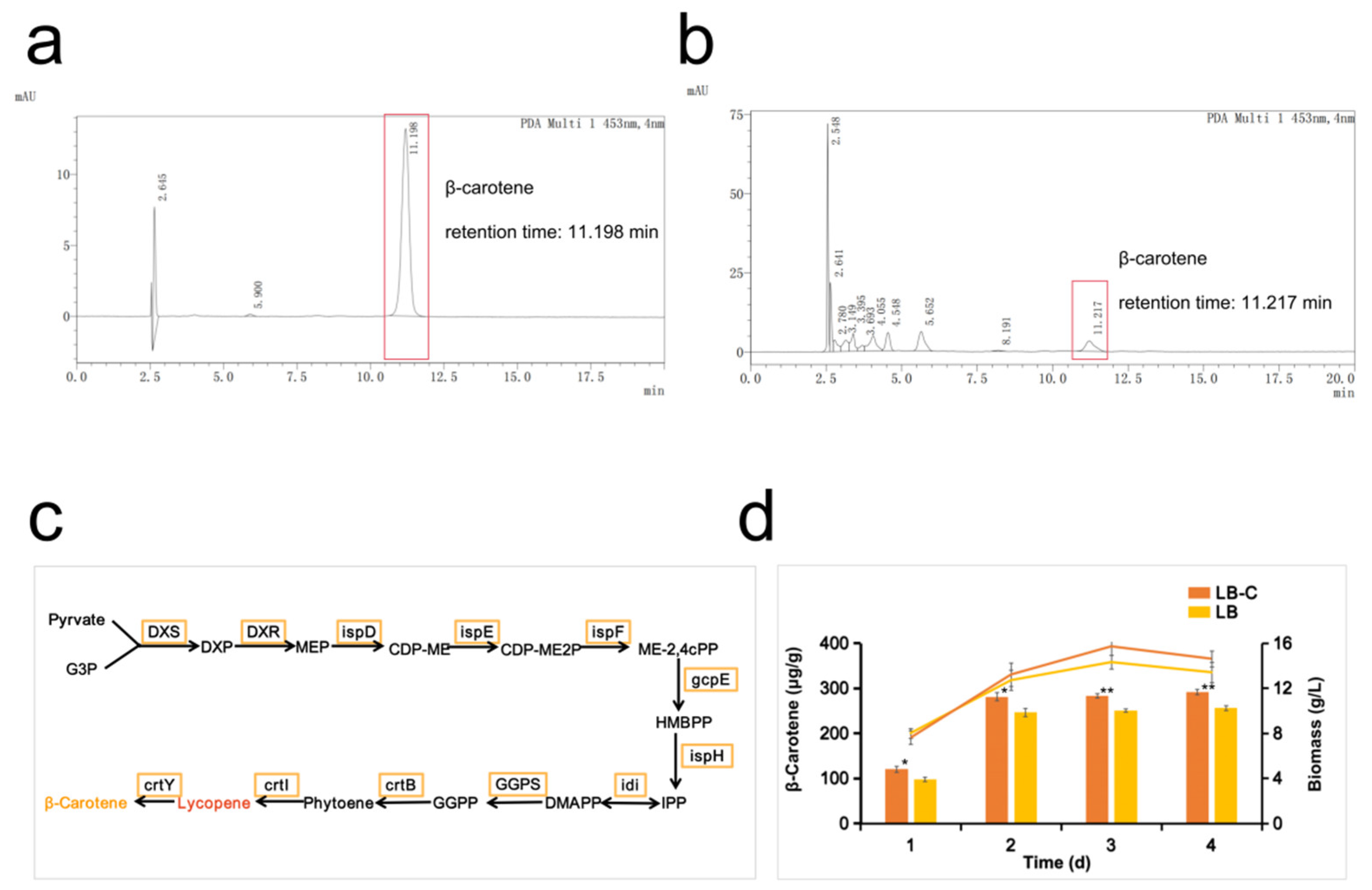
3.1. Analysis of the Pathway and Ability of Strain MSC14 to Produce β-Carotene and the Positive Effect of Corn Steep Liquor Powder on β-Carotene Synthesis
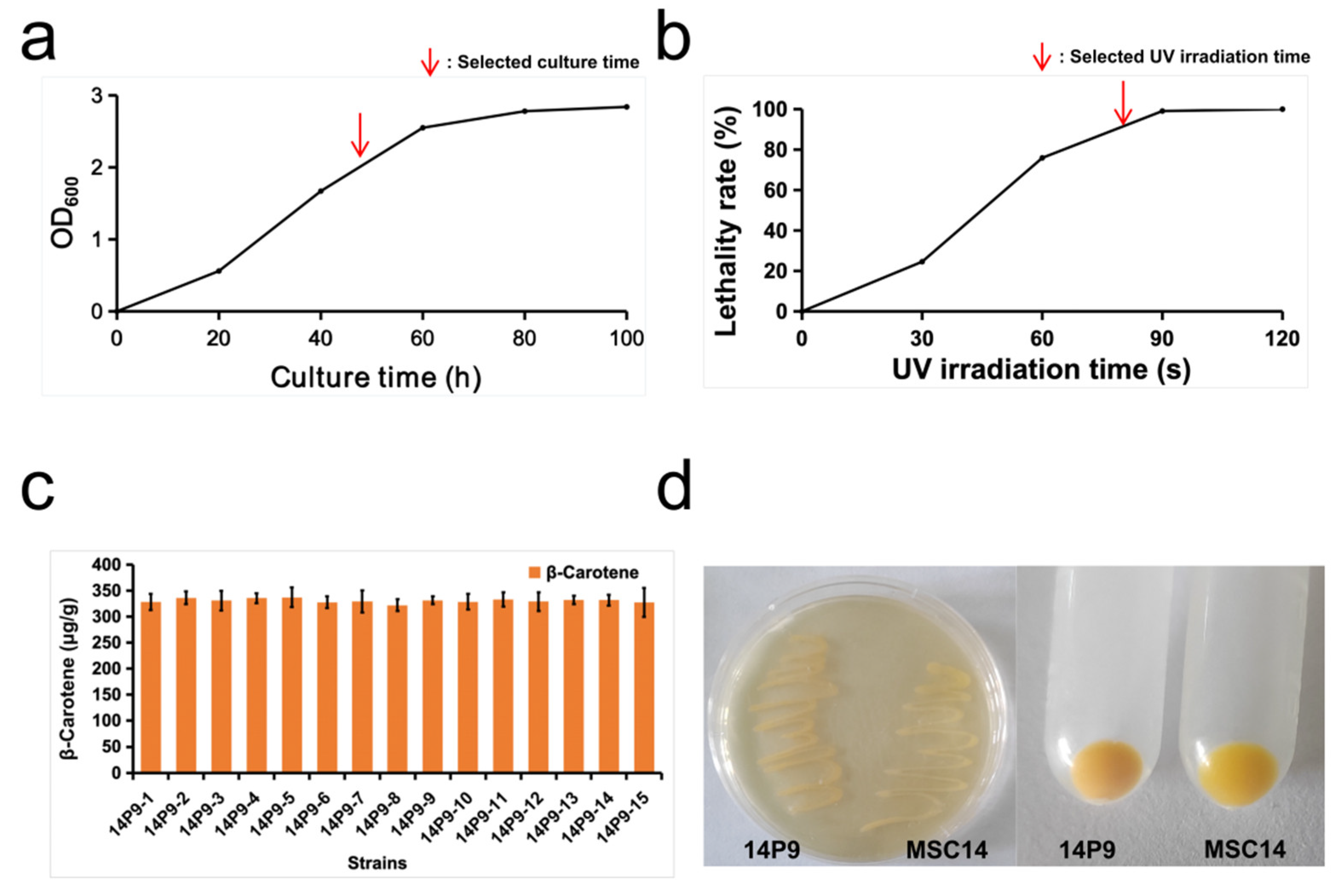
3.2. The Ability of the Mutagenized Strain 14P9 to Produce β-Carotene Was Significantly Increased
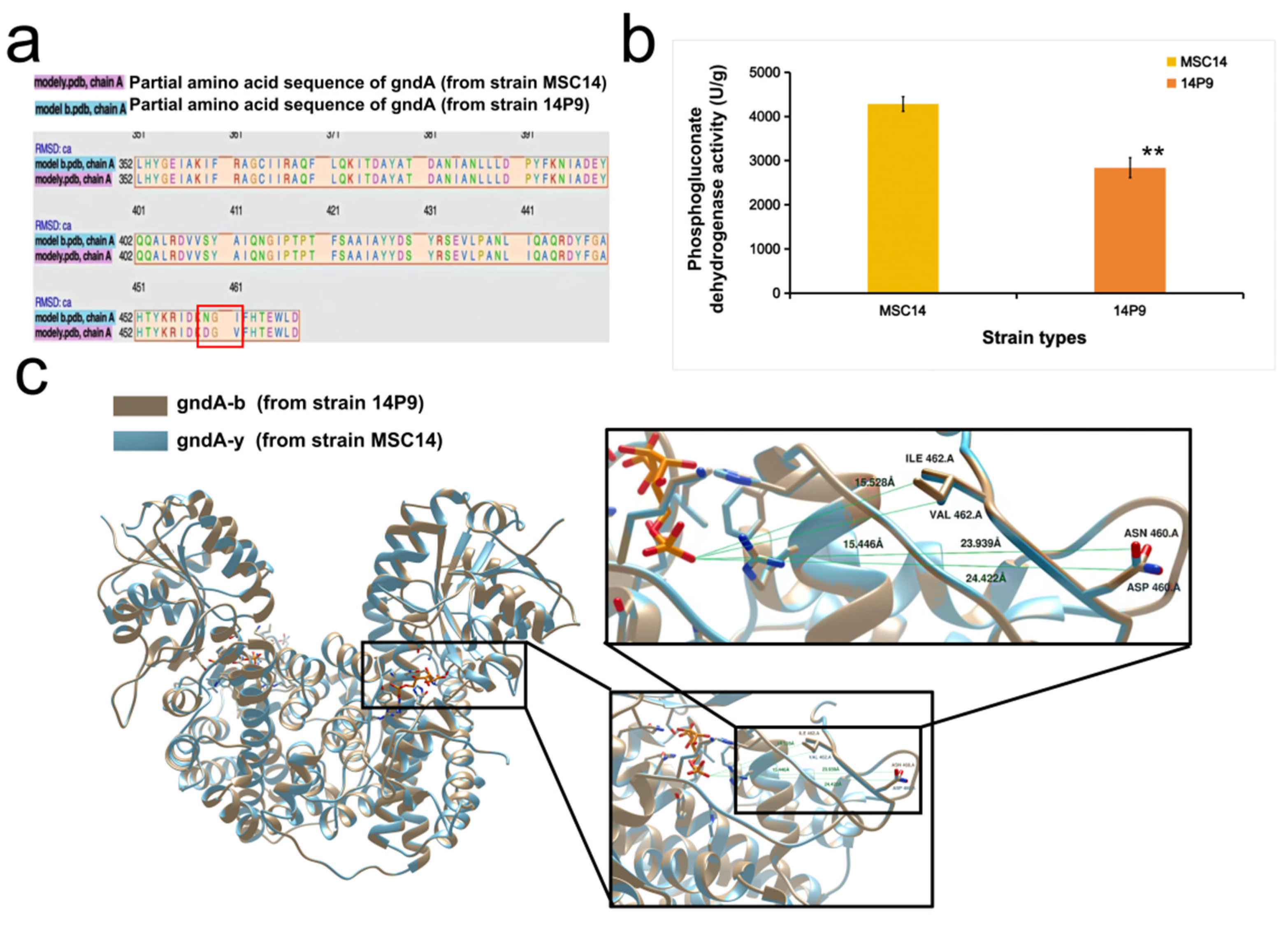
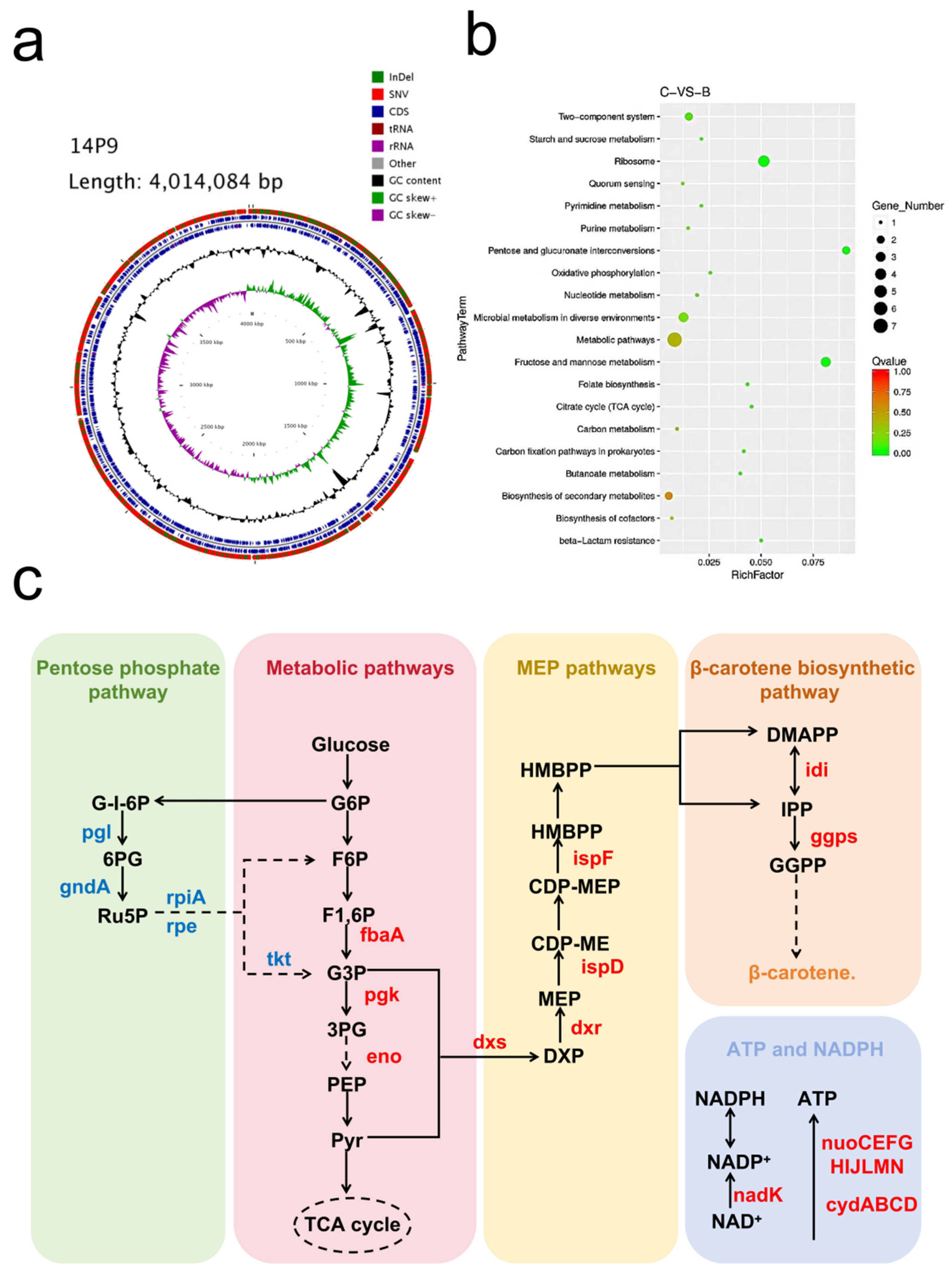
3.3. Genomic Analysis and Transcriptomics Analysis between Strain MSC14 and Strain 14P9
3.4. Transcriptomic Analysis of Strain MSC14 in Different Media
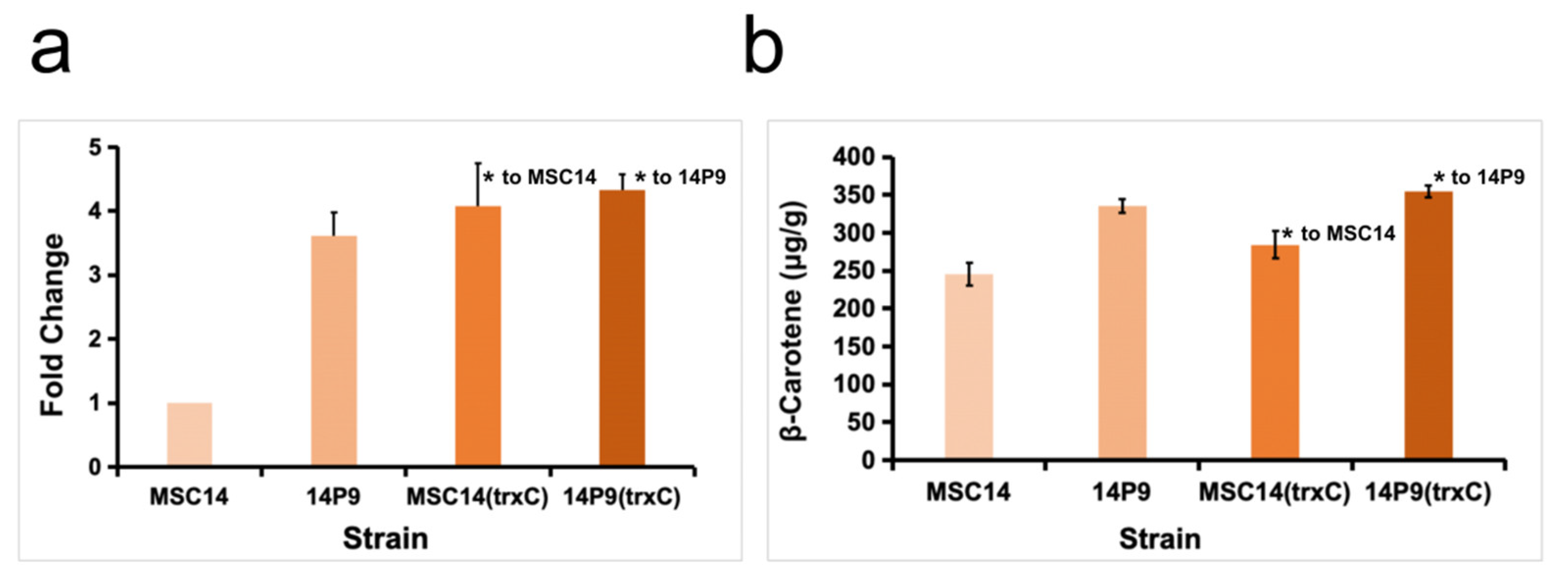
3.5. Effect of trxC Gene on Growth Performance and Ability to Synthesize β-Carotene in P. dispersa
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albermann, C. High versus low level expression of the lycopene biosynthesis genes from Pantoea ananatis in Escherichia coli. Biotechnol. Lett. 2011, 33, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ananda, N.; Vadlani, P.V. Production and optimization of carotenoid-enriched dried distiller’s grains with solubles by Phaffia rhodozyma and Sporobolomyces roseus fermentation of whole stillage. J. Ind. Microbiol. Biotechnol. 2010, 37, 1183–1192. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Schürmann, P.; Jacquot, J.-P. Thioredoxin and metabolic regulation. Semin. Cell Biol. 1994, 5, 285–293. [Google Scholar] [CrossRef]
- Burton, G.W.; Mogg, T.J.; Riley, W.W.; Nickerson, J.G. β-Carotene oxidation products—Function and safety. Food Chem. Toxicol. 2021, 152, 112207. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Kang, B.; Lee, Y.; Lee, Y.; Kim, J. Pantoea ananatis carotenoid production confers toxoflavin tolerance and is regulated by Hfq-controlled quorum sensing. Microbiologyopen 2021, 10, e1143. [Google Scholar] [CrossRef] [PubMed]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Fiedor, J.; Burda, K. Potential Role of Carotenoids as Antioxidants in Human Health and Disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef]
- Fulco, A.J. Fatty acid metabolism in bacteria. Prog. Lipid Res. 1983, 22, 133–160. [Google Scholar] [CrossRef] [PubMed]
- Gassel, S.; Breitenbach, J.; Sandmann, G. Genetic engineering of the complete carotenoid pathway towards enhanced astaxanthin formation in Xanthophyllomyces dendrorhous starting from a high-yield mutant. Appl. Microbiol. Biotechnol. 2014, 98, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Gómez, P.I.; Inostroza, I.; Pizarro, M.; Pérez, J. From genetic improvement to commercial-scale mass culture of a Chilean strain of the green microalga Haematococcus pluvialis with enhanced productivity of the red ketocarotenoid astaxanthin. AoB Plants 2013, 5, plt026. [Google Scholar] [CrossRef] [PubMed]
- Gromer, S.; Urig, S.; Becker, K. The thioredoxin system—From science to clinic. Med. Res. Rev. 2004, 24, 40–89. [Google Scholar] [CrossRef] [PubMed]
- Harker, M.; Bramley, P. Expression of prokaryotic 1-deoxy-d-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett. 1999, 448, 115–119. [Google Scholar] [CrossRef]
- Kar, A.; Paramasivam, B.; Jayakumar, D.; Swaroop, A.K.; Jubie, S. Thioredoxin Interacting Protein Inhibitors in Diabetes Mellitus: A Critical Review. Curr. Drug Res. Rev. 2023, 15, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-W.; Keasling, J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001, 72, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Momen, A.Z.R.; Mijts, B.N.; Schmidt-Dannert, C. Biosynthesis of Structurally Novel Carotenoids in Escherichia coli. Chem. Biol. 2003, 10, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, Y.; Li, L.; Ma, Y.; Huang, J.; Ye, J. Engineering Sphingobium sp. to Accumulate Various Carotenoids Using Agro-Industrial Byproducts. Front. Bioeng. Biotechnol. 2021, 9, 784559. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yi, H.; Zhan, H.; Wang, L.; Wang, J.; Li, Y.; Liu, B. Gibberellic acid-induced fatty acid metabolism and ABC transporters promote astaxanthin production in Phaffia rhodozyma. J. Appl. Microbiol. 2022, 132, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Mantzouridou, F.; Roukasa, T.; Kotzekidoua, P.; Liakopoulou, M. Optimization of β-Carotene Production from Synthetic Medium by Blakeslea trispora: A Mathematical Modeling. Appl. Biochem. Biotechnol. 2002, 101, 153–175. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2019, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, A. Thioredoxin and redox signaling: Roles of the thioredoxin system in control of cell fate. Arch. Biochem. Biophys. 2017, 617, 101–105. [Google Scholar] [CrossRef]
- Mehta, B.J.; Obraztsova, I.N.; Cerdá-Olmedo, E. Mutants and Intersexual Heterokaryons of Blakeslea trispora for Production of β-Carotene and Lycopene. Appl. Environ. Microbiol. 2003, 69, 4043–4048. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, E.H.; Liakopoulou-Kyriakides, M. Agro-food wastes utilization by Blakeslea trispora for carotenoids production. Acta Biochim. Pol. 2012, 59, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Nanou, K.; Roukas, T.; Papadakis, E.; Kotzekidou, P. Carotene production from waste cooking oil by Blakeslea trispora in a bubble column reactor: The role of oxidative stress. Eng. Life Sci. 2017, 17, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; He, X.; Zhang, H.; Guo, X.; Cheng, Y.; Liu, X.; Wang, Z.; He, X. Whole Genome Sequencing and RNA-seq-Driven Discovery of New Targets That Affect Carotenoid Synthesis in Phaffia rhodozyma. Front. Microbiol. 2022, 13, 837894. [Google Scholar] [CrossRef]
- Oka, S.-I.; Titus, A.S.; Zablocki, D.; Sadoshima, J. Molecular properties and regulation of NAD+ kinase (NADK). Redox Biol. 2023, 59, 102561. [Google Scholar] [CrossRef]
- Park, S.Y.; Binkley, R.M.; Kim, W.J.; Lee, M.H.; Lee, S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab. Eng. 2018, 49, 105–115. [Google Scholar] [CrossRef]
- Rohdich, F.; Hecht, S.; Gärtner, K.; Adam, P.; Krieger, C.; Amslinger, S.; Arigoni, D.; Bacher, A.; Eisenreich, W. Studies on the nonmevalonate terpene biosynthetic pathway: Metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. USA 2002, 99, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Rohdich, F.; Zepeck, F.; Adam, P.; Hecht, S.; Kaiser, J.; Laupitz, R.; Gräwert, T.; Amslinger, S.; Eisenreich, W.; Bacher, A.; et al. The deoxyxylulose phosphate pathway of isoprenoid biosynthesis: Studies on the mechanisms of the reactions catalyzed by IspG and IspH protein. Proc. Natl. Acad. Sci. USA 2003, 100, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Carotenoids of biotechnological importance. In Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 148, pp. 449–467. [Google Scholar]
- Wu, Y.; Yan, P.; Li, Y.; Liu, X.; Wang, Z.; Chen, T.; Zhao, X. Enhancing β-Carotene Production in Escherichia coli by Perturbing Central Carbon Metabolism and Improving the NADPH Supply. Front. Bioeng. Biotechnol. 2020, 8, 585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xing, J.; Chen, H. Progress in metabolic engineering of β-carotene synthesis. Sheng Wu Gong Cheng Xue Bao 2017, 33, 578–590. [Google Scholar] [CrossRef] [PubMed]
- Song, G.H.; Kim, S.H.; Choi, B.H.; Han, S.J.; Lee, P.C. Heterologous Carotenoid-Biosynthetic Enzymes: Functional Complementation and Effects on Carotenoid Profiles in Escherichia coli. Appl. Environ. Microbiol. 2013, 79, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.-L.; Chen, K.-Q.; Dai, W.-Y.; Ma, J.-F.; Zhang, M.; Jiang, M.; Wei, P.; Ouyang, P.-K. Succinic acid production by Actinobacillus succinogenes NJ113 using corn steep liquor powder as nitrogen source. Bioresour. Technol. 2013, 136, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Lv, X.; Yu, H. Engineering microbes for isoprene production. Metab. Eng. 2016, 38, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Kim, J.-E.; Lee, S.-H.; Park, H.-M.; Choi, M.-S.; Kim, J.-Y.; Lee, S.-H.; Shin, Y.-C.; Keasling, J.D.; Kim, S.-W. Engineering the lycopene synthetic pathway in E. coli by comparison of the carotenoid genes of Pantoea agglomerans and Pantoea ananatis. Appl. Microbiol. Biotechnol. 2007, 74, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Zeller, T.; Li, K.; Klug, G. Expression of the trxC Gene of Rhodobacter capsulatus: Response to Cellular Redox Status Is Mediated by the Transcriptional Regulator OxyR. J. Bacteriol. 2006, 188, 7689–7695. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.-G.; Han, M.; Zhang, W.-G.; Qian, H. Carotene production from agro-industrial wastes by Arthrobacter globiformis in shake-flask culture. Prep. Biochem. Biotechnol. 2014, 44, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Nambou, K.; Wei, L.; Cao, J.; Imanaka, T.; Hua, Q. Lycopene production in recombinant strains of Escherichia coli is improved by knockout of the central carbon metabolism gene coding for glucose-6-phosphate dehydrogenase. Biotechnol. Lett. 2013, 35, 2137–2145. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Cui, T. Using Omics Techniques to Analyze the Effects of Gene Mutations and Culture Conditions on the Synthesis of β-Carotene in Pantoea dispersa. Fermentation 2024, 10, 83. https://doi.org/10.3390/fermentation10020083
Liu N, Cui T. Using Omics Techniques to Analyze the Effects of Gene Mutations and Culture Conditions on the Synthesis of β-Carotene in Pantoea dispersa. Fermentation. 2024; 10(2):83. https://doi.org/10.3390/fermentation10020083
Chicago/Turabian StyleLiu, Na, and Tangbing Cui. 2024. "Using Omics Techniques to Analyze the Effects of Gene Mutations and Culture Conditions on the Synthesis of β-Carotene in Pantoea dispersa" Fermentation 10, no. 2: 83. https://doi.org/10.3390/fermentation10020083
APA StyleLiu, N., & Cui, T. (2024). Using Omics Techniques to Analyze the Effects of Gene Mutations and Culture Conditions on the Synthesis of β-Carotene in Pantoea dispersa. Fermentation, 10(2), 83. https://doi.org/10.3390/fermentation10020083






