New Solutions in Single-Cell Protein Production from Methane: Construction of Glycogen-Deficient Mutants of Methylococcus capsulatus MIR
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains Used in This Study
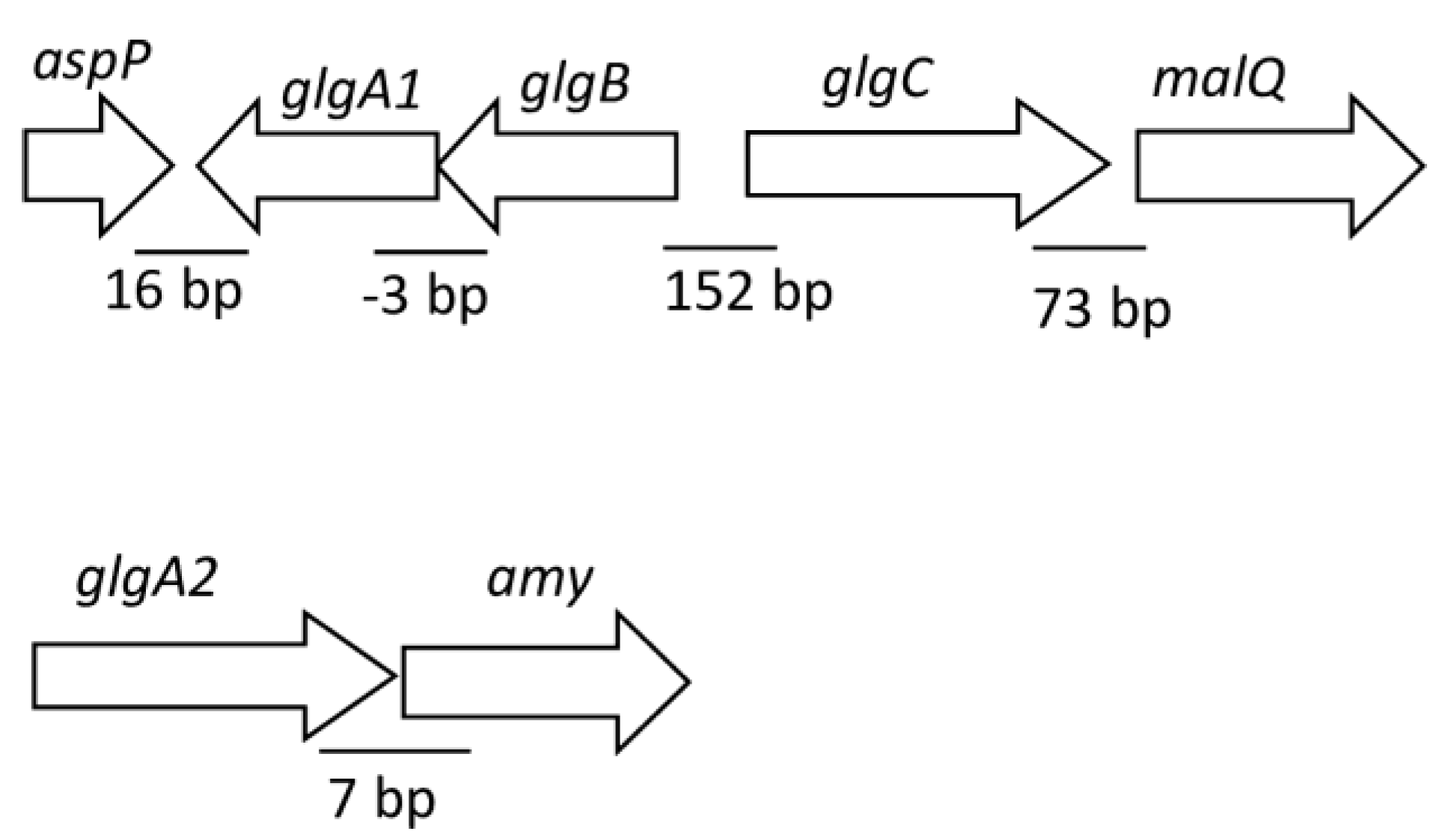
2.2. Construction of Mutant Strains Defective in Glycogen Biosynthesis
2.3. Glycogen and Glucose Extraction and Quantification
2.4. Protein Content Assay
2.5. Electron Microscopy
2.6. RNA Isolation and Real-Time PCR
2.7. Cultivation in a Bioreactor
3. Results
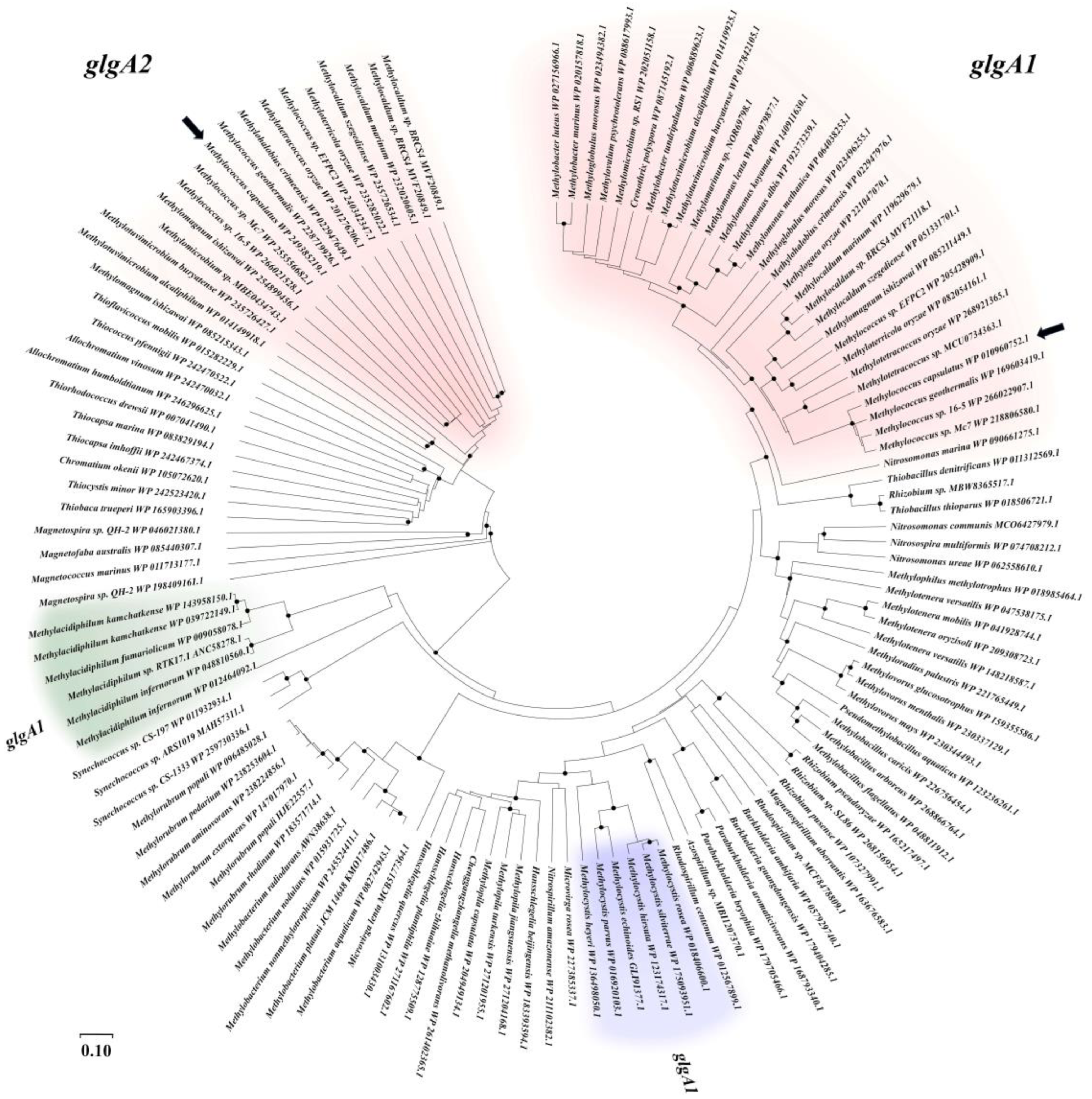
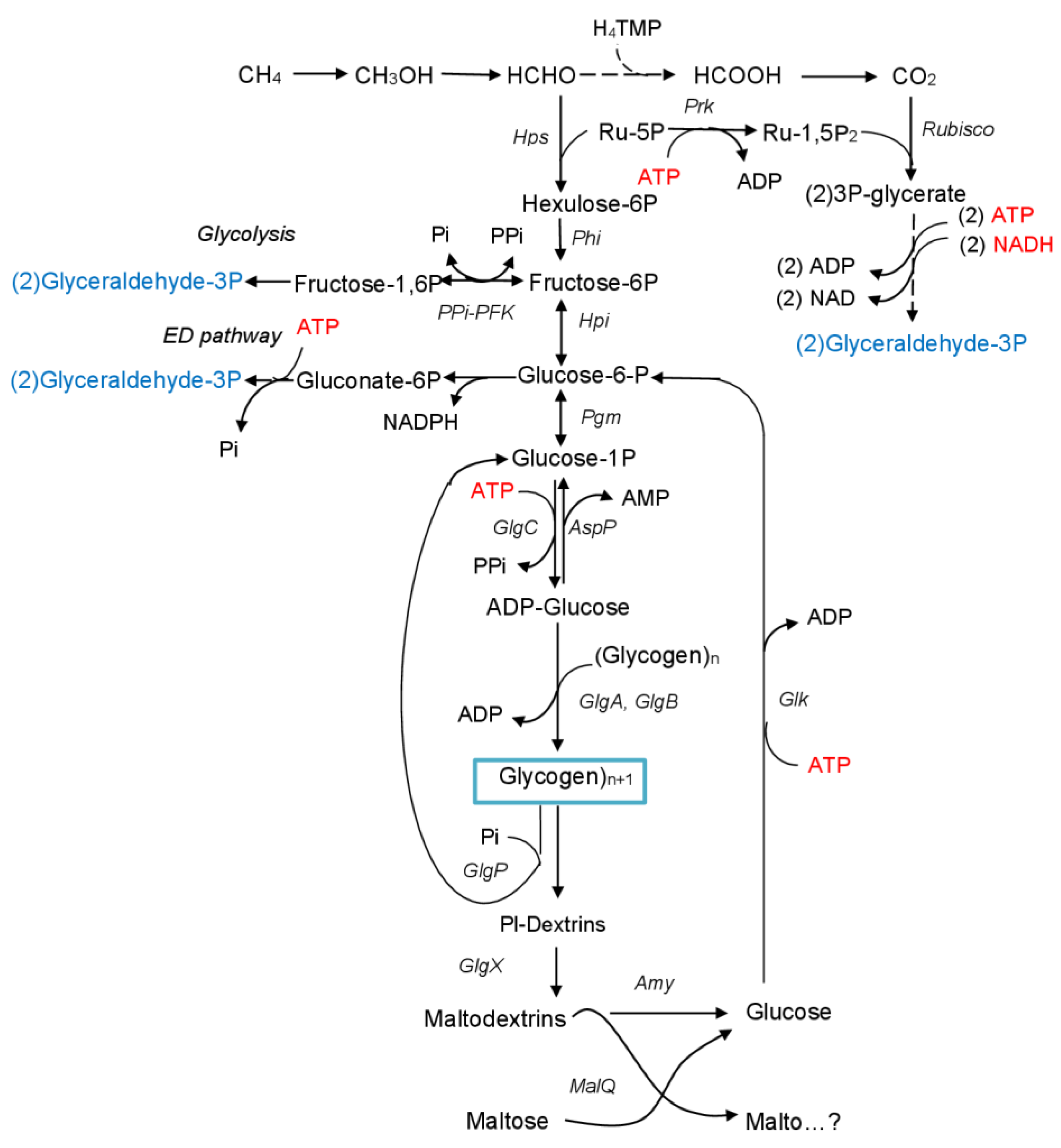
3.1. Identification of Glycogen Synthase Genes in the Genome of Mc. capsulatus MIR
3.2. Construction of Mutant Strains and Phenotypic Characterization
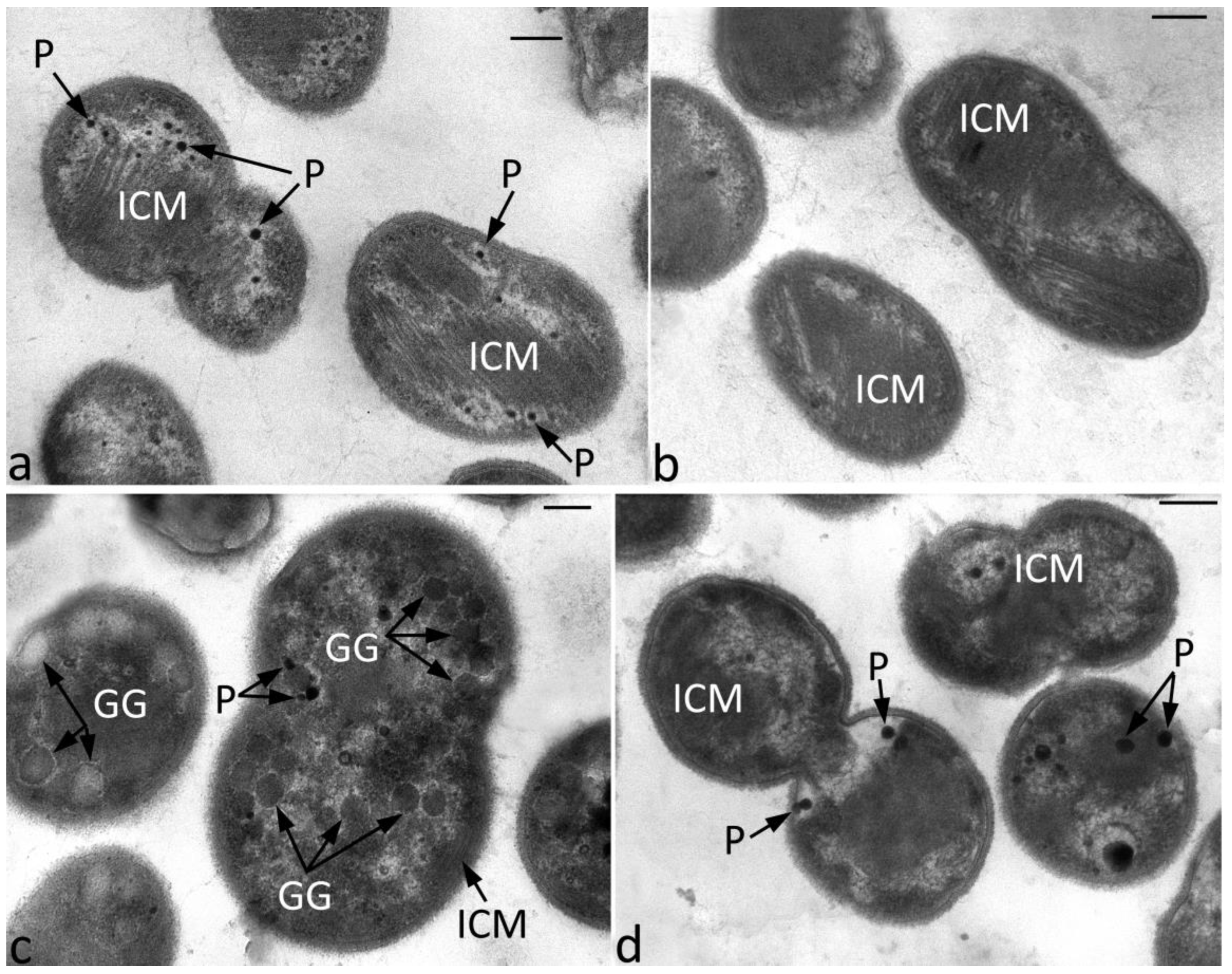
3.3. Cell Ultrastructure of WT and Mutant Strains under Nitrogen-Limited and Nitrogen-Sufficient Growth Conditions
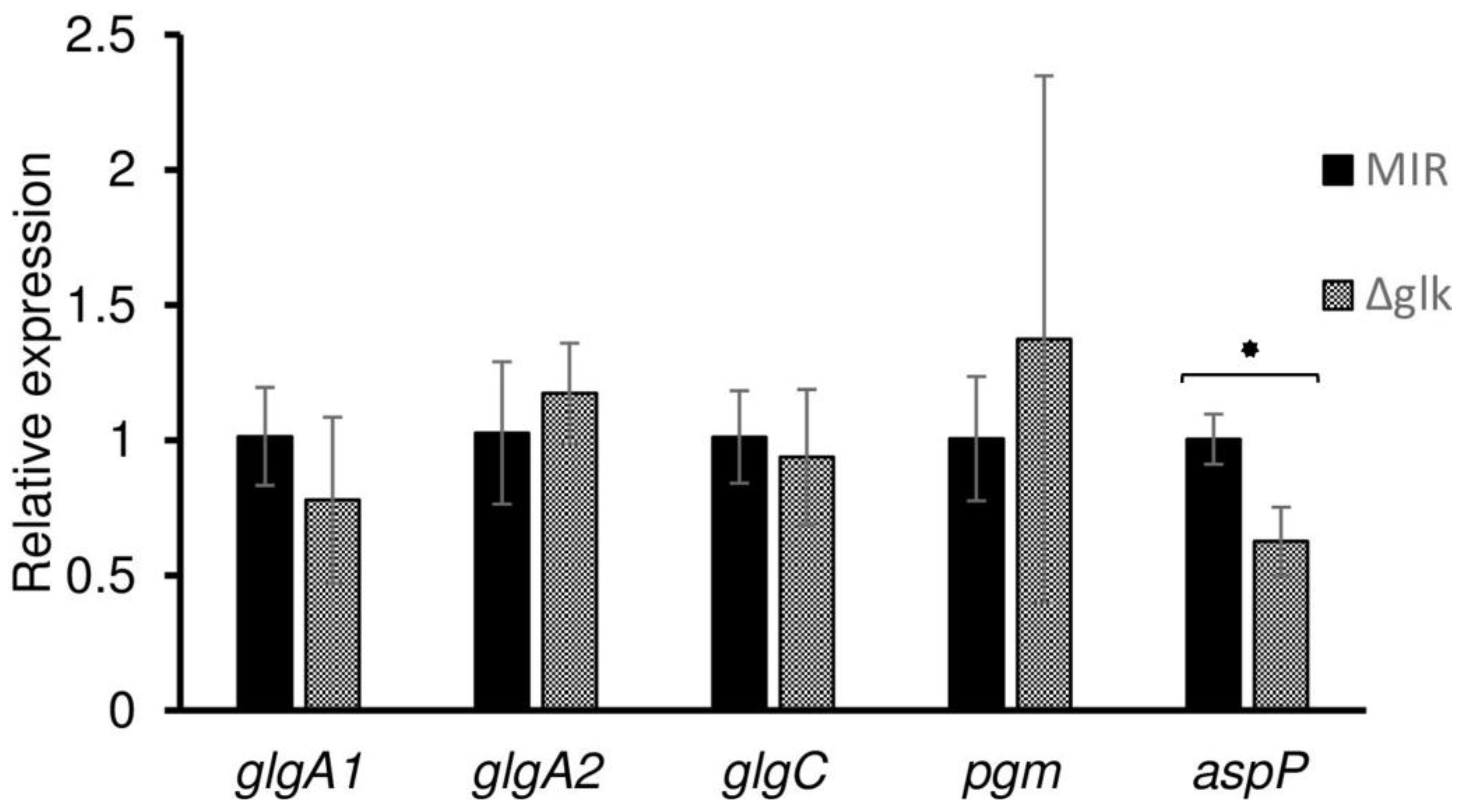
3.4. Real-Time PCR Analysis of the Genes Responsible for Glycogen Synthesis
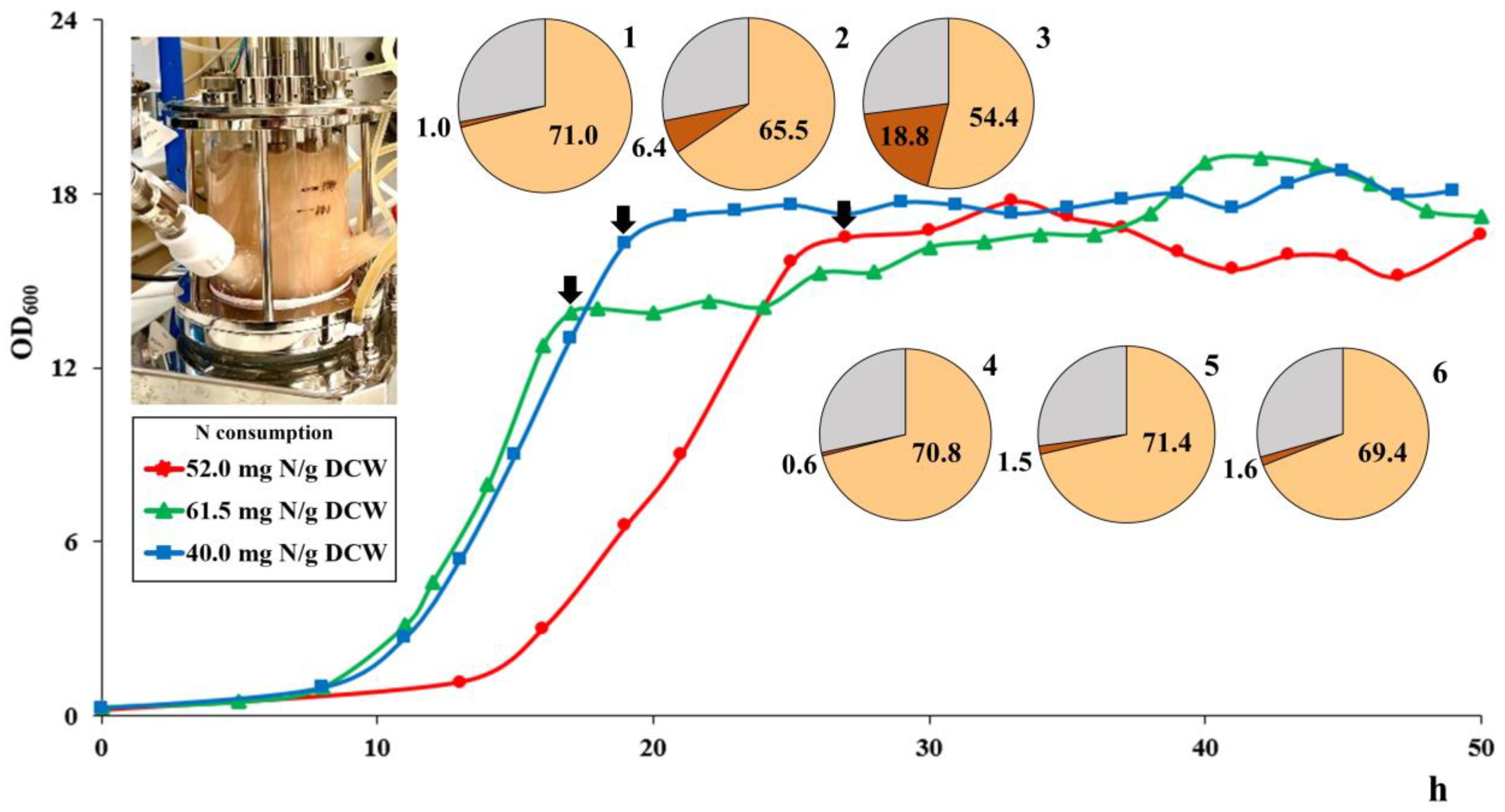
3.5. Growth Experiments in a Bioreactor
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ritala, A.; Häkkinen, S.T.; Toivari, M.; Wiebe, M.G. Single Cell Protein-State-of-the-Art, Industrial Landscape and Patents 2001–2016. Front. Microbiol. 2017, 8, 2009. [Google Scholar] [CrossRef] [PubMed]
- García Martínez, J.B.; Pearce, J.M.; Throup, J.; Cates, J.; Lackner, M.; Denkenberger, D.C. Methane Single Cell Protein: Potential to Secure a Global Protein Supply Against Catastrophic Food Shocks. Front. Bioeng. Biotechnol. 2022, 10, 906704. [Google Scholar] [CrossRef] [PubMed]
- Matassa, S.; Papirio, S.; Pikaar, I.; Hülsen, T.; Leijenhorst, E.; Esposito, G.; Pirozzi, F.; Verstraete, W. Upcycling of Biowaste Carbon and Nutrients in Line with Consumer Confidence: The “Full Gas” Route to Single Cell Protein. Green Chem. 2020, 22, 4912–4929. [Google Scholar] [CrossRef]
- Risso, C.; Choudhary, S.; Johannessen, A.; Silverman, J. Methanotrophy Goes Commercial: Challenges, Opportunities, and Brief History BT. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 293–298. ISBN 978-3-319-74866-5. [Google Scholar]
- Henard, C.A.; Smith, H.; Dowe, N.; Kalyuzhnaya, M.G.; Pienkos, P.T.; Guarnieri, M.T. Bioconversion of Methane to Lactate by an Obligate Methanotrophic Bacterium. Sci. Rep. 2016, 6, 21585. [Google Scholar] [CrossRef]
- Strong, P.J.; Kalyuzhnaya, M.; Silverman, J.; Clarke, W.P. A Methanotroph-Based Biorefinery: Potential Scenarios for Generating Multiple Products from a Single Fermentation. Bioresour. Technol. 2016, 215, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Strong, P.J.; Xie, S.; Clarke, W.P. Methane as a Resource: Can the Methanotrophs Add Value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef]
- Le, H.T.Q.; Lee, E.Y. Methanotrophs: Metabolic Versatility from Utilization of Methane to Multi-Carbon Sources and Perspectives on Current and Future Applications. Bioresour. Technol. 2023, 384, 129296. [Google Scholar] [CrossRef]
- Mühlemeier, I.M.; Speight, R.; Strong, P.J. Biogas, Bioreactors and Bacterial Methane Oxidation BT. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 213–235. ISBN 978-3-319-74866-5. [Google Scholar]
- Hanson, R.; Hanson, T. Methanotrophic Bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef]
- Hakemian, A.S.; Rosenzweig, A.C. The Biochemistry of Methane Oxidation. Annu. Rev. Biochem. 2007, 76, 223–241. [Google Scholar] [CrossRef]
- Rosenzweig, A.C. Biochemistry: Breaking Methane. Nature 2015, 518, 309–310. [Google Scholar] [CrossRef]
- Hamer, G.; Harrison, D.E.F. Single Cell Protein: The Technology, Economics and Future Potential. In Hydrocarbons in Biotechnology; Harrison, D.E.F., Higgins, I.J., Watkinson, W., Eds.; Heyden: London, UK, 1980; pp. 59–73. ISBN 9789811305016. [Google Scholar]
- Zhivotchenko, A.G.; Nikonova, E.S.; Jørgensen, M.H. Copper Effect on the Growth Kinetics of Methylococcus capsulatus (Bath). Biotechnol Technol. 1995, 9, 163–168. [Google Scholar] [CrossRef]
- Øverland, M.; Tauson, A.H.; Shearer, K.; Skrede, A. Evaluation of Methane-Utilising Bacteria Products as Feed Ingredients for Monogastric Animals. Arch. Anim. Nutr. 2010, 64, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.J.; Aufderheide, B.; Ramjattan, D.M.; Dass, R. Enhanced Production of Single Cell Protein from M. capsulatus (Bath) Growing in Mixed Culture. J. Microbiol. Biotechnol. Food Sci. 2016, 6, 894–899. [Google Scholar] [CrossRef]
- Linton, J.D.; Cripps, R.E. The Occurrence and Identification of Intracellular Polyglucose Storage Granules in Methylococcus NCIB 11083 Grown in Chemostat Culture on Methane. Arch. Microbiol. 1978, 117, 41–48. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Kalyuzhnaya, M.G.; Sakharovsky, V.G.; Snzina, N.E.; Trotsenko, Y.A.; Gottschalk, G. Osmoadaptation in Halophilic and Alkaliphilic Methanotrophs. Arch. Microbiol. 1999, 172, 321–329. [Google Scholar] [CrossRef] [PubMed]
- But, S.Y.; Dedysh, S.N.; Popov, V.O.; Pimenov, N.V.; Khmelenina, V.N. Construction of a Type-I Metanotroph with Reduced Capacity for Glycogen and Sucrose Accumulation. Appl. Biochem. Microbiol. 2020, 56, 538–543. [Google Scholar] [CrossRef]
- Ballicora, M.A.; Iglesias, A.A.; Preiss, J. ADP-Glucose Pyrophosphorylase, a Regulatory Enzyme for Bacterial Glycogen Synthesis. Microbiol. Mol. Biol. Rev. 2003, 67, 213–225. [Google Scholar] [CrossRef]
- Preiss, J.; Romeo, T. Molecular Biology and Regulatory Aspects of Glycogen Biosynthesis in Bacteria. Prog. Nucleic Acid Res. Mol. Biol. 1994, 47, 299–329. [Google Scholar] [CrossRef]
- Morán-Zorzano, M.T.; Alonso-Casajús, N.; Muñoz, F.J.; Viale, A.M.; Baroja-Fernández, E.; Eydallin, G.; Pozueta-Romero, J. Occurrence of More than One Important Source of ADPglucose Linked to Glycogen Biosynthesis in Escherichia Coli and Salmonella. FEBS Lett. 2007, 581, 4423–4429. [Google Scholar] [CrossRef]
- Morán-Zorzano, M.T.; Viale, A.M.; Muñoz, F.J.; Alonso-Casajús, N.; Eydallín, G.G.; Zugasti, B.; Baroja-Fernández, E.; Pozueta-Romero, J. Escherichia Coli AspP Activity Is Enhanced by Macromolecular Crowding and by Both Glucose-1,6-Bisphosphate and Nucleotide-Sugars. FEBS Lett. 2007, 581, 1035–1040. [Google Scholar] [CrossRef]
- Eydallin, G.; Morán-Zorzano, M.T.; Muñoz, F.J.; Baroja-Fernández, E.; Montero, M.; Alonso-Casajús, N.; Viale, A.M.; Pozueta-Romero, J. An Escherichia Coli Mutant Producing a Truncated Inactive Form of GlgC Synthesizes Glycogen: Further Evidences for the Occurrence of Various Important Sources of ADPglucose in Enterobacteria. FEBS Lett. 2007, 581, 4417–4422. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, X.; Zhu, P.; Cheng, M.; Hong, Q.; Yan, X. PheSAG Based Rapid and Efficient Markerless Mutagenesis in Methylotuvimicrobium. Front. Microbiol. 2020, 11, 441. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Ren, J.; Yu, M.S.; Kim, H.; Kim, W.Y.; Shen, J.; Yoo, S.M.; Eyun, S.I.; Na, D. Construction of a Tunable Promoter Library to Optimize Gene Expression in Methylomonas sp. DH-1, a Methanotroph, and Its Application to Cadaverine Production. Biotechnol. Biofuels 2021, 14, 228. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Kumaresan, D.; Heimann, K.; Caetano, N.S.; Visvanathan, C.; Parthiba Karthikeyan, O. Editorial: Methane: A Bioresource for Fuel and Biomolecules. Front. Environ. Sci. 2020, 8, 9. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Hwang, I.Y.; Lee, O.K.; Kim, D.; Kalyuzhnaya, M.G.; Mariyana, R.; Hadiyati, S.; Kim, M.S.; Lee, E.Y. Systematic Metabolic Engineering of Methylomicrobium alcaliphilum 20Z for 2,3-Butanediol Production from Methane. Metab. Eng. 2018, 47, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.N.; Lee, O.K.; Hadiyati, S.; Affifah, A.N.; Kim, M.S.; Lee, E.Y. Metabolic Engineering of the Type I Methanotroph Methylomonas sp. DH-1 for Production of Succinate from Methane. Metab. Eng. 2019, 54, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Oshkin, I.Y.; Suleimanov, R.Z.; Khmelenina, V.N.; Mardanov, A.V.; Pimenov, N.V.; Dedysh, S.N. Complete Genome Sequence of Methylococcus capsulatus MIR, a Methanotroph Capable of Growth on Methanol. Microbiol. Resour. Announc. 2022, 11, 97–98. [Google Scholar] [CrossRef] [PubMed]
- Mamiatis, T.; Fritsch, E.F.; Sambrook, J.; Engel, J. Molecular Cloning–A Laboratory Manual. New York: Cold Spring Harbor Laboratory. 1982, 545 S., 42 $. Acta Biotechnol. 1985, 5, 104. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small Mobilizable Multi-Purpose Cloning Vectors Derived from The. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Rozova, O.N.; Ekimova, G.A.; Molochkov, N.V.; Reshetnikov, A.S.; Khmelenina, V.N.; Mustakhimov, I.I. Enzymes of an Alternative Pathway of Glucose Metabolism in Obligate Methanotrophs. Sci. Rep. 2021, 11, 8795. [Google Scholar] [CrossRef]
- Yee, Y.C.; Hashim, R.; Mohd Yahya, A.R.; Bustami, Y. Colorimetric Analysis of Glucose Oxidase-magnetic Cellulose Nanocrystals (CNCS) for Glucose Detection. Sensors 2019, 19, 2511. [Google Scholar] [CrossRef]
- Schacterle, G.R.; Pollack, R.L. A Simplified Method for the Quantitative Assay of Small Amounts of Protein in Biologic Material. Anal. Biochem. 1973, 51, 654–655. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E.S. The Use of Lead Citrate at High PH as an Electron-Opaque Stain in Electron Microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Bessman, M.J.; Frick, D.N.; O’Handley, S.F. The MutT Proteins or “Nudix” Hydrolases, a Family of Versatile, Widely Distributed, “Housecleaning” Enzymes. J. Biol. Chem. 1996, 271, 25059–25062. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Bruna, B.; Baroja-Fernández, E.; Muñoz, F.J.; Bastarrica-Berasategui, A.; Zandueta-Criado, A.; Rodríguez-López, M.; Lasa, I.; Akazawa, T.; Pozueta-Romero, J. Adenosine Diphosphate Sugar Pyrophosphatase Prevents Glycogen Biosynthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2001, 98, 8128–8132. [Google Scholar] [CrossRef] [PubMed]
- Mustakhimov, I.I.; Rozova, O.N.; Solntseva, N.P.; Khmelenina, V.N.; Reshetnikov, A.S.; Trotsenko, Y.A. The Properties and Potential Metabolic Role of Glucokinase in Halotolerant Obligate Methanotroph Methylomicrobium alcaliphilum 20Z. Antonie Van Leeuwenhoek 2017, 110, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tiago Guerra, L.; Li, Z.; Ludwig, M.; Charles Dismukes, G.; Bryant, D.A. Altered Carbohydrate Metabolism in Glycogen Synthase Mutants of Synechococcus sp. Strain PCC 7002: Cell Factories for Soluble Sugars. Metab. Eng. 2013, 16, 56–67. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Gayazov, R.R.; Suzina, N.E.; Doronina, N.V.; Mshenskii, Y.N.; Trotsenko, Y.A. The Synthesis of Polysaccharides by Methylococcus capsulatus under Various Conditions of Cultivation. Mikrobiologiya 1992, 61, 404–410. [Google Scholar]
- Hernández, M.A.; Alvarez, H.M. Glycogen Formation by Rhodococcus Species and the Effect of Inhibition of Lipid Biosynthesis on Glycogen Accumulation in Rhodococcus opacus PD630. FEMS Microbiol. Lett. 2010, 312, 93–99. [Google Scholar] [CrossRef]
- Palmer, T.N.; Wöber, G.; Whelan, W.J. The Pathway of Exogenous and Endogenous Carbohydrate Utilization in Escherichia coli: A Dual Function for the Enzymes of the Maltose Operon. Eur. J. Biochem. 1973, 39, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Park, J.T.; Shim, J.H.; Tran, P.L.; Hong, I.H.; Yong, H.U.; Oktavina, E.F.; Nguyen, H.D.; Kim, J.W.; Lee, T.S.; Park, S.H.; et al. Role of Maltose Enzymes in Glycogen Synthesis by Escherichia coli. J. Bacteriol. 2011, 193, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B. The Energy Spilling Reactions of Bacteria and Other Organisms. J. Mol. Microbiol. Biotechnol. 2007, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Yoshikawa, K.; Toya, Y.; Matsuda, F.; Shimizu, H. Metabolic Flux Analysis of the Synechocystis sp. PCC 6803 ΔnrtABCD Mutant Reveals a Mechanism for Metabolic Adaptation to Nitrogen-Limited Conditions. Plant Cell Physiol. 2017, 58, 537–545. [Google Scholar] [CrossRef]
- Lengsfeld, C.; Schönert, S.; Dippel, R.; Boos, W. Glucose- and Glucokinase-Controlled Mal Gene Expression in Escherichia coli. J. Bacteriol. 2009, 191, 701–712. [Google Scholar] [CrossRef]

| Name | Primer (5′-3′) | Target |
|---|---|---|
| Primers for cloning | ||
| MIR glg1-up-F | ATATCTAGAACCGGCATTACCATCACGA | Homology region (HR) upstream of glgA1 gene |
| MIR glg1-up-R | CAAGCATGCAGGAGTGGCGGACGGTGCGA | |
| MIR glg1-dw-F | CAAGCATGCGCTGCTCCATCGCCGAC | HR downstream of glgA1 gene |
| MIR glg1-dw-R | TCAAAGCTTGCCAAGGAGATCGTGAATTA | |
| MIR glg2-up-F | AGTGAATTCGACCACGACCAGGCCGAGCA | HR upstream of glgA2 gene |
| MIR glg2-up-R | TCAGGTACCTCCAGTACCGCCGACACCTA | |
| MIR glg2-dw-F | CAAGCATGCCGCTACGACTATTCCTGGA | HR downstream of glgA2 gene |
| MIR glg2-dw-R | CTTAAGCTTTGAGCCTGGGCGTGTCGTG | |
| 20Z-glg2C-F | AGTGAATTCGGAGGAGACACATGGCAAAGCAAACTACAAC | glgA2 gene from Mm. alcaliphilum 20Z |
| 20Z-glg2C-R | CAAGGATCCTTATTTGTTACGAATATAGTCATAGAT | |
| GLK_MIR-F | TTGGATCCTGATCGGCGGCTATCGCAT | glk gene |
| GLK_MIR-R | TTCGATCGTCCTGCGGCTTTTCGTAGT | |
| Primers for real-time PCR | ||
| MIR qrpoB-F | CTGGATGCCCTGGTGGAAAT | rpoB gene |
| MIR qrpoB-R | ATTCTCCACCATCTCCCCCA | |
| q-pgmMIR-F | CTACGCAAGAAGGTGAAGGTT | pgm gene |
| q-pgmMIR-R | TGTTTACGGATTACGCAGGA | |
| q-glgCMIR-F | ACTTCCCGCTGTCCAACTG | glgC gene |
| q-glgCMIR-R | CCAAGTTCTGATACACCGCAT | |
| q-glgA1MIR-F | TACCCCTCTGGCTGCTGGA | glgA1 gene |
| q-glgA1MIR-R | TTCGGCTCGTCGCTCAAAA | |
| q-glgA2MIR-F | GCCAAGAGCCCCCCAGT | glgA2 gene |
| Growth Conditions | Strain | Glycogen, mg/g DCW | Glucose, µg/g DCW | Protein, mg/g DCW |
|---|---|---|---|---|
| Nitrogen excess (1 g L−1 KNO3) | WT | 14.3 ± 3.8 | 32 ± 3 | 496 ± 15 |
| ΔglgA1 | 9.4 ± 2.5 | 26 ± 3 | 420 ±22 | |
| ΔglgA2 | 17.6 ± 3.2 | 28 ± 1 | 420 ± 17 | |
| ΔglgA1ΔglgA2 | 1.1 ± 0.4 | 35 ± 2 | 477 ± 16 | |
| Δglk | 1.3 ± 0.1 | 75 ± 19 | 498 ± 27 | |
| Nitrogen limit (0.29 g L−1 KNO3) | WT | 202 ± 54 | nd | 297 ± 63 |
| ΔglgA1 | 61.0 ± 9 | nd | 510 ± 24 | |
| ΔglgA2 | 21.0 ± 3 | nd | 511 ± 52 | |
| ΔglgA1ΔglgA2 | 3.0 ± 0.4 | nd | 532 ± 13 | |
| Δglk | 142.0 ± 5 | nd | 482 ± 36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
But, S.Y.; Suleimanov, R.Z.; Oshkin, I.Y.; Rozova, O.N.; Mustakhimov, I.I.; Pimenov, N.V.; Dedysh, S.N.; Khmelenina, V.N. New Solutions in Single-Cell Protein Production from Methane: Construction of Glycogen-Deficient Mutants of Methylococcus capsulatus MIR. Fermentation 2024, 10, 265. https://doi.org/10.3390/fermentation10050265
But SY, Suleimanov RZ, Oshkin IY, Rozova ON, Mustakhimov II, Pimenov NV, Dedysh SN, Khmelenina VN. New Solutions in Single-Cell Protein Production from Methane: Construction of Glycogen-Deficient Mutants of Methylococcus capsulatus MIR. Fermentation. 2024; 10(5):265. https://doi.org/10.3390/fermentation10050265
Chicago/Turabian StyleBut, Sergey Y., Ruslan Z. Suleimanov, Igor Y. Oshkin, Olga N. Rozova, Ildar I. Mustakhimov, Nikolai V. Pimenov, Svetlana N. Dedysh, and Valentina N. Khmelenina. 2024. "New Solutions in Single-Cell Protein Production from Methane: Construction of Glycogen-Deficient Mutants of Methylococcus capsulatus MIR" Fermentation 10, no. 5: 265. https://doi.org/10.3390/fermentation10050265
APA StyleBut, S. Y., Suleimanov, R. Z., Oshkin, I. Y., Rozova, O. N., Mustakhimov, I. I., Pimenov, N. V., Dedysh, S. N., & Khmelenina, V. N. (2024). New Solutions in Single-Cell Protein Production from Methane: Construction of Glycogen-Deficient Mutants of Methylococcus capsulatus MIR. Fermentation, 10(5), 265. https://doi.org/10.3390/fermentation10050265






