Penicillium janthinellum: A Potential Producer of Natural Products
Abstract
:1. Introduction
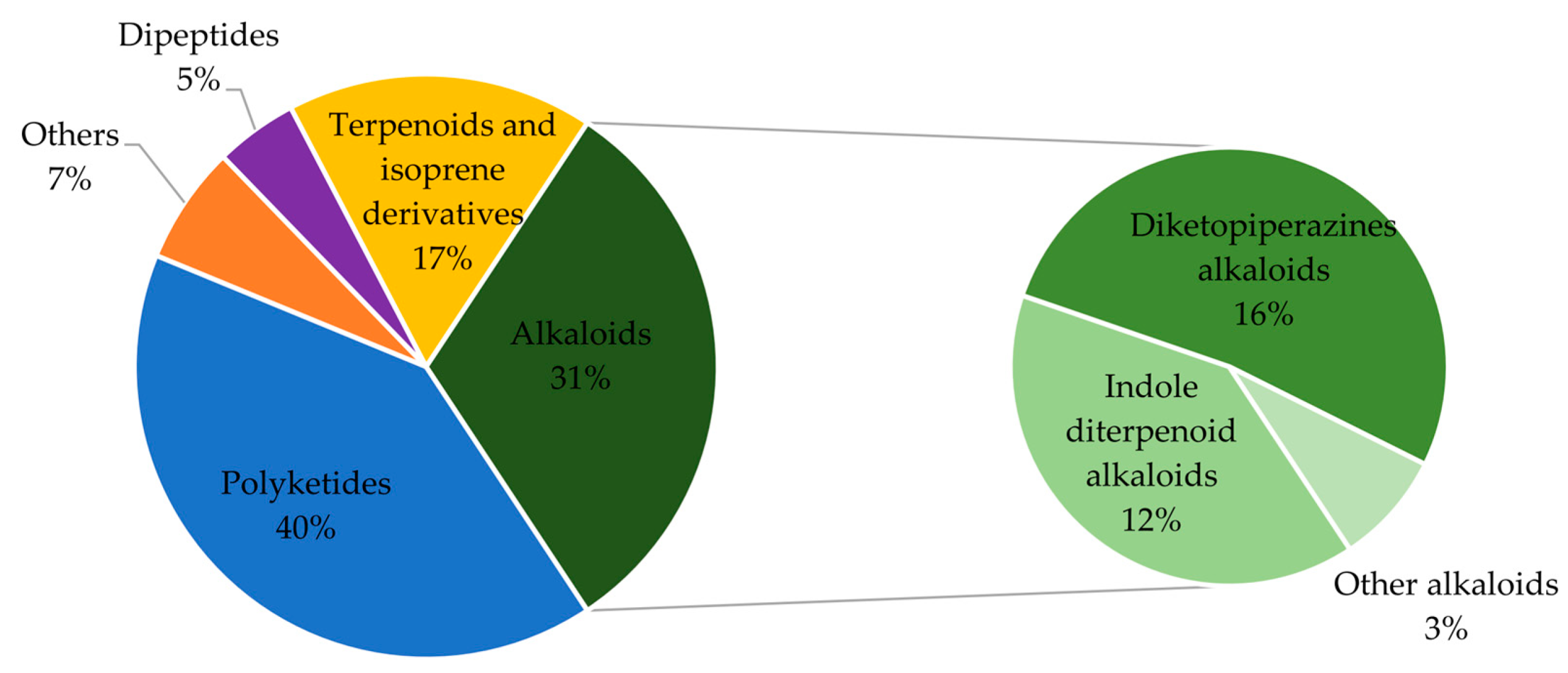
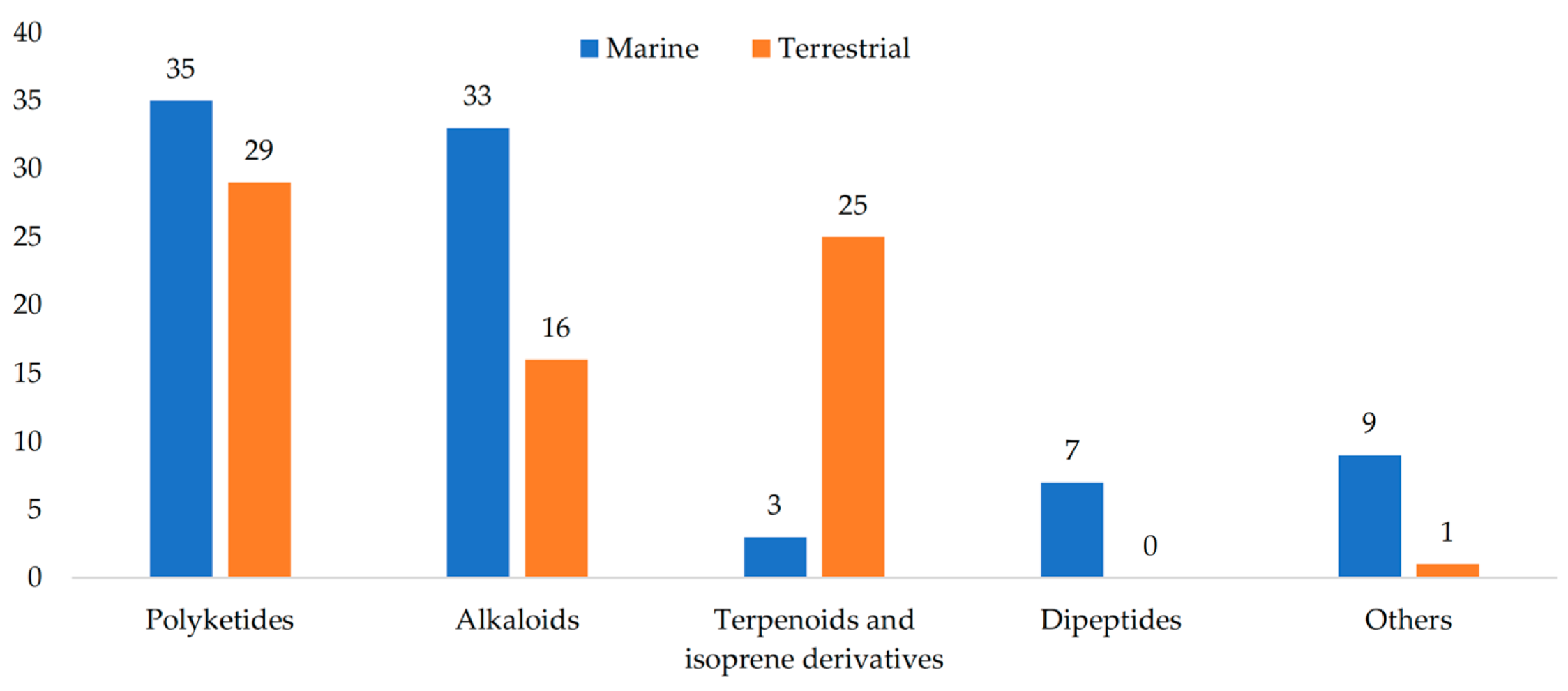
2. Secondary Metabolites of Penicillium janthinellum
2.1. Polyketides
2.2. Alkaloids
2.3. Terpenoids and Isoprene Derivatives
2.4. Dipeptides
2.5. Others
3. Biological Activities
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Koul, M.; Singh, S. Penicillium spp.: Prolific producer for harnessing cytotoxic secondary metabolites. Anticancer Drugs 2017, 28, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fu, Y.; Song, F.; Xu, X. Recent updates on the antimicrobial compounds from marine-derived Penicillium fungi. Chem. Biodivers. 2023, 20, e202301278. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, A.C.; Ogawa, C.Y.; Rodrigues, L.D.; de Medeiros, L.S.; Veiga, T.A.M. Penicillium genus as a source for anti-leukemia compounds: An overview from 1984 to 2020. Leuk. Lymphoma 2021, 62, 2079–2093. [Google Scholar] [CrossRef] [PubMed]
- Matsukuma, S.; Nishizuka, Y.; Watanabe, J.; Okuda, T. Identification of Penicillium sp. NR 6564 and taxonomic notes on P. janthinellum. Mycoscience 1994, 35, 183–186. [Google Scholar] [CrossRef]
- Hu, X.-M.; Dong, S.-J.; Han, H.-Y.; Gao, Y.-F.; Zhang, B.-X. Screening of highly efficient fungi for the degradation of lignocelluloses by ionic liquids-assisted cellulase. BioResources 2021, 17, 355–368. [Google Scholar] [CrossRef]
- Zheng, C.J.; Xu, L.L.; Li, Y.Y.; Han, T.; Zhang, Q.Y.; Ming, Q.L.; Rahman, K.; Qin, L.P. Cytotoxic metabolites from the cultures of endophytic fungi from Panax ginseng. Appl. Microbiol. Biotechnol. 2013, 97, 7617–7625. [Google Scholar] [CrossRef]
- Marinho, A.M.D.; Rodrigues, E.; Moitinho, M.D.R.; Santos, L.S. Biologically active polyketides produced by Penicillium janthinellum isolated as an endophytic fungus from fruits of Melia azedarach. J. Braz. Chem. Soc. 2005, 16, 280–283. [Google Scholar] [CrossRef]
- Tapfuma, K.I.; Sebola, T.E.; Uche-Okereafor, N.; Koopman, J.; Hussan, R.; Makatini, M.M.; Mekuto, L.; Mavumengwana, V. Anticancer activity and metabolite profiling data of Penicillium janthinellum KTMT5. Data Brief 2020, 28, 104959. [Google Scholar] [CrossRef]
- Widden, P.; Parkinson, A.D. The effects of temperature on growth of four high Arctic soil fungi in a three-phase system. Can. J. Microbiol. 1978, 24, 415–421. [Google Scholar] [CrossRef]
- Sun, T.-T.; Yang, J.-K.; Zhu, H.-J.; Pan, L.; Cao, F. Antibacterial secondary metabolites from the marine-derived fungus Penicillium janthinellum. Chem. Nat. Compd. 2020, 56, 968–970. [Google Scholar] [CrossRef]
- Cui, H.; Li, X.D.; Li, M.Q.; Lu, F.M.; Wang, Y.H.; Liu, D.; Kang, J.G. Study on anti-HBV secondary metabolites from sponge-associated fungus Penicillium janthinellum LZDX-32-1. Chin. J. Mar. Drugs 2017, 36, 41–46. [Google Scholar] [CrossRef]
- Smetanina, O.F.; Kalinovsky, A.I.; Khudyakova, Y.V.; Pivkin, M.V.; Dmitrenok, P.S.; Fedorov, S.N.; Ji, H.; Kwak, J.Y.; Kuznetsova, T.A. Indole alkaloids produced by a marine fungus isolate of Penicillium janthinellum Biourge. J. Nat. Prod. 2007, 70, 906–909. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zong, L.; Zhu, Y.; Li, Y.; Zhou, Y.; Zhou, H. Penicillium janthinellum pneumonia in an SLE patient: A case study. Infect. Drug Resist. 2020, 13, 2745–2749. [Google Scholar] [CrossRef] [PubMed]
- Mernitz, G.; Koch, A.; Henrissat, B.; Schulz, G. Endoglucanase II (EGII) of Penicillium janthinellum: cDNA sequence, heterologous expression and promotor analysis. Curr. Genet. 1996, 29, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, Y.; Long, X.; Meng, X.; Liu, Z. Cloning of exoinulinase gene from Penicillium janthinellum strain B01 and its high-level expression in Pichia pastoris. J. Appl. Microbiol. 2011, 111, 1371–1380. [Google Scholar] [CrossRef]
- Chi, B.B.; Lu, Y.N.; Yin, P.C.; Liu, H.Y.; Chen, H.Y.; Shan, Y. Sequencing and comparative genomic analysis of a highly metal-tolerant Penicillium janthinellum P1 provide insights into its metal tolerance. Front. Microbiol. 2021, 12, 663217. [Google Scholar] [CrossRef]
- Christopher, M.; Sreeja-Raju, A.; Kooloth-Valappil, P.; Gokhale, D.V.; Sukumaran, R.K. Correction to: Cellulase hyper-producing fungus Penicillium janthinellum NCIM 1366 elaborates a wider array of proteins involved in transport and secretion, potentially enabling a diverse substrate range. BioEnergy Res. 2022, 16, 74. [Google Scholar] [CrossRef]
- Keskar, S.S. Cellulase production by Penicillium janthinellum. World J. Microbiol. Biotechnol. 1992, 8, 534–535. [Google Scholar] [CrossRef]
- Rao, M.; Varma, A.J.; Deshmukh, S.S. Production of single cell protein, essential amino acids, and xylanase by Penicillium janthinellum. Bioresources 2010, 5, 2470–2477. [Google Scholar] [CrossRef]
- Bao, S.; Mu, J.; Yin, P.; Chen, H.; Zhou, S. Exploration of anti-chromium mechanism of marine Penicillium janthinellum P1 through combinatorial transcriptomic analysis and WGCNA. Ecotoxicol. Environ. Saf. 2022, 233, 113326. [Google Scholar] [CrossRef] [PubMed]
- Seydametova, E. Novel pravastatin-producing Penicillium janthinellum strain isolated from soil. Int. J. Biosci. Biochem. Bioinf. 2015, 5, 80–90. [Google Scholar] [CrossRef]
- Mai, Z.-P.; Xin, X.-L.; Sun, Z.; Zhang, N.; Huang, S.-S.; Wang, C.; Chen, L.; Li, Y.; Huo, X.-K.; Fan, G.-J. Biotransformation of alisol G by Penicillium janthinellum and the hCE2 inhibitory effects of its metabolites. Phytochem. Lett. 2015, 13, 228–233. [Google Scholar] [CrossRef]
- Lv, X.; Liu, D.; Hou, J.; Dong, P.; Zhan, L.; Wang, L.; Deng, S.; Wang, C.; Yao, J.; Shu, X.; et al. Biotransformation of imperatorin by Penicillium janthinellum. Anti-osteoporosis activities of its metabolites. Food Chem. 2013, 138, 2260–2266. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Khan, A.R.; Hussain, J.; Kang, S.M.; Gilani, S.A.; Hamayun, M.; Shin, J.H.; Kamran, M.; Al-Harrasi, A.; et al. Fungal endophyte Penicillium janthinellum LK5 improves growth of ABA-deficient tomato under salinity. World J. Microbiol. Biotechnol. 2013, 29, 2133–2144. [Google Scholar] [CrossRef]
- Itabashi, T.; Hosoe, T.; Wakana, D.; Fukushima, K.; Takizawa, K.; Yaguchi, T.; Okada, K.; Takaki, G.M.; Kawai, K. A new indoloditerpene derivative, penijanthine A, isolated from Penicillium janthinellum. J. Nat. Med. 2009, 63, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Musgrave, O.C. 828. Curvularin. Part I. Isolation and partial characterisation of a metabolic product from a new species of Curvularia. J. Chem. Soc. 1956, 1956, 4301–4305. [Google Scholar] [CrossRef]
- Birch, A.J.; Musgrave, O.C.; Rickards, R.W.; Smith, H. 638. Studies in relation to biosynthesis. Part XX. The structure and biosynthesis of curvularin. J. Chem. Soc. 1959, 1959, 3146–3152. [Google Scholar] [CrossRef]
- Mohapatra, D.; Rahaman, H.; Pal, R.; Gurjar, M. Total synthesis of (S)-(-)-curvularin: A ring-closing-metathesis-based construction of the macrocyclic framework. Synlett 2008, 2008, 1801–1804. [Google Scholar] [CrossRef]
- Zhan, J.X.; Gunatilaka, A.A.L. Microbial transformation of curvularin. J. Nat. Prod. 2005, 68, 1271–1273. [Google Scholar] [CrossRef]
- Raistrick, H.; Rice, F.H. 2,3-Dihydro-3,6-dihydroxy-2-methyl-4-pyrone and curvularin from Penicillium gilmanii. J. Chem. Soc. Perkin Trans. 1 1971, 18, 3069–3070. [Google Scholar] [CrossRef]
- Ghisalberti, E.L.; Rowland, C.Y. 6-chlorodehydrocurvularin, a new metabolite from Cochliobolus spicifer. J. Chem. Soc. 1993, 56, 2175–2177. [Google Scholar] [CrossRef]
- Ye, X.; Anjum, K.; Song, T.; Wang, W.; Yu, S.; Huang, H.; Lian, X.Y.; Zhang, Z. A new curvularin glycoside and its cytotoxic and antibacterial analogues from marine actinomycete Pseudonocardia sp. HS7. Nat. Prod. Res. 2015, 30, 1156–1161. [Google Scholar] [CrossRef]
- Paek, S.M. Recent synthesis and discovery of Brefeldin A analogs. Mar. Drugs 2018, 16, 133. [Google Scholar] [CrossRef]
- Hu, Z.F.; Qin, L.L.; Ding, W.J.; Liu, Y.; Ma, Z.J. New analogues of brefeldin A from sediment-derived fungus Penicillium sp. DT-F29. Nat. Prod. Res. 2016, 30, 2311–2315. [Google Scholar] [CrossRef]
- Cheng, X.; Yu, L.; Wang, Q.; Ding, W.; Chen, Z.; Ma, Z. New brefeldins and penialidins from marine fungus Penicillium janthinellum DT-F29. Nat. Prod. Res. 2018, 32, 282–286. [Google Scholar] [CrossRef]
- Wang, W.J.; Liao, L.X.; Huang, Z.D.; Wei, F.T.; Yang, X.L. Thiazolo[5,4-b]pyridine alkaloid and seven ar-bisabol sesquiterpenes produced by the endophytic fungus Penicillium janthinellum. ACS Omega 2022, 7, 35280–35287. [Google Scholar] [CrossRef]
- Zeng, F.; Chen, C.; Al Chnani, A.A.; Zhou, Q.; Tong, Q.; Wang, W.; Zang, Y.; Gong, J.; Wu, Z.; Liu, J.; et al. Dibrefeldins A and B, a pair of epimers representing the first brefeldin A dimers with cytotoxic activities from Penicillium janthinellum. Bioorg. Chem. 2019, 86, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Härri, E.; Loeffler, W.; Sigg, H.P.; Stähelin, H.; Tamm, C. Über die Isolierung neuer Stoffwechselprodukte aus Penicillium brefeldianum DODGE. Helv. Chim. Acta 1963, 64, 1235–1243. [Google Scholar] [CrossRef]
- Wang, C.F.; Ma, J.; Jing, Q.Q.; Cao, X.Z.; Chen, L.; Chao, R.; Zheng, J.Y.; Shao, C.L.; He, X.X.; Wei, M.Y. Integrating activity-guided strategy and fingerprint analysis to target potent cytotoxic brefeldin A from a fungal library of the medicinal mangrove Acanthus ilicifolius. Mar. Drugs 2022, 20, 432. [Google Scholar] [CrossRef] [PubMed]
- Gais, H.-J.; Lied, T. Asymmetric total synthesis of macrolides brefeldin A and 7-epi-brefeldin A. Angew. Chem. 1984, 96, 143–145. [Google Scholar] [CrossRef]
- Hazalin, N.A.; Lim, S.M.; Cole, A.L.; Majeed, A.B.; Ramasamy, K. Apoptosis induced by desmethyl-lasiodiplodin is associated with upregulation of apoptotic genes and downregulation of monocyte chemotactic protein-3. Anticancer Drugs 2013, 24, 852–861. [Google Scholar] [CrossRef] [PubMed]
- Driouich, A.; Jauneau, A.; Staehelin, L.A. 7-Dehydrobrefeldin A, a naturally occurring brefeldin A derivative, inhibits secretion and causes a cis-to-trans breakdown of Golgi stacks in plant cells. Plant Physiol. 1997, 113, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.W.; Nagasawa, H.; Nagura, F.; Mohamad, S.B.; Uto, Y.; Ohkura, K.; Hori, H. Elucidation of strict structural requirements of brefeldin A as an inducer of differentiation and apoptosis. Bioorg. Med. Chem. 2000, 8, 455–463. [Google Scholar] [CrossRef]
- Aoki, Y.; Yamazaki, T.; Kondoh, M.; Sudoh, Y.; Nakayama, N.; Sekine, Y.; Shimada, H.; Arisawa, M. A new series of natural antifungals that inhibit P450 lanosterol C-14 demethylase. II. Mode of action. J. Antibiot. 1992, 45, 160–170. [Google Scholar] [CrossRef]
- Schwartz, R.E.; Dufresne, C.; Flor, J.E.; Kempf, A.J.; Wilson, K.E.; Lam, T.; Onishi, J.; Milligan, J.; Fromtling, R.A.; Abruzzo, G.K. Restricticin, a novel glycine-containing antifungal agent. J. Antibiot. 1991, 44, 463–471. [Google Scholar] [CrossRef]
- Marinho, A.M.D.; Rodrigues-Fo, E. Dicitrinol, a citrinin dimer, produced by Penicillium janthinellum. Helv. Chim. Acta 2011, 94, 835–841. [Google Scholar] [CrossRef]
- Molee, W.; Phanumartwiwath, A.; Kesornpun, C.; Sureram, S.; Ngamrojanavanich, N.; Ingkaninan, K.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Naphthalene derivatives and quinones from Ventilago denticulata and their nitric oxide radical scavenging, antioxidant, cytotoxic, antibacterial, and phosphodiesterase inhibitory activities. Chem. Biodivers. 2018, 15, e1700537. [Google Scholar] [CrossRef]
- Amin, I.M.; Kamaludin, R.; Yeap, S.K.; Isa, M.R.; Rosdy, N.; Siran, R.; Kadir, S.; Hasani, N.A.H. Aloe emodin induces apoptosis in ER+-breast cancer cells; MCF-7 through IGF-1R signalling pathway. Sains Malays. 2015, 44, 1137–1143. [Google Scholar] [CrossRef]
- Choi, S.G.; Kim, J.; Sung, N.D.; Son, K.H.; Cheon, H.G.; Kim, K.R.; Kwon, B.M. Anthraquinones, Cdc25B phosphatase inhibitors, isolated from the roots of Polygonum multiflorum Thunb. Nat. Prod. Res. 2007, 21, 487–493. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Sun, C.M.; Ou, J.C.; Tsai, W.J. A tumor cell growth inhibitor from Polygonum hypoleucum Ohwi. Life Sci. 1997, 61, 2335–2344. [Google Scholar] [CrossRef]
- Abdulkadir Emam, J.; Ele Yaya, E.; Iqbal Choudhary, M.; Yousuf, S.; Mehari Gebremedihn, T. In vitro antifungal, anti-inflammatory and cytotoxic activities of Rumex abyssinicus rhizome extract and bioassay-guided isolation of cytotoxic compounds from Rumex abyssinicus. Bull. Chem. Soc. Ethiop. 2022, 36, 879–892. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Tsai, W.J.; Meng, H.C.; Chen, W.P.; Yang, L.Y.; Lin, C.Y. Immune reponses in human mesangial cells regulated by emodin from Polygonum hypoleucum Ohwi. Life Sci. 2001, 68, 1271–1286. [Google Scholar] [CrossRef]
- Guetchueng, S.T.; Djouonzo, P.T.; Lame, Y.; Kopa Kowa, T.; Dotse, E.; Tchokouaha, L.R.Y.; Kamdem Wabo, H.; Appiah-Opong, R.; Agbor, G.A. Antileishmanial anthraquinones from the rhyzomes of Rumex abyssinicus Jacq (Polygonaceae). Nat. Prod. Res. 2023, 37, 2935–2939. [Google Scholar] [CrossRef]
- Huang, P.H.; Huang, C.Y.; Chen, M.C.; Lee, Y.T.; Yue, C.H.; Wang, H.Y.; Lin, H. Emodin and aloe-emodin suppress breast cancer cell proliferation through ER alpha inhibition. J. Evid.-Based Complement. Altern. Med. 2013, 2013, 376123. [Google Scholar] [CrossRef]
- Li, W.Y.; Ng, Y.F.; Zhang, H.; Guo, Z.D.; Guo, D.J.; Kwan, Y.W.; Leung, G.P.; Lee, S.M.; Yu, P.H.; Chan, S.W. Emodin elicits cytotoxicity in human lung adenocarcinoma A549 cells through inducing apoptosis. Inflammopharmacology 2014, 22, 127–134. [Google Scholar] [CrossRef]
- Li, W.Y.; Chan, R.Y.; Yu, P.H.; Chan, S.W. Emodin induces cytotoxic effect in human breast carcinoma MCF-7 cell through modulating the expression of apoptosis-related genes. Pharm. Biol. 2013, 51, 1175–1181. [Google Scholar] [CrossRef]
- Liu, L.; Zou, J.; Liu, X.; Jiang, L.H.; Li, J. Inhibition of ATP-induced macrophage death by emodin via antagonizing P2X7 receptor. Eur. J. Pharmacol. 2010, 640, 15–19. [Google Scholar] [CrossRef]
- Hwangbo, K.; Zheng, M.S.; Kim, Y.J.; Im, J.Y.; Lee, C.S.; Woo, M.H.; Jahng, Y.; Chang, H.W.; Son, J.K. Inhibition of DNA topoisomerases I and II of compounds from Reynoutria japonica. Arch. Pharmacal Res. 2012, 35, 1583–1589. [Google Scholar] [CrossRef]
- Khalil, A.A.K.; Park, W.S.; Kim, H.J.; Akter, K.M.; Ahn, M.-J. Anti-Helicobacter pylori compounds from Polygonum cuspidatum. Nat. Prod. Sci. 2016, 22, 220–224. [Google Scholar] [CrossRef]
- Marinho, A.M.R.; Marinho, P.S.B.; Santos, L.S.; Rodrigues, E.; Ferreira, I.C.P. Active polyketides isolated from Penicillium herquei. An. Acad. Bras. Cienc. 2013, 85, 909–912. [Google Scholar] [CrossRef]
- Ióca, L.P.; Romminger, S.; Santos, M.F.C.; Bandeira, K.F.; Rodrigues, F.T.; Kossuga, M.H.; Nicacio, K.J.; Ferreira, E.L.F.; Morais-Urano, R.P.; Passos, M.S.; et al. A strategy for the rapid identification of fungal metabolites and the discovery of the antiviral activity of pyrenocine a and harzianopyridone. Quim. Nova 2016, 39, 720–731. [Google Scholar] [CrossRef]
- Ooi, S.C.; Ho, C.C.; Seow, H.F. Isolation of a potential anticancer agent with protein phosphatase inhibitory activity from soil-derived Penicillium sp strain H9318. Trop. J. Pharm. Res. 2016, 15, 1423–1429. [Google Scholar] [CrossRef]
- Chai, Y.J.; Cui, C.B.; Li, C.W.; Wu, C.J.; Tian, C.K.; Hua, W. Activation of the dormant secondary metabolite production by introducing gentamicin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2012, 10, 559–582. [Google Scholar] [CrossRef]
- Xue, C.M.; Li, T.; Deng, Z.W.; Fu, H.Z.; Lin, W.H. Janthinolide A-B, two new 2,5-piperazinedione derivatives from the endophytic Penicillium janthinellum isolated from the soft coral Dendronephthya sp. Pharmazie 2006, 61, 1041–1044. [Google Scholar] [CrossRef]
- Oxford, A.E.; Raistrick, H.; Simonart, P. Studies in the biochemistry of micro-organisms: Griseofulvin, C17H17O6Cl, a metabolic product of Penicillium griseo-fulvum Dierckx. Biochem. J. 1939, 33, 240–248. [Google Scholar] [CrossRef]
- De-sheng, L.; Yu-ling, H.; Li-ying, M.; Chang-jun, L.; Wei-zhong, L. Chemical constituents and their cytotoxic activities of the secondary metabolites of Penicillium janthinellum. Chin. Tradit. Pat. Med. 2016, 38, 830–834. [Google Scholar]
- Guo, J.; Ran, H.; Zeng, J.; Liu, D.; Xin, Z. Tafuketide, a phylogeny-guided discovery of a new polyketide from Talaromyces funiculosus Salicorn 58. Appl. Microbiol. Biotechnol. 2016, 100, 5323–5338. [Google Scholar] [CrossRef]
- Sheng, J.W.; Liu, D.M.; Jing, L.; Xia, G.X.; Zhang, W.F.; Jiang, J.R.; Tang, J.B. Striatisporolide A, a butenolide metabolite from Athyrium multidentatum (Doll.) Ching, as a potential antibacterial agent. Mol. Med. Rep. 2019, 20, 198–204. [Google Scholar] [CrossRef]
- Park, B.K.; Nakagawa, M.; Hirota, A.; Nakayama, M. Methylenolactocin, a novel antitumor antibiotic from Penicillium sp. J. Antibiot. 1988, 41, 751–758. [Google Scholar] [CrossRef]
- Yang, Z.; Bao, L.; Yin, Y.; Ding, G.; Ge, M.; Chen, D.; Qian, X. Pyrenocines N-O: Two novel pyrones from Colletotrichum sp. HCCB03289. J. Antibiot. 2014, 67, 791–793. [Google Scholar] [CrossRef]
- Nilanonta, C.; Isaka, M.; Kittakoop, P.; Saenboonrueng, J.; Rukachaisirikul, V.; Kongsaeree, P.; Thebtaranonth, Y. New diketopiperazines from the entomopathogenic fungus Verticillium hemipterigenum BCC 1449. J. Antibiot. 2003, 56, 647–651. [Google Scholar] [CrossRef]
- Sparace, S.A.; Reeleder, R.D.; Khanizadeh, S. Antibiotic activity of the pyrenocines. Can. J. Microbiol. 1987, 33, 327–330. [Google Scholar] [CrossRef]
- Rukachaisirikul, V.; Kaeobamrung, J.; Panwiriyarat, W.; Saitai, P.; Sukpondma, Y.; Phongpaichit, S.; Sakayaroj, J. A new pyrone derivative from the endophytic fungus Penicillium paxilli PSU-A71. Chem. Pharm. Bull. 2007, 55, 1383–1384. [Google Scholar] [CrossRef] [PubMed]
- Jouda, J.B.; Tamokou, J.D.; Mbazoa, C.D.; Sarkar, P.; Bag, P.K.; Wandji, J. Anticancer and antibacterial secondary metabolites from the endophytic fungus Penicillium sp. CAM64 against multi-drug resistant Gram-negative bacteria. Afr. Health Sci. 2016, 16, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Jouda, J.B.; Kusari, S.; Lamshoft, M.; Mouafo Talontsi, F.; Douala Meli, C.; Wandji, J.; Spiteller, M. Penialidins A-C with strong antibacterial activities from Penicillium sp., an endophytic fungus harboring leaves of Garcinia nobilis. Fitoterapia 2014, 98, 209–214. [Google Scholar] [CrossRef]
- Jouda, J.B.; Mawabo, I.K.; Notedji, A.; Mbazoa, C.D.; Nkenfou, J.; Wandji, J.; Nkenfou, C.N. Anti-mycobacterial activity of polyketides from Penicillium sp. endophyte isolated from Garcinia nobilis against Mycobacterium smegmatis. Int. J. Mycobact. 2016, 5, 192–196. [Google Scholar] [CrossRef]
- Fu, J.; Hu, L.; Shi, Z.; Sun, W.; Yue, D.; Wang, Y.; Ma, X.; Ren, Z.; Zuo, Z.; Peng, G.; et al. Two metabolites isolated from endophytic fungus Coniochaeta sp. F-8 in Ageratina adenophora exhibit antioxidative activity and cytotoxicity. Nat. Prod. Res. 2021, 35, 2840–2848. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Intereya, K.; Kocharin, K. New diphenyl ethers from the insect pathogenic fungus Cordyceps sp. BCC 1861. Chem. Pharm. Bull. 2007, 55, 304–307. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, W.; Ge, H.; Zhang, Z.; Wu, B. A new antimicrobial indoloditerpene from a marine-sourced fungus Aspergillus versicolor ZZ761. Nat. Prod. Res. 2021, 35, 3114–3119. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1106–1107, 71–83. [Google Scholar] [CrossRef]
- Deng, L.; Zhong, M.; Li, Y.; Hu, G.; Zhang, C.; Peng, Q.; Zhang, Z.; Fang, J.; Yu, X. High hydrostatic pressure harnesses the biosynthesis of secondary metabolites via the regulation of polyketide synthesis genes of hadal sediment-derived fungi. Front. Microbiol. 2023, 14, 1207252. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Smetanina, O.F.; Kalinovsky, A.I.; Pivkin, M.V.; Dmitrenok, P.S.; Kuznetsova, T.A. A new meroterpenoid from the marine fungus Aspergillus versicolor (Vuill.) Tirab. Russ. Chem. Bull. 2010, 59, 852–856. [Google Scholar] [CrossRef]
- Cimmino, A.; Maddau, L.; Masi, M.; Evidente, M.; Linaldeddu, B.T.; Evidente, A. Further secondary metabolites produced by Diplodia corticola, a fungal pathogen involved in cork oak decline. Tetrahedron 2016, 72, 6788–6793. [Google Scholar] [CrossRef]
- Takahashi, K.; Sakai, K.; Fukasawa, W.; Nagano, Y.; Sakaguchi, S.O.; Lima, A.O.; Pellizari, V.H.; Iwatsuki, M.; Takishita, K.; Yoshida, T.; et al. Quellenin, a new anti-Saprolegnia compound isolated from the deep-sea fungus, Aspergillus sp. YK-76. J. Antibiot. 2018, 71, 741–744. [Google Scholar] [CrossRef]
- Nguyen, M.V.; Han, J.W.; Kim, H.; Choi, G.J. Phenyl ethers from the marine-derived fungus Aspergillus tabacinus and their antimicrobial activity against plant pathogenic fungi and bacteria. ACS Omega 2022, 7, 33273–33279. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Yan, Q.X.; Zhang, Y.; Wu, T.Z.; Zou, Z.B.; Liu, Q.M.; Jiang, J.Y.; Xie, M.M.; Xu, L.; Hao, Y.J.; et al. Citriquinolinones A and B: Rare isoquinolinone-embedded citrinin analogues and related metabolites from the deep-sea-derived Aspergillus versicolor 170217. Mar. Drugs 2023, 21, 504. [Google Scholar] [CrossRef]
- Chen, M.; Shen, N.X.; Chen, Z.Q.; Zhang, F.M.; Chen, Y. Penicilones A-D, anti-MRSA azaphilones from the marine-derived fungus Penicillium janthinellum HK1-6. J. Nat. Prod. 2017, 80, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zheng, Y.Y.; Chen, Z.Q.; Shen, N.X.; Shen, L.; Zhang, F.M.; Zhou, X.J.; Wang, C.Y. NaBr-induced production of brominated azaphilones and related tricyclic polyketides by the marine-derived fungus Penicillium janthinellum HK1-6. J. Nat. Prod. 2019, 82, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, X.; Chen, L.; Shen, L.; Fu, X.; Chen, Q.; Chen, M.; Wang, C. Isolation and neuroprotective activity of phenolic derivatives from the marine-derived fungus Penicillium janthinellum. J. Ocean Univ. China 2020, 19, 700–706. [Google Scholar] [CrossRef]
- Arai, N.; Shiomi, K.; Tomoda, H.; Tabata, N.; Da Jun, Y.; Masuma, R.; Kawakubo, T.; Omura, S. Isochromophilones III-VI, inhibitors of acyl-CoA:cholesterol acyltransferase produced by Penicillium multicolor FO-3216. J. Antibiot. 1995, 48, 696–702. [Google Scholar] [CrossRef]
- Niu, G.; Kalani, K.; Wang, X.; Li, J. Sterigmatocystin limits Plasmodium falciparum proliferation and transmission. Pharmaceuticals 2021, 14, 1238. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-D.; Su, J.-C.; Jiang, B.-Z.; Li, Y.-L.; Guo, Y.-Q.; Zhang, P. Janthinoid A, an unprecedented tri-nor-meroterpenoid with highly modified bridged 4a,1-(epoxymethano)phenanthrene scaffold, produced by the endophyte of Penicillium janthinellum TE-43. Org. Chem. Front. 2021, 8, 6196–6202. [Google Scholar] [CrossRef]
- Mouafo Talontsi, F.; Kongue Tatong, M.D.; Dittrich, B.; Douanla-Meli, C.; Laatsch, H. Structures and absolute configuration of three α-pyrones from an endophytic fungus Aspergillus niger. Tetrahedron 2013, 69, 7147–7151. [Google Scholar] [CrossRef]
- Zhu, M.; Zhang, X.; Feng, H.; Dai, J.; Li, J.; Che, Q.; Gu, Q.; Zhu, T.; Li, D. Penicisulfuranols A-F, alkaloids from the mangrove endophytic fungus Penicillium janthinellum HDN13-309. J. Nat. Prod. 2017, 80, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.C.; Zhang, C.Z.; Liu, D. Study on secondary metabolites of sponge-associated fungus Penicillium janthinellum. Chin. J. Mar. Drugs 2019, 38, 21–26. [Google Scholar] [CrossRef]
- Cui, C.B.; Kakeya, H.; Okada, G.; Onose, R.; Osada, H. Novel mammalian cell cycle inhibitors, tryprostatins A, B and other diketopiperazines produced by Aspergillus fumigatus. I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. 1996, 49, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-C.; Ma, Y.-M.; Liu, R.; Zhou, F. Endophytic fungus Aspergillus tamarii from Ficus carica L., a new source of indolyl diketopiperazines. Biochem. Syst. Ecol. 2012, 45, 31–33. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.-M.; Meng, L.-H.; Wang, B.-G. N-Formyllapatin A, a new N-formylspiroquinazoline derivative from the marine-derived fungus Penicillium adametzioides AS-53. Phytochem. Lett. 2014, 10, 145–148. [Google Scholar] [CrossRef]
- Malekinejad, H.; Fani, F.; Shafie-Irannejad, V.; Fink-Gremmel, F. Aspergillus fumigatus toxins cause cytotoxic and apoptotic effects on human T lymphocytes (Jurkat cells). World Mycotoxin J. 2013, 6, 65–71. [Google Scholar] [CrossRef]
- Dai, J.M.; Mi, Q.L.; Li, X.M.; Gang, D.; Yang, G.Y.; Zhang, J.D.; Wang, J.; Li, Y.K.; Yang, H.Y.; Miao, D.; et al. The anti-TMV potency of the tobacco-derived fungus Aspergillus versicolor and its active alkaloids, as anti-TMV activity inhibitors. Phytochemistry 2023, 205, 113485. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Shen, N.X.; Liang, Z.Y.; Shen, L.; Chen, M.; Wang, C.Y. Paraherquamide J, a new prenylated indole alkaloid from the marine-derived fungus Penicillium janthinellum HK1-6. Nat. Prod. Res. 2020, 34, 378–384. [Google Scholar] [CrossRef] [PubMed]
- López-Gresa, M.P.; González, M.C.; Ciavatta, L.; Ayala, I.; Moya, P.; Primo, J. Insecticidal activity of paraherquamides, including paraherquamide H and paraherquamide I, two new alkaloids isolated from Penicillium cluniae. J. Agric. Food Chem. 2006, 54, 2921–2925. [Google Scholar] [CrossRef] [PubMed]
- Banks, R.M.; Blanchflower, S.E.; Everett, J.R.; Manger, B.R.; Reading, C. Novel anthelmintic metabolites from an Aspergillus species; the aspergillimides. J. Antibiot. 1997, 50, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.T.; Latch, G.C.; Keogh, R.G. The janthitrems: Fluorescent tremorgenic toxins produced by Penicillium janthinellum isolates from ryegrass pastures. Appl. Environ. Microbiol. 1980, 39, 272–273. [Google Scholar] [CrossRef] [PubMed]
- Penn, J.; Swift, R.; Wigley, L.J.; Mantle, P.G.; Sheppard, R.N. Janthitrems B and C, two principal indole-diterpenoids produced by Penicillium janthinellum. Phytochemistry 1993, 32, 1431–1434. [Google Scholar] [CrossRef]
- Babu, J.V.; Popay, A.J.; Miles, C.O.; Wilkins, A.L.; di Menna, M.E.; Finch, S.C. Identification and structure elucidation of janthitrems A and D from Penicillium janthinellum and determination of the tremorgenic and anti-insect activity of janthitrems A and B. J. Agric. Food Chem. 2018, 66, 13116–13125. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, A.E.; Steyn, P.S.; Van Heerden, F.R.; Vleggaar, R. Structure elucidation of the janthitrems, novel tremorgenic mycotoxins from Penicillium janthinellum. J. Chem. Soc. Perkin Trans. 1 1984, 697–701. [Google Scholar] [CrossRef]
- Heine, D.; Holmes, N.A.; Worsley, S.F.; Santos, A.C.A.; Innocent, T.M.; Scherlach, K.; Patrick, E.H.; Yu, D.W.; Murrell, J.C.; Vieria, P.C.; et al. Chemical warfare between leafcutter ant symbionts and a co-evolved pathogen. Nat. Commun. 2018, 9, 2208. [Google Scholar] [CrossRef]
- Liu, S.; Fan, W.; Ren, J.; Wang, W.; Liu, X.; Liang, Y.; Wei, T.; Li, E. Peniterpenoids A-C, new sesquiterpenoid metabolites from a wheat cyst nematode Penicillium janthinellum. Fitoterapia 2021, 148, 104801. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, Y.; Liu, P.; Fu, P.; Zhu, T.; Wang, W.; Zhu, W. Indole-diterpenoids with anti-H1N1 activity from the aciduric fungus Penicillium camemberti OUCMDZ-1492. J. Nat. Prod. 2013, 76, 1328–1336. [Google Scholar] [CrossRef]
- Chaiyosang, B.; Kanokmedhakul, K.; Yodsing, N.; Boonlue, S.; Yang, J.X.; Wang, Y.A.; Andersen, R.J.; Yahuafai, J.; Kanokmedhakul, S. Three new indole diterpenoids from Aspergillus aculeatus KKU-CT2. Nat. Prod. Res. 2022, 36, 4973–4981. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cheung, D.W. Effects of paxilline on K+ channels in rat mesenteric arterial cells. Eur. J. Pharmacol. 1999, 372, 103–107. [Google Scholar] [CrossRef]
- Bilmen, J.G.; Wootton, L.L.; Michelangeli, F. The mechanism of inhibition of the sarco/endoplasmic reticulum Ca2+ ATPase by paxilline. Arch. Biochem. Biophys. 2002, 406, 55–64. [Google Scholar] [CrossRef]
- Kulawiak, B.; Szewczyk, A. Glutamate-induced cell death in HT22 mouse hippocampal cells is attenuated by paxilline, a BK channel inhibitor. Mitochondrion 2012, 12, 169–172. [Google Scholar] [CrossRef]
- Lin, W.; Li, H.; Wu, Z.; Su, J.; Zhang, Z.; Yang, L.; Deng, X.; Xu, Q. Paspalines C-D and paxillines B-D: New indole diterpenoids from Penicillium brefeldianum WZW-F-69. Mar. Drugs 2022, 20, 684. [Google Scholar] [CrossRef] [PubMed]
- Nagumo, Y.; Motoyama, T.; Hayashi, T.; Hirota, H.; Aono, H.; Kawatani, M.; Osada, H.; Usui, T. Structure-activity relationships of terpendole E and its natural derivatives. ChemistrySelect 2017, 2, 1533–1536. [Google Scholar] [CrossRef]
- Sallam, A.A.; Houssen, W.E.; Gissendanner, C.R.; Orabi, K.Y.; Foudah, A.I.; El Sayed, K.A. Bioguided discovery and pharmacophore modeling of the mycotoxic indole diterpene alkaloids penitrems as breast cancer proliferation, migration, and invasion inhibitors. Medchemcomm 2013, 4, 1360. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wu, Y.Y.; Zhang, T.Y.; Zhang, M.Y.; Peng, F.; Lin, B.; Zhang, Y.X. New antimicrobial compounds produced by endophytic Penicillium janthinellum isolated from Panax notoginseng as potential inhibitors of FtsZ. Fitoterapia 2018, 131, 35–43. [Google Scholar] [CrossRef]
- McTavish, D.; Sorkin, E.M. Pravastatin: A review of its pharmacological properties and therapeutic potential in hypercholesterolaemia. Drugs 1991, 42, 65–89. [Google Scholar] [CrossRef]
- Tong, S.; Kaitu’u-Lino, T.J.; Hastie, R.; Brownfoot, F.; Cluver, C.; Hannan, N. Pravastatin, proton-pump inhibitors, metformin, micronutrients, and biologics: New horizons for the prevention or treatment of preeclampsia. Am. J. Obstet. Gynecol. 2022, 226, S1157–S1170. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Yuan, B.; Liu, D.; Zhu, K.; Huang, J.; Proksch, P.; Lin, W. DMOA-based meroterpenoids with diverse scaffolds from the sponge-associated fungus Penicillium brasilianum. Tetrahedron 2019, 75, 2193–2205. [Google Scholar] [CrossRef]
- Mo, S.; Yin, J.; Ye, Z.; Li, F.; Lin, S.; Zhang, S.; Yang, B.; Yao, J.; Wang, J.; Hu, Z.; et al. Asperanstinoids A-E: Undescribed 3,5-dimethylorsellinic acid-based meroterpenoids from Aspergillus calidoustus. Phytochemistry 2021, 190, 112892. [Google Scholar] [CrossRef]
- Orfali, R.; Perveen, S. New bioactive metabolites from the thermophilic fungus Penicillium sp. isolated from Ghamiqa hot spring in Saudi Arabia. J. Chem. 2019, 2019, 7162948. [Google Scholar] [CrossRef]
- Li, G.; Kusari, S.; Lamshoft, M.; Schuffler, A.; Laatsch, H.; Spiteller, M. Antibacterial secondary metabolites from an endophytic fungus, Eupenicillium sp. LG41. J. Nat. Prod. 2014, 77, 2335–2341. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, Z.; Feng, H.; Gan, Q.; Che, Q.; Zhu, T.; Gu, Q.; Han, B.; Li, D. Trichodermamides D–F, heterocyclic dipeptides with a highly functionalized 1,2-oxazadecaline core isolated from the endophytic fungus Penicillium janthinellum HDN13-309. RSC Adv. 2017, 7, 48019–48024. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, M.L.; Li, D.H.; Zeng, R.; Han, B.N. N-Me-trichodermamide B isolated from Penicillium janthinellum, with antioxidant properties through Nrf2-mediated signaling pathway. Bioorg. Med. Chem. 2017, 25, 6614–6622. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Gu, Q.-Q.; Zhu, W.-M.; Cui, C.-B.; Fan, G.-T. Trichodermamide A and aspergillazine A, two cytotoxic modified dipeptides from a marine-derived fungus Spicaria elegans. Arch. Pharmacal Res. 2005, 28, 1042–1046. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Kimishima, A.; Ishida, R.; Setiawan, A.; Arai, M. Selective cytotoxicity of epidithiodiketopiperazine DC1149B, produced by marine-derived Trichoderma lixii on the cancer cells adapted to glucose starvation. J. Nat. Med. 2020, 74, 153–158. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Z.; Sun, K.; Zhu, W. Effects of high salt stress on secondary metabolite production in the marine-derived fungus Spicaria elegans. Mar. Drugs 2011, 9, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Garo, E.; Starks, C.M.; Jensen, P.R.; Fenical, W.; Lobkovsky, E.; Clardy, J. Trichodermamides A and B, cytotoxic modified dipeptides from the marine-derived fungus Trichoderma virens. J. Nat. Prod. 2003, 66, 423–426. [Google Scholar] [CrossRef]
- Davis, R.A.; Longden, J.; Avery, V.M.; Healy, P.C. The isolation, structure determination and cytotoxicity of the new fungal metabolite, trichodermamide C. Bioorg. Med. Chem. Lett. 2008, 18, 2836–2839. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Xiao, S.-Q.; Guan, X.; He, P.; Yin, Z. Isolation and identification of an endophytic fungus from cumin and GC analysis for its metabolites. J. Shenyang Agric. Univ. 2018, 49, 730–734. [Google Scholar]
- Kalin, S.N.; Altay, A.; Budak, H. Inhibition of thioredoxin reductase 1 by vulpinic acid suppresses the proliferation and migration of human breast carcinoma. Life Sci. 2022, 310, 121093. [Google Scholar] [CrossRef]
- Roser, L.A.; Erkoc, P.; Ingelfinger, R.; Henke, M.; Ulshofer, T.; Schneider, A.K.; Laux, V.; Geisslinger, G.; Schmitt, I.; Furst, R.; et al. Lecanoric acid mediates anti-proliferative effects by an M phase arrest in colon cancer cells. Biomed. Pharmacother. 2022, 148, 112734. [Google Scholar] [CrossRef] [PubMed]
- Ristic, S.; Rankovic, B.; Kosanic, M.; Stanojkovic, T.; Stamenkovic, S.; Vasiljevic, P.; Manojlovic, I.; Manojlovic, N. Phytochemical study and antioxidant, antimicrobial and anticancer activities of Melanelia subaurifera and Melanelia fuliginosa lichens. J. Food Sci. Technol. 2016, 53, 2804–2816. [Google Scholar] [CrossRef] [PubMed]
- Lunne, F.; Niehaus, E.M.; Lipinski, S.; Kunigkeit, J.; Kalinina, S.A.; Humpf, H.U. Identification of the polyketide synthase PKS7 responsible for the production of lecanoric acid and ethyl lecanorate in Claviceps purpurea. Fungal Genet. Biol. 2020, 145, 103481. [Google Scholar] [CrossRef]
- Ivanova, V.; Backor, M.; Dahse, H.M.; Graefe, U. Molecular structural studies of lichen substances with antimicrobial, antiproliferative, and cytotoxic effects from Parmelia subrudecta. Prep. Biochem. Biotechnol. 2010, 40, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak-Celinska, A.; Kleszcz, R.; Studzinska-Sroka, E.; Lukaszyk, A.; Szoszkiewicz, A.; Stelcer, E.; Jopek, K.; Rucinski, M.; Cielecka-Piontek, J.; Krajka-Kuzniak, V. Lichen secondary metabolites inhibit the Wnt/beta-catenin pathway in glioblastoma cells and improve the anticancer effects of temozolomide. Cells 2022, 11, 1084. [Google Scholar] [CrossRef]
- Lü, F.; Li, X.; Chi, L.; Meng, L.; Wang, B. A new acyclic peroxide from Aspergillus nidulans SD-531, a fungus obtained from deep-sea sediment of cold spring in the South China Sea. J. Oceanol. Limnol. 2020, 38, 1225–1232. [Google Scholar] [CrossRef]
- Nugraha, A.S.; Untari, L.F.; Laub, A.; Porzel, A.; Franke, K.; Wessjohann, L.A. Anthelmintic and antimicrobial activities of three new depsides and ten known depsides and phenols from lndonesian lichen: Parmelia cetrata Ach. Nat. Prod. Res. 2021, 35, 5001–5010. [Google Scholar] [CrossRef]
- Thadhani, V.M.; Choudhary, M.I.; Khan, S.; Karunaratne, V. Antimicrobial and toxicological activities of some depsides and depsidones. J. Natl. Sci. Found. Sri Lanka 2012, 40, 43–48. [Google Scholar] [CrossRef]
- Rajendran, K.; Ponmurugan, P.; Gnanamangai, B.M.; Karuppiah, P.; Shaik, M.R.; Khan, M.; Khan, M.; Shaik, B. Bioefficacy of lecanoric acid produced by Parmotrema austrosinense (Zahlbr.) Hale against tea fungal pathogens. Horticulturae 2023, 9, 705. [Google Scholar] [CrossRef]
- Paguirigan, J.A.; Liu, R.; Im, S.M.; Hur, J.S.; Kim, W. Evaluation of antimicrobial properties of lichen substances against plant pathogens. Plant Pathol. J. 2022, 38, 25–32. [Google Scholar] [CrossRef]
- Zorrilla, J.G.; D’Addabbo, T.; Roscetto, E.; Varriale, C.; Catania, M.R.; Zonno, M.C.; Altomare, C.; Surico, G.; Nimis, P.L.; Evidente, A. Antibiotic and nematocidal metabolites from two lichen species collected on the island of Lampedusa (Sicily). Int. J. Mol. Sci. 2022, 23, 8471. [Google Scholar] [CrossRef]
- Biskup, I.; Zaczynska, E.; Krauze-Baranowska, M.; Fecka, I. Evaluation of cytotoxicity of 5-n-alkylresorcinol homologs and fraction on mouse fibroblast cell line L929. Eur. Food Res. Technol. 2017, 243, 1137–1148. [Google Scholar] [CrossRef]
- Adsul, M.G.; Bastawde, K.B.; Varma, A.J.; Gokhale, D.V. Strain improvement of Penicillium janthinellum NCIM 1171 for increased cellulase production. Bioresour. Technol. 2007, 98, 1467–1473. [Google Scholar] [CrossRef]
- Ma, Y.P.; Sun, S.W.; Hao, H.; Xu, C.P. Production, purification and characterization of an exo-polygalacturonase from Penicillium janthinellum sw09. An. Acad. Bras. Cienc. 2016, 88, 479–487. [Google Scholar] [CrossRef]
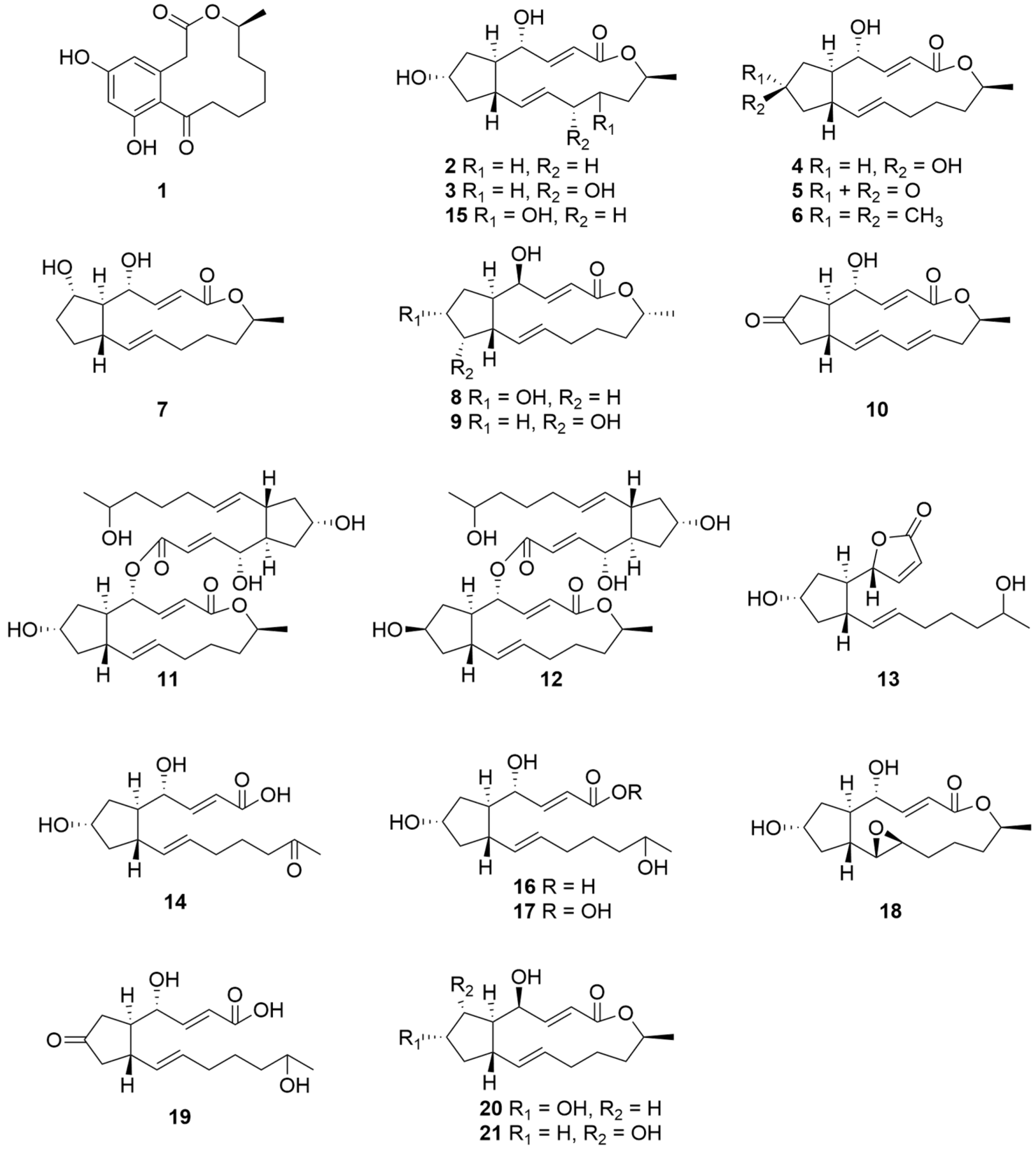
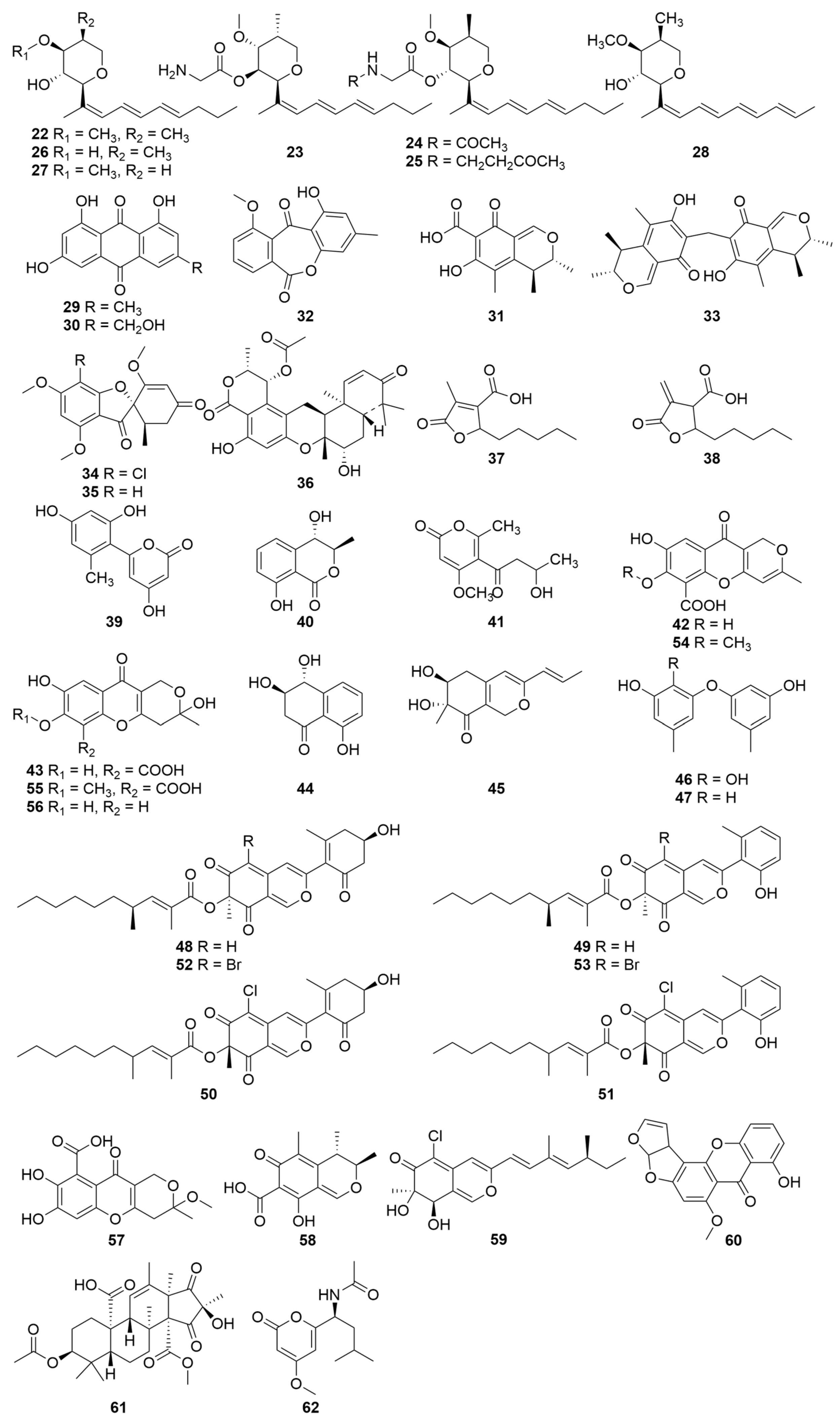

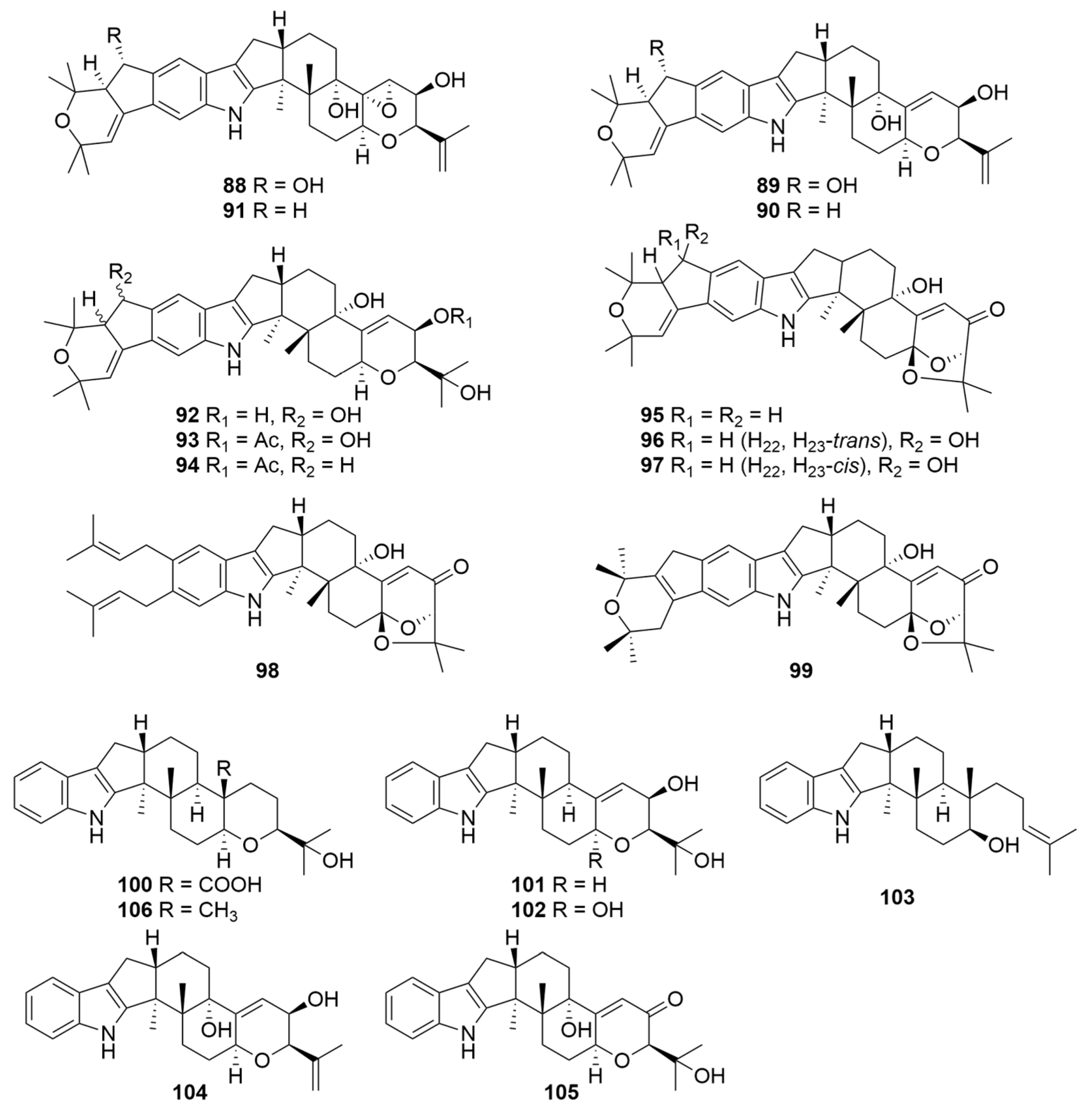
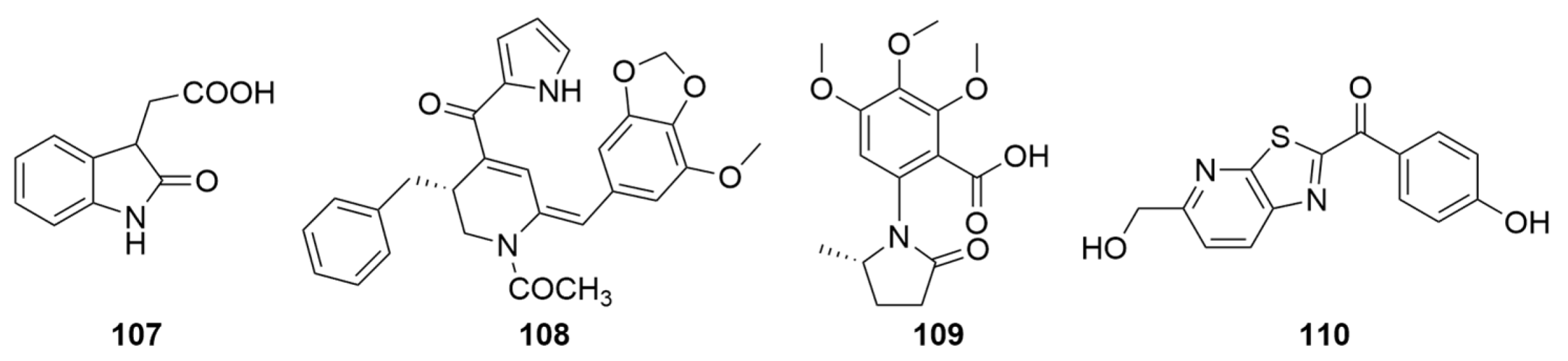


| Compounds | Sources | Media | Distribution | Years | Refs. |
|---|---|---|---|---|---|
| 1 | P. janthinellum IFM 55557 | Moist rice for mass culture | Japan | 2009 | [25] |
| 2 | P. janthinellum AJ608945 | PDA medium for seed stage cultures and potato dextrose broth medium for fermentation | Jilin Province China | 2013 | [6] |
| 2–9 | Marine-derived P. janthinellum DT-F29 GenBank No. KT443922.1 | Solid rice medium for mass culture | China | 2016 | [34] |
| 2, 5, 10 | Marine-derived P. janthinellum DT-F29 GenBank No. KT443922.1 | Solid rice medium for mass culture | China | 2018 | [35] |
| 2, 5 | Taxus wallichiana endophytic P. janthinellum MPT-25 GenBank No.MZ048774 | PDA medium for seed stage cultures and rice medium for fermentation | Hebei Province China | 2022 | [36] |
| 2, 11–21 | Soil-divided P. Janthinellum | PDA medium for seed stage cultures and rice medium for fermentation | Chongqing China | 2019 | [30] |
| 22–28 | Soil-derived P. janthinellum NR6564 | Glucose, glycerol, polypeptone, yeast extract, etc., for fermentation | Hong Kong China | 1992 | [44] |
| 29–32 | Melia azedarach endophytic P. janthinellum LaBioMi-018 | PDA medium for seed stage cultures and white corn medium for fermentation | São Carlos Brazil | 2005 | [7] |
| 31, 33 | Melia azedarach endophytic P. janthinellum LaBioMi-018 | PDA medium for seed stage cultures and white corn medium for fermentation | São Carlos Brazil | 2011 | [46] |
| 34, 35 | Soft coral-isolated P. janthinellum | Seawater-based medium for mass culture | South China Sea | 2006 | [64] |
| 36–38 | Heavily saline–alkali soil-isolated P. janthinellum | Seawater culture medium for fermentation | Binzhou China | 2016 | [66] |
| 39–47 | Sponge-associated P. janthinellumLZDX-32-1 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | South China Sea | 2017 | [11] |
| 48–51 | Soil-derived P. janthinellum HK1-6 GenBank No. KY412802 | Potato glucose liquid medium for mass culture | Hainan Island China | 2017 | [87] |
| 48, 49, 52, 53 | Soil-derived P. janthinellum HK1-6 GenBank No. KY412802 | Potato dextrose broth medium (supplemented NaBr) for mass culture | Hainan Island China | 2019 | [88] |
| 52, 53 | Soil-derived P. janthinellum HK1-6 GenBank No. KY412802 | Potato dextrose broth medium (supplemented NaBr) for mass culture | Hainan Island China | 2020 | [89] |
| 43, 54, 55, 56, 57 | Marine-derived P. janthinellum DT-F29 GenBank No. KT443922.1 | Solid rice medium for mass culture | China | 2018 | [35] |
| 58, 59 | Marine-derived P. janthinellum JK07-5 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | Bohai Sea | 2020 | [10] |
| 60 | P. janthinellum GenBank No. GU565141.1 | MEA agar medium for seed stage cultures MEA liquid medium for fermentation | China | 2021 | [91] |
| 61 | Nicotiana tabacum endophytic P. janthinellum TE-43 GenBank No. MZ310442 | PDA medium for seed stage cultures and modified PDB liquid medium for fermentation | Qingdao China | 2021 | [92] |
| 62 | Taxus wallichiana endophytic P. janthinellum MPT-25 GenBank No.MZ048774 | PDA medium for seed stage cultures and rice medium for fermentation | Hebei Province China | 2022 | [93] |
| 63–65 | Soft coral-isolated P. janthinellum | Seawater-based medium for mass culture | South China Sea | 2006 | [64] |
| 66–71 | Mangrove endophytic P. janthinellum HDN13-309 GenBank No. KM659023 | PDA medium for preparation culture and seawater culture medium for mass culture | Hainan Province China | 2016 | [94] |
| 72–75 | Heavily saline–alkali soil-isolated P. janthinellum | Seawater culture medium for fermentation | Binzhou China | 2016 | [66] |
| 76–79 | Sponge-associated P. janthinellumLZDX-32-1 | Rice solid culture medium for fermentation | South China Sea | 2019 | [95] |
| 80–82 | Marine-derived P. janthinellum JK07-5 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | Bohai Sea | 2020 | [10] |
| 83–87 | Soil-derived P. janthinellum HK1-6 GenBank No. KY412802 | Rice medium for mass culture | Hainan Island China | 2020 | [101] |
| 88–90 | Pasture-isolated P. janthinellum | Potato broth and potato dextrose agar-tryptophan medium for mass culture | New Zealand | 1980 | [104] |
| 89, 90 | Pasture-isolated P. janthinellum (FRR 3777) | CDYE medium for seed stage cultures and CDYE medium supplemented with CaCl2 (2%) for mass culture | New Zealand | 1993 | [105] |
| 88–91 | Pasture-isolated P. janthinellum | Potato/milk/sucrose broth for mass culture | New Zealand | 2018 | [106] |
| 92–94 | Pasture-isolated P. janthinellum TDD4 | Modified Czapek medium for mass culture | New Zealand | 1984 | [107] |
| 95–98 | Marine sediment-isolated P. janthinellum | Nutrient medium RM14 for mass culture | Amursky Bay Japan | 2007 | [12] |
| 99–103 | Entomogenous fungus P. janthinellum (LB1.20090001) GenBank No. KY427360.1 | Culture dish of potato dextrose agar for seed stage cultures and potato dextrose broth for mass culture and rice medium for fermentation | Anhui Province China | 2021 | [109] |
| 104–106 | P. janthinellum IFM 55557 | Moist rice for mass culture | Japan | 2009 | [25] |
| 106 | Sponge-associated P. janthinellumLZDX-32-1 | Rice solid culture medium for fermentation | South China Sea | 2019 | [95] |
| 107 | Sponge-associated P. janthinellumLZDX-32-1 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | South China Sea | 2017 | [118] |
| 108–109 | Panax notoginseng endophytic P. janthinellum SYPF 7899 GenBank No. KU360251 | PDA medium for strain isolation and rice medium for mass culture | Yunnan Province China | 2018 | [118] |
| 110 | Taxus wallichiana endophytic P. janthinellum MPT-25 GenBank No.MZ048774 | PDA medium for seed stage cultures and rice medium for fermentation | Hebei Province China | 2022 | [36] |
| 111 | Soil fungal P. janthinellum ESF20P GenBank No. JX456373 | PDA medium for seed stage cultures | Malaysia | 2015 | [21] |
| 112–119 | Panax notoginseng endophytic P. janthinellum SYPF 7899 GenBank No. KU360251 | PDA medium for strain isolation and rice medium for mass culture | Yunnan Province China | 2018 | [118] |
| 113, 120 | Marine-derived P. janthinellum JK07-5 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | Bohai Sea | 2020 | [10] |
| 121–124 | Entomogenous fungus P. janthinellum (LB1.20090001) GenBank No. KY427360.1 | Culture dish of potato dextrose agar for seed stage cultures and potato dextrose broth for mass culture and rice medium for fermentation | Anhui Province China | 2021 | [118] |
| 125 | Nicotiana tabacum endophytic P. janthinellum TE-43 GenBank No. MZ310442 | PDA medium for seed stage cultures and modified PDB liquid medium for fermentation | Qingdao China | 2021 | [92] |
| 126–132 | Taxus wallichiana endophytic P. janthinellum MPT-25 GenBank No.MZ048774 | PDA medium for seed stage cultures and rice medium for fermentation | Hebei Province China | 2022 | [36] |
| 133–136 | Penicillium janthinellum | PDA medium for seed stage cultures and white corn medium for fermentation: | São Carlos Brazil | 2005 | [7] |
| 133 | Heavily saline–alkali soil-isolated P. janthinellum | Seawater culture medium for fermentation | Binzhou China | 2016 | [66] |
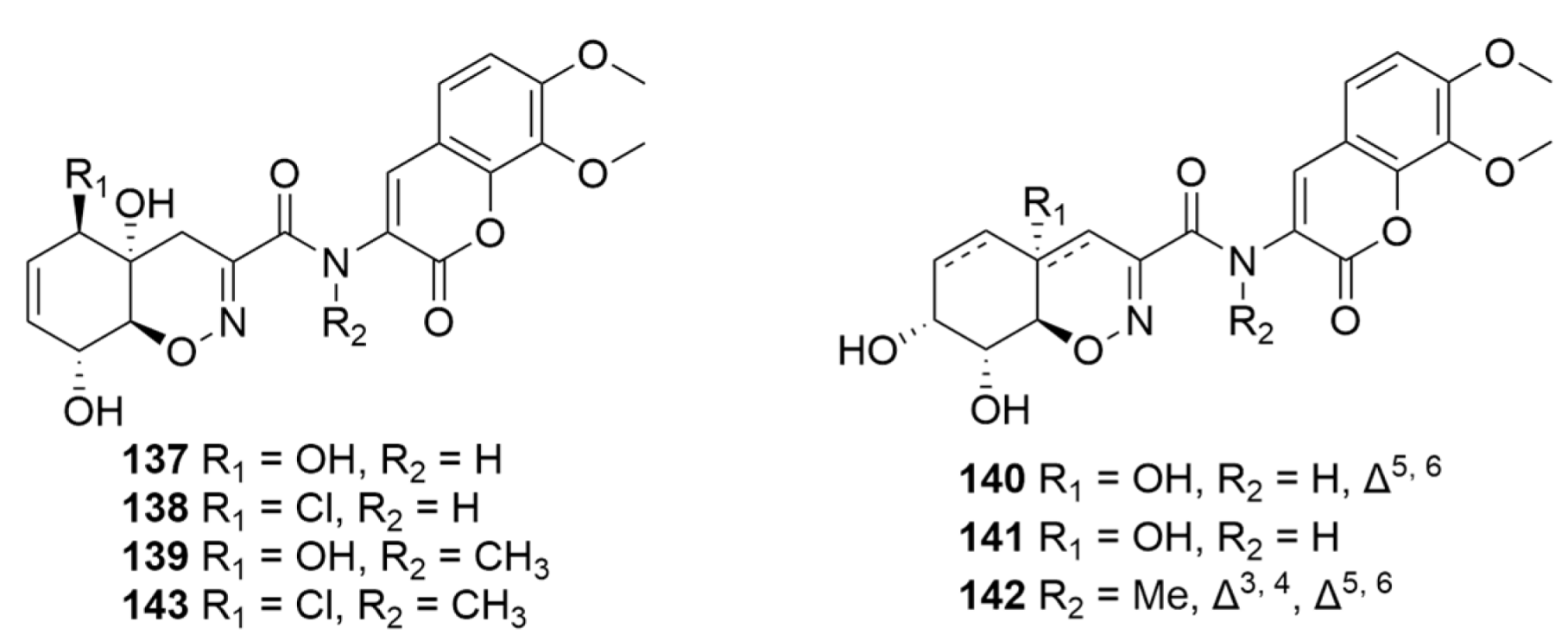
| 137–142 | Mangrove endophytic P. janthinellum HDN13-309 GenBank No. KM659023 | PDA medium for preparation culture and seawater culture medium for mass culture | Hainan Province China | 2017 | [125] |
| 143 | Mangrove endophytic P. janthinellum HDN13-309 GenBank No. KM659023 | PDA medium for preparation culture and seawater culture medium for mass culture | Hainan Province China | 2017 | [126] |
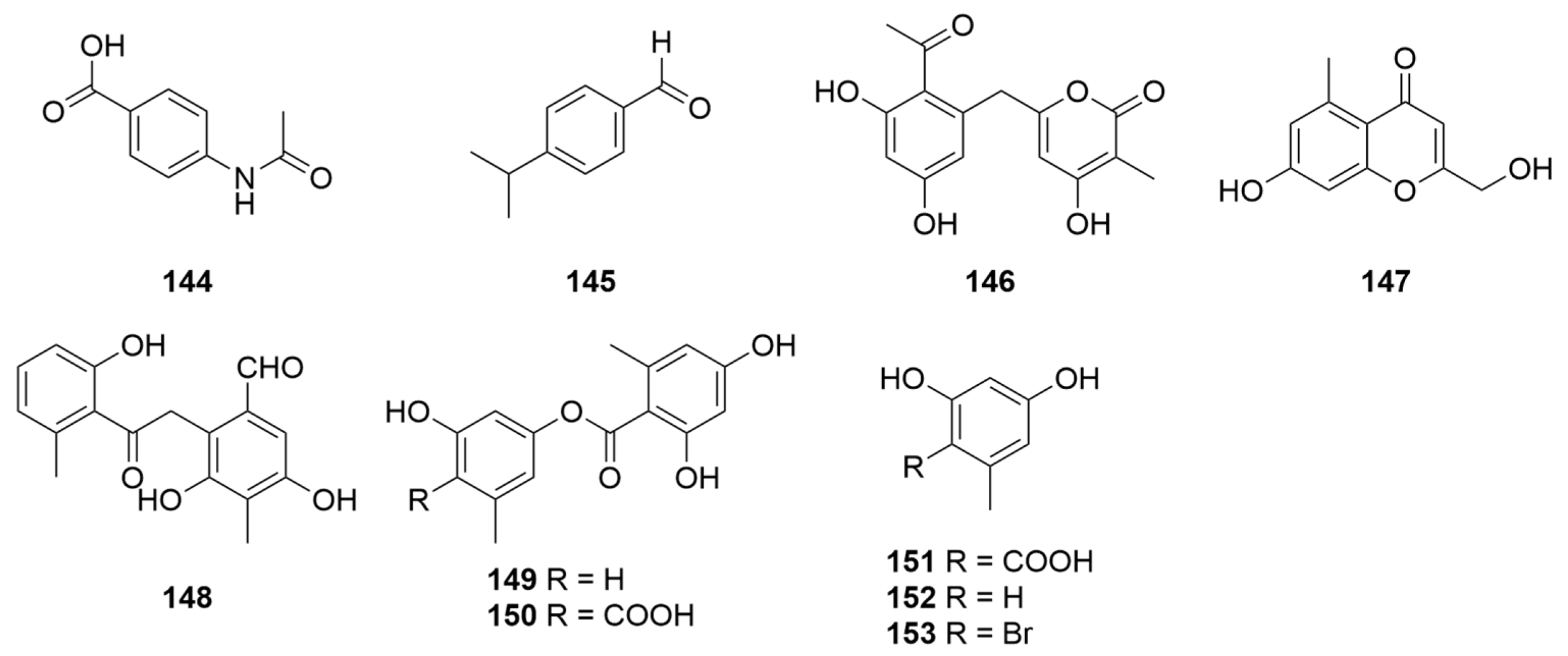
| 144 | Sponge-associated P. janthinellumLZDX-32-1 | PDA medium for seed stage cultures and rice solid culture medium for fermentation | South China Sea | 2017 | [11] |
| 145 | Endophytic fungi ZR-003 | PDA medium for preparation culture and PD liquid medium for mass culture | China | 2018 | [132] |
| 146–153 | Soil-derived P. janthinellum HK1-6 GenBank No. KY412802 | Potato dextrose broth medium (supplemented NaBr) for mass culture | Hainan Island China | 2020 | [89] |
| Compounds | Tested Strains | IC50 Values (μM) | IC50 Values of Positive Controls (μM) | Pros and Cons | Refs. |
|---|---|---|---|---|---|
| 2 | MKN45 | <0.004 | 0.114 | One commonly used reversible protein transport inhibitor | [6] |
| LoVo | 0.428 | 0.024 | |||
| A549 | 0.143 | 0.046 | |||
| MDA-MB-435 | <0.004 | 0.044 | |||
| HepG2 | <0.004 | 0.064 | |||
| HL-60 | <0.004 | 0.002 | |||
| H1975 | <0.2 | 4.8 ± 0.2 | [34] | ||
| J-Lat clones C11 cells | 3.3 ± 0.3 × 10−2 | 0.8 ± 0.2 | |||
| A549 | 0.101 | [39] | |||
| HeLa | 0.172 | ||||
| HepG2 | 0.239 | ||||
| HL-60 | 0.11 ± 0.02 | [37] | |||
| U87MG | 0.01 ± 0.00 | ||||
| MDA-MB-231 | 0.03 ± 0.00 | ||||
| A549 | 0.05 ± 0.00 | ||||
| HEP-3B | 0.04 ± 0.00 | ||||
| SW480 | 0.04 ± 0.01 | ||||
| NCM460 | 0.04 ± 0.00 | ||||
| 4 | H1975 | 5.2 ± 0.6 | 4.8 ± 0.2 | Comparable to the positive control | [34] |
| WRL68 | 0.05 | [41] | |||
| MCF-7 | 0.35 | 0.11 | |||
| 11/12/18 | HL-60 | 2.67 ± 0.14/2.55 ± 0.12/4.45 ± 0.05 | A broad-spectrum anticancer agent | [37] | |
| U87MG | 0.1 ± 0.00/0.3 ± 0.00/3.75 ± 0.01 | ||||
| MDA-MB-231 | 1.11 ± 0.34/1.05 ± 0.26/3.82 ± 0.03 | ||||
| A549 | 0.68 ± 0.08/0.75 ± 0.10/3.98 ± 0.06 | ||||
| HEP-3B | 0.54 ± 0.10/0.63 ± 0.10/3.91 ± 0.09 | ||||
| SW480 | 0.83 ± 0.12/0.77 ± 0.01/4.10 ± 0.01 | ||||
| NCM460 | 0.97 ± 0.07/0.88 ± 0.09/4.10 ± 0.03 | ||||
| 29 | MCF-7 | 80 | Broad-spectrum cytotoxicity with selectivity | [48] | |
| SW620 | 22.57 | [49] | |||
| K562 | 5.55 ± 0.74 | [50] | |||
| HeLa | 31.08 ± 5.92 | ||||
| Calu-1 | 32.93 ± 3.70 | ||||
| Wish | 32.19 ± 1.85 | ||||
| Vero | 12.95 ± 0.44 | ||||
| Raji | 10.36 ± 1.48 | ||||
| Human mesangial cells | 17.9 ± 1.2 | [52] | |||
| HeLa | 8.94 | [51] | |||
| A549 | 62.35 | [55] | |||
| MCF-7 | 26.72 | [56] | |||
| HuCCA-1 | 73.71 ± 4.29 | 1.23 ± 0.09 | [47] | ||
| A549 | 143.61 ± 4.26 | 0.49 ± 0.02 | |||
| HepG2 | 150.01 ± 5.55 | 0.48 ± 0.06 | |||
| MOLT-3 | 18.47 ± 0.89 | 0.04 ± 0.002 | |||
| 30 | A549 | Weak | Weak activity | [58] | |
| SK-OV-3 | |||||
| HepG2 | |||||
| HT-29 | |||||
| 31 | HT-29 | 71.92 | Moderate activity | [62] | |
| 32 | K562 | 34.6% (inhibition rate at 100 μg/mL) | Weak activity | [63] | |
| 36/37/38 | A549 | 88.7/36.5/45.4 | 12.4 | Moderate activity | [66] |
| 41 | A549 | 31.83 | 577 | Significant cytotoxicity | [70] |
| MDA-MB-231 | 114 | 361 | |||
| PANC-1 | 62.33 | 500 | |||
| KB | 14.14 | [71] | |||
| BC-1 | 5.30 | ||||
| Vero cells | 10.17 | ||||
| 42/43 | HeLa cells | 28.01 ± 0.62/20.54 ± 2.14 | 2.79 ± 0.16 | Weak cytotoxicity and cellular protection effects. | [74] |
| Vero cells | 803.74 ± 12.85/404.62 ± 4.12 | 335.32 ± 0.94 | |||
| Balb/c3T3 | -/95.35 ± 3.69% (survival rate at 50 μg/mL) | [77] | |||
| -/90.60 ± 4.85% (survival rate at 400 μg/mL) | |||||
| 46 | BC-cells | 35.13 | Significant cytotoxicity | [78] | |
| NCI-H187 cells | 15.12 | ||||
| Vero cells | 53.20 | ||||
| 47 | U87MG | 4.4 | 1.6 ± 0.3 | It exhibits cytotoxicity against multiple cell lines, but the activity is not satisfactory. | [79] |
| U251 | 6.2 | 6.8 ± 1.6 | |||
| K562 | Inhibited the growth at 30 μM | [80] | |||
| NCI-H460 | 90% (inhibition rate at 200 μM) | [81] | |||
| HePG-2 | 55% (inhibition rate at 200 μM) | ||||
| MCF-7 | 57% (inhibition rate at 200 μM) | ||||
| MDA-MB-231 | 78% (inhibition rate at 200 μM) | ||||
| mouse splenic cells | 110 | [82] | |||
| 59 | B-16 | 36 | Weak activity | [90] | |
| 60 | HEK293 | Significantly reduced the density at 128 μM | Weak activity | [91] | |
| 61 | A549 | Dose-dependent manner | Significantly | Dose-dependent | [92] |
| 62 | Brine shrimp larvae (A. salina) | Weak toxicity at a concentration of 10 μg/mL | Weak activity | [93] | |
| 66/67/68 | HeLa | 0.5/3.9/0.3 | 0.5 | Significant cytotoxicity | [94] |
| HL-60 | 0.1/1.6/1.2 | 0.2 | |||
| 76/77/78 | A549 | 8.60 ± 0.67%/36.14 ± 2.09%/5.77 ± 1.72% (inhibition rate at 10 μM) | 0.23 ± 0.11 | Weak activity | [95] |
| HCT-8 | 8.41 ± 0.93%/23.73 ± 0.97%/6.82 ± 1.03% (inhibition rate at 10 μM) | 0.71 ± 0.29 | |||
| MCF-7 | 8.74 ± 0.78%/36.88 ± 1.88%/18.24 ± 1.68% (inhibition rate at 10 μM) | 0.20 ± 0.06 | |||
| tsFT210 | -/-/MIC = 26.1 μM | [96] | |||
| 79 | A549 | 36.65 ± 2.99% (inhibition rate at 10 μM) | 0.23 ± 0.11 | Weak activity | [94] |
| HCT-8 | 22.76 ± 2.01% (inhibition rate at 10 μM) | 0.71 ± 0.29 | |||
| MCF-7 | 23.84 ± 0.90% (inhibition rate at 10 μM) | 0.20 ± 0.06 | |||
| Jurkat | 68.17 ± 6.10 μM | [99] | |||
| 95/96/97 | HL-60 | 10%/39%/34% (inhibition rate at 100 μM) | Weak activity | [12] | |
| 103 | HelaS3 | 44.47 | 0.13 | It exhibits cytotoxicity against multiple cell lines, but the activity is not satisfactory. | [111] |
| KB | 35.77 | 0.11 | |||
| HepG2 | 48.25 | 0.22 | |||
| MCF-7 | 16.19 | 0.53 | |||
| A549 | 51.91 | 0.58 | |||
| Vero | 48.63 | 0.95 | |||
| 106 | A549 | 70.45 ± 0.97% (inhibition rate at 10 μM) | 0.23 ± 0.11 | It exhibits cytotoxicity against multiple cell lines, but the activity is not satisfactory. | [95] |
| HCT-8 | 67.03 ± 0.40% (inhibition rate at 10 μM) | 0.71 ± 0.29 | |||
| MCF-7 | 88.54 ± 0.34% (inhibition rate at 10 μM) | 0.20 ± 0.06 | |||
| HeLa | 5.7 ± 0.1 | 11.3 ± 2.5 | [116] | ||
| HepG-2 | 52.4% (inhibition rate at 1 μM) | [115] | |||
| U2OS | 83.4% (inhibition rate at 1 μM) | ||||
| MCF-7 | 47.5% (inhibition rate at 1 μM) | ||||
| JeKo-1 | 72.4% (inhibition rate at 1 μM) | ||||
| HL-60 | 60.3% (inhibition rate at 1 μM) | ||||
| 112 | HL-60 | 27.8 | 0.461 | Weak activity | [122] |
| SU-DHL-4 | 23.5 | 0.264 | |||
| RKO | 21.5 | 0.521 | |||
| 113/116 | RAW264.7 | 68.95/194.48 | Weak activity | [121] | |
| IEC-6 | 32.13/18.26 | ||||
| A549 | 263.39/376.23 | ||||
| HTB-176 | -/10 ± 3.92 | 4.3 ± 0.25 | [123] | ||
| 125 | A549 | Suppressed the proliferation and metastasis in a dose-dependent manner | Significantly | Dose-dependent | [92] |
| 137 | HL-60 | 89 | Weak activity | [127] | |
| PANC-1 Glucose (−) | 270 | 0.0003 | [128] | ||
| 138 | K562 | 8.0 | Significant activity but no positive control | [125] | |
| HL-60 | 1.8 | ||||
| HO-8910 | 1.9 | ||||
| MGC803 | 1.6 | ||||
| HCT-116 | 0.71 | [131] | |||
| 139 | HCT116 | 1.52 | Significant activity but no positive control | [131] | |
| A549 | 9.59 | ||||
| 150 | MCF-7 | Very weak cytotoxicity | A broad-spectrum anticancer agent | [133] | |
| HCT-116 | Significantly reduced the viability at 30 μg/mL | [134] | |||
| Significantly reduced the formation of cells at 0.03 μg/mL | |||||
| 20% (inhibition rate at 30 μg/mL) | |||||
| HEK293 | Responsive at 0.3 μg/mL | ||||
| HeLa | Responsive at 3 μg/mL | ||||
| NIH3T3 | Responsive at 3 μg/mL | ||||
| RAW264.7 | Responsive at 3 μg/mL | ||||
| HeLa | 389.50 ± 6.47 | [135] | |||
| HepG2 | Concentration-dependent | [136] | |||
| CCF | 40% (inhibition rate at 40 μM) | ||||
| 50% (inhibition rate from 60 μM to 100 μM) | |||||
| L-929 | 157.09 | [137] | |||
| K562 | 157.09 | ||||
| HeLa | 157.09 | ||||
| 157.09 | |||||
| A-172 | 30% (inhibition rate at 100 μM) | [138] | |||
| 152 | L929 | 6918 | Weak activity | [145] |
| Compounds | Tested Strains | MIC Values (μg/mL) | MIC Values of Positive Controls (μg/mL) | Pros and Cons | Refs. |
|---|---|---|---|---|---|
| 29 | E. coli | 500 | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [7] | |
| P. aeruginosa | 62.50 | ||||
| B. subtilis | 7.81 | ||||
| B. cereus | 25.0 | [47] | |||
| S. aureus | 25.0 | ||||
| 30 | H. pylori | 1.79 | 15 | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [59] |
| E. coli | 31.25 | [60] | |||
| P. aeruginosa | 62.50 | ||||
| B. subtilis | 250 | ||||
| 31/32/33 | E. coli | 500,000/500,000/31,250 | Weak activity | [7,46] | |
| P. aeruginosa | 62,500/500,000/125,000 | ||||
| B. subtilis | 31,250/500,000/31,250 | ||||
| 36 | E. coli | 20.8 ± 0.25 | 6.7 ± 0.17 | Weak activity | [67] |
| 37 | E. coli | 32.74 ± 0.058% (inhibition rate at 200 μM) | 100 ± 0.07% (inhibition rate at 0.5 μg/mL) | Weak activity | [68] |
| 31.24 ± 0.065% (inhibition rate at 400 μM) | |||||
| 38 | S. aureus IFO 3060 | 6 | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [69] | |
| M. roseus IFO 3764 | 6 | ||||
| M. luteus IFO 3333 | 6 | ||||
| C. xerosis IFO 12684 | 6 | ||||
| B. brevis IFO 3331 | 25 | ||||
| B. cereus IFO 3514 | 50 | ||||
| B. subtilis IFO 12210 | 100 | ||||
| M. luteus IFO 12708 | 100 | ||||
| A. simplex IFO 12069 | 100 | ||||
| P. vulgaris IFO 3851 | 100 | ||||
| P. chrysogenum IFO 4897 | 100 | ||||
| P. notatum | 100 | ||||
| P. urticae IFO 7011 | 100 | ||||
| P. experimentwn | 100 | ||||
| 42/43 | S. aureus subsp. aureus (DSM 799) | 5.0/- | 5.0/- | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [75] |
| E. coli (DSM 1116) | 10.0/- | 1.0/- | |||
| E. coli (DSM 682) | 10.0/- | 1.0/- | |||
| B. subtilis (DSM 1088) | 5.0/- | 5.0/- | |||
| M. smegmatis | 15.6/62.5 | 0.62 | [76] | ||
| V. cholerae SG24 (1) | 0.50/8 | 16 | [74] | ||
| V. cholerae CO6 | 16/16 | 16 | |||
| V. cholerae NB2 | 8/32 | 8 | |||
| V. cholerae PC2 | 0.50/32 | 1 | |||
| S. flexneri SDINT | 8/16 | 64 | |||
| 46 | TB | 200 | Weak activity | [78] | |
| 47 | B. subtilis | 11.8 mm (inhibition zones at 30 μg/disc) | 27 mm (inhibition zones at 10 μg/disc) | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [84] |
| C. violaceum | 57% (inhibition rate at 10 mg/mL) | [81] | |||
| 12.50 | 6.25 | ||||
| S. aureus | 59% (inhibition rate at 10 mg/mL) | ||||
| 12.50 | 6.25 | ||||
| E. faecalis | 60% (inhibition rate at 10 mg/mL) | ||||
| 6.25 | 6.25 | ||||
| S. choleraesuis | 57% (inhibition rate at 10 mg/mL) | ||||
| 25.00 | 3.13 | ||||
| M. smegmatis | 48% (inhibition rate at 10 mg/mL) | ||||
| E. coli | 45% (inhibition rate at 10 mg/mL) | ||||
| A. avenae subsp. Cattleyae | 25 | 6 | [85] | ||
| A. tumefaciens | 100 | 1 | |||
| B. glumae | 200 | 0.4 | |||
| C. michiganensis subsp. Michiganensis | 200 | 0.4 | |||
| D. chrysanthemi | 200 | 0.4 | |||
| P. carotovorum subsp. Carotovorum | 200 | 0.4 | |||
| R. solanacearum | 50 | 0.4 | |||
| V. parahemolyticus | 29 | 46 | [86] | ||
| S. aureus | 1001.6 | [82] | |||
| B. subtilis | 1001.6 | ||||
| 49/50/51/52/53 | S. aureus ATCC 43300 | 3.13/6.25/6.25/50/12.5 | 0.39 | Potent antibacterial activities against MRSA ATCC 43300 and ATCC 33591 | [87,88] |
| S. aureus ATCC 33591 | 3.13/6.25/6.25/12.5/3.13 | 0.78 | |||
| S. aureus ATCC 25923 | 3.13/12.5/12.5/12.5/6.25 | 0.78 | |||
| S. aureus ATCC 29213 | 3.13/6.25/3.13/25/12.5 | 3.13 | |||
| E. faecalis ATCC 51299 | 3.13/12.5/6.25/25/3.13 | 6.25 | |||
| E. faecium ATCC 35667 | 3.13/12.5/12.5/25/12.5 | 0.39 | |||
| 58 | S. typhi | 0.1 | 1.7 | Potent antibacterial activity but no more experimental data | [10] |
| 59 | S. aureus FDA 209P | Inhibited at 50 μg/disk | [90] | ||
| B. fragilis ATCC 23745 | Inhibited at 50 μg/disk | ||||
| 78/79 | V. harveyi | 32/32 | 8 | Weak activity | [98] |
| 81 | M. lysodeikticus | 5.5 | [100] | ||
| 82 | B. subtilis | 2.1 | Significant antimicrobial activity but no positive control | [100] | |
| V. parahemolyticus | 4.3 | ||||
| 96 | P. echinatior Ae706 | 5 | Significant antimicrobial activity but no positive control | [108] | |
| P. octospinosus Ae707 | 5 | ||||
| 100/101/102 | S. aureus | 25.0/50.0/12.5 | 0.78 | Weak activity | [109] |
| 103 | B. cereus | 128 | 1.0 | Weak activity | [111] |
| 108/109 | B. subtilis | 15/35 | Weak activity | [118] | |
| S. aureus | 18/39 | ||||
| 116 | S. aureus | 1.4 ± 2.4 | 0.523 | Antibacterial activities against both Gram-positive and Gram-negative bacteria | [123] |
| E. fergusonii | 2.5 ± 1.7 | 0.523 | |||
| P. aeruginosa | 0.13 ± 0.4 | 0.523 | |||
| 117 | B. subtilis | 50 | Weak activity | [105] | |
| S. aureus | 60 | ||||
| 124 | E. coli (DSM 1116) | 10.0 | 1.0 | Comparable to the positive control | [124] |
| Acinetobacter sp. BD4 (DSM 586) | 5.0 | 5.0 | |||
| 137/138 | E. aerogenes | 24.2/25.2 | 0.4 | Weak activity | [129] |
| E. coli | 96.4/100.1 | 1.7 | |||
| P. aeruginosa | 963.0/100.1 | 12.6 | |||
| S. aureus | 96.4/100.1 | 0.4 | |||
| 150 | A. hydrophilia | 32 | 0.5 | A broad spectrum of antibacterial activities against both Gram-positive and Gram-negative bacteria | [139] |
| E. ictarda | 32 | 2 | |||
| E. coli | 16 | 1 | |||
| V. harveyi | 8 | 1 | |||
| V. parahaemolyticus | 4 | 0.5 | |||
| S. aureus | 1000 | 31 | [135] | ||
| B. subtilis | 1000 | 16 | |||
| B. cereus | 500 | 16 | |||
| E. coli | 1000 | 62 | |||
| P. mirabilis | 1000 | 62 | |||
| M. mucedo | 1000 | 156 | |||
| T. viride | 1000 | 78 | |||
| S. aureus SG 511 | 200 | [139] | |||
| S. aureus MRSA | 1000 | ||||
| M. tuberculosis | 100 | ||||
| A. fischeri | 100% (inhibition rate at 100 μM) | 100% | [141] | ||
| C. michiganensis subsp. michiganensis | Antibacterial | [143] | |||
| 152 | MRSA 43300 | 18.75 | 1 | Stronger activity against Gram-positive bacteria | [144] |
| E. faecalis 29212 | 9.37 | 0.5 | |||
| E. coli 25922 | 9.37 | 1 | |||
| P. aeruginosa 27853 | 300 | 4 |
| Compounds | Tested Strains | MIC Values (μg/mL) | MIC Values of Positive Controls (μg/mL) | Pros and Cons | Refs. |
|---|---|---|---|---|---|
| 2/5 | A. fragriae | 12.5/25 | <0.78 | Weak activity | [36] |
| 23 | C. neoformans MY1051 | 2.0 | A broad-spectrum antifungal activity | [45] | |
| C. neoformans MY1146 | 4.0 | ||||
| C. albicans MY1058 | 0.5 | ||||
| C. albicans MY0992 | 4.0 | ||||
| C. parapsilosis MY1009 | 2.0 | ||||
| C. parapsilosis MY1010 | 2.0 | ||||
| C. pseudotropicalis MY1040 | 32.0 | ||||
| C. krusei MY1020 | 8.0 | ||||
| C. rugosa MY1022 | 0.5 | ||||
| C. guilliermondii MY1019 | 16.0 | ||||
| T. glabrata MY1059 | 32.0 | ||||
| P. italicum MY2819 | 2.0 | ||||
| S. cerevisiae ATCC9763 | IC50 = 1.5 μg/mL | Significant activity | [44] | ||
| 24 | S. cerevisiae ATCC9763 | IC50 = 46 μg/mL | Weak activity | [44] | |
| 25 | S. cerevisiae ATCC9763 | IC50 = 1.2 μg/mL | Significant activity | [44] | |
| 34 | A. solani | 2.75 | [64] | ||
| P. oryzae | 20 | ||||
| 41 | R. stolonifer | Significant inhibition of spore germination at 250 μg/mL | Weak activity against multiple fungi | [72] | |
| M. hiemalis | |||||
| F. solani | |||||
| F. oxysporum | |||||
| M. gypseum SH-MU-4 | 32 | [73] | |||
| 47 | A. rolfsii | 100% (inhibition rate at 0.01 mg/plug) | A broad-spectrum antifungal activity | [83] | |
| L. mediterranea | 100% (inhibition rate at 0.01 mg/plug) | ||||
| P. cinnamomi | 100% (inhibition rate at 0.01 mg/plug) | ||||
| F. avenaceum | 100% (inhibition rate at 0.2 mg/plug) | ||||
| 72.1% (inhibition rate at 0.1 mg/plug) | |||||
| 47.3% (inhibition rate at 0.05 mg/plug) | |||||
| S. parasitica | 17.5 mm (inhibition zones at 30 μg/disc) | 36 mm (inhibition zones at 10 μg/disc) | [84] | ||
| Pythium sp. | 13.0 mm (inhibition zones at 30 μg/disc) | 38 mm (inhibition zones at 10 μg/disc) | |||
| A. brassicicola | 100 | 3.1 | [85] | ||
| B. cinerea | 100 | 25 | |||
| C. cucumerinum | 100 | 50 | |||
| C. coccodes | 100 | 6.3 | |||
| C. destructans | 100 | 100 | |||
| F. oxysporum | 100 | 25 | |||
| M. oryzae | 6.3 | 6.3 | |||
| P. infestans | 25 | 1.6 | |||
| C. albicans | 794 | [82] | |||
| C. albican | 63% (inhibition rate at 10 mg/mL) | [81] | |||
| 6.25 | 0.28 | ||||
| 59 | P. oryzae KF 180 | Inhibited the growth at 50 μg/disk | [90] | ||
| 78 | P. oryzae | Comparable to the positive control nystatin | Comparable to the positive control | [97] | |
| F. graminearum | |||||
| B. cinerea | |||||
| P. capsici | |||||
| 137/138 | C. albicans | 241/125 | 2.66 | Weak activity | [129] |
| 150 | C. cladosporioides | 1000 | 39 | A broad-spectrum antifungal activity but suboptimal | [135] |
| F. oxysporum | 1000 | 78 | |||
| A. alternata | 1000 | 78 | |||
| A. flavus | 1000 | 312 | |||
| A. niger | 1000 | 78 | |||
| C. albicans | 500 | 39 | |||
| P. expansum | 1000 | 156 | |||
| P. chrysogenum | 1000 | 78 | |||
| T. longifusus | 40% (inhibition rate at 200 μg/mL) | 70 | [140] | ||
| A. flavus | 40% (inhibition rate at 200 μg/mL) | 20 | |||
| F. solani | 50% (inhibition rate at 200 μg/mL) | 74 | |||
| H. serpens | 77.7 ± 1.3% (inhibition rate at 15 μL) | [141] | |||
| M. theicola | 76.5 ± 1.5% (inhibition rate at 15 μL) | ||||
| P. theae | 80.5 ± 1.3% (inhibition rate at 15 μL) | ||||
| T. aculeate | 75.0 ± 1.4% (inhibition rate at 15 μL) | ||||
| Cercosporatheae | 91.5 ± 2.0% (inhibition rate at 15 μL) | ||||
| G. cingulata | 86.5 ± 2.1% (inhibition rate at 15 μL) | ||||
| P. theae | 90.0 ± 2.3% (inhibition rate at 15 μL) | ||||
| P. hypolateritia | 73.3 ± 1.5% (inhibition rate at 15 μL) | ||||
| R. solani | A broad spectrum of fungal growth inhibition | [143] | |||
| B. cinerea | |||||
| S. sclerotiorum | |||||
| D. eres | |||||
| D. actinidiae | |||||
| R. cerealis | |||||
| A. mali |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Li, Y.; Wang, Y.; Shi, T.; Wang, B. Penicillium janthinellum: A Potential Producer of Natural Products. Fermentation 2024, 10, 157. https://doi.org/10.3390/fermentation10030157
Wang H, Li Y, Wang Y, Shi T, Wang B. Penicillium janthinellum: A Potential Producer of Natural Products. Fermentation. 2024; 10(3):157. https://doi.org/10.3390/fermentation10030157
Chicago/Turabian StyleWang, Han, Yanjing Li, Yifei Wang, Ting Shi, and Bo Wang. 2024. "Penicillium janthinellum: A Potential Producer of Natural Products" Fermentation 10, no. 3: 157. https://doi.org/10.3390/fermentation10030157
APA StyleWang, H., Li, Y., Wang, Y., Shi, T., & Wang, B. (2024). Penicillium janthinellum: A Potential Producer of Natural Products. Fermentation, 10(3), 157. https://doi.org/10.3390/fermentation10030157






