Microbial Characterisation of a Two-Stage Anaerobic Digestion Process for Conversion of Agri-Based Feedstock in Biogas and Long-Chain Fatty Acids in a Circular Economy Framework
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reactor Set-Up
2.2. Sampling, Biomass Characterisation and Process Analyses
2.3. Microbial Communities’ Characterisation
2.3.1. DNA Extraction and Amplicon Sequencing
2.3.2. Bioinformatic and Statistical Analyses
3. Results and Discussion
3.1. Process Performance
3.2. Microbial Characterisation
3.2.1. Microbial Diversity
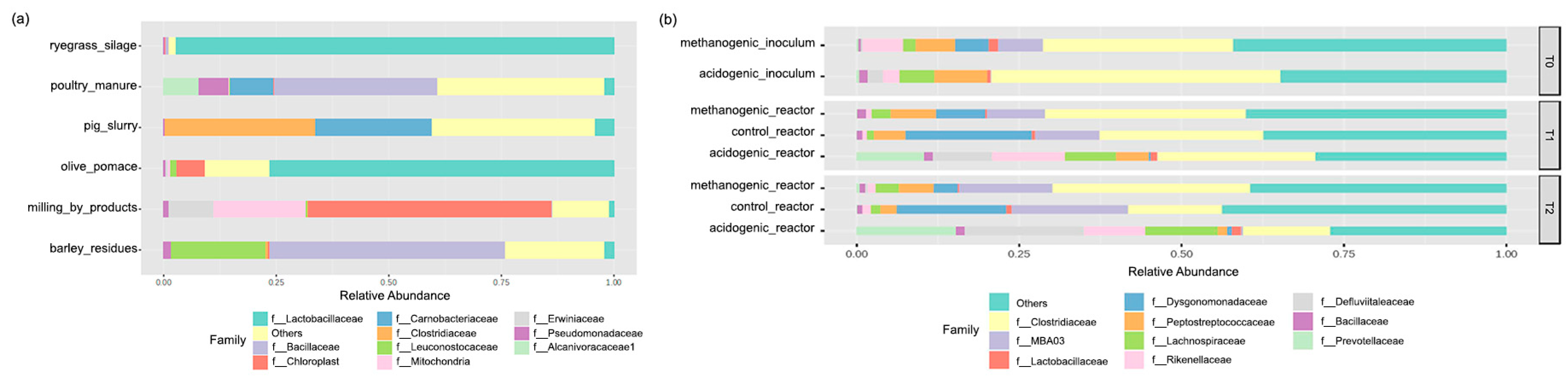
3.2.2. Feedstock and Inoculum Effects on the Dominant Reactors’ Steady-State Microbial Families
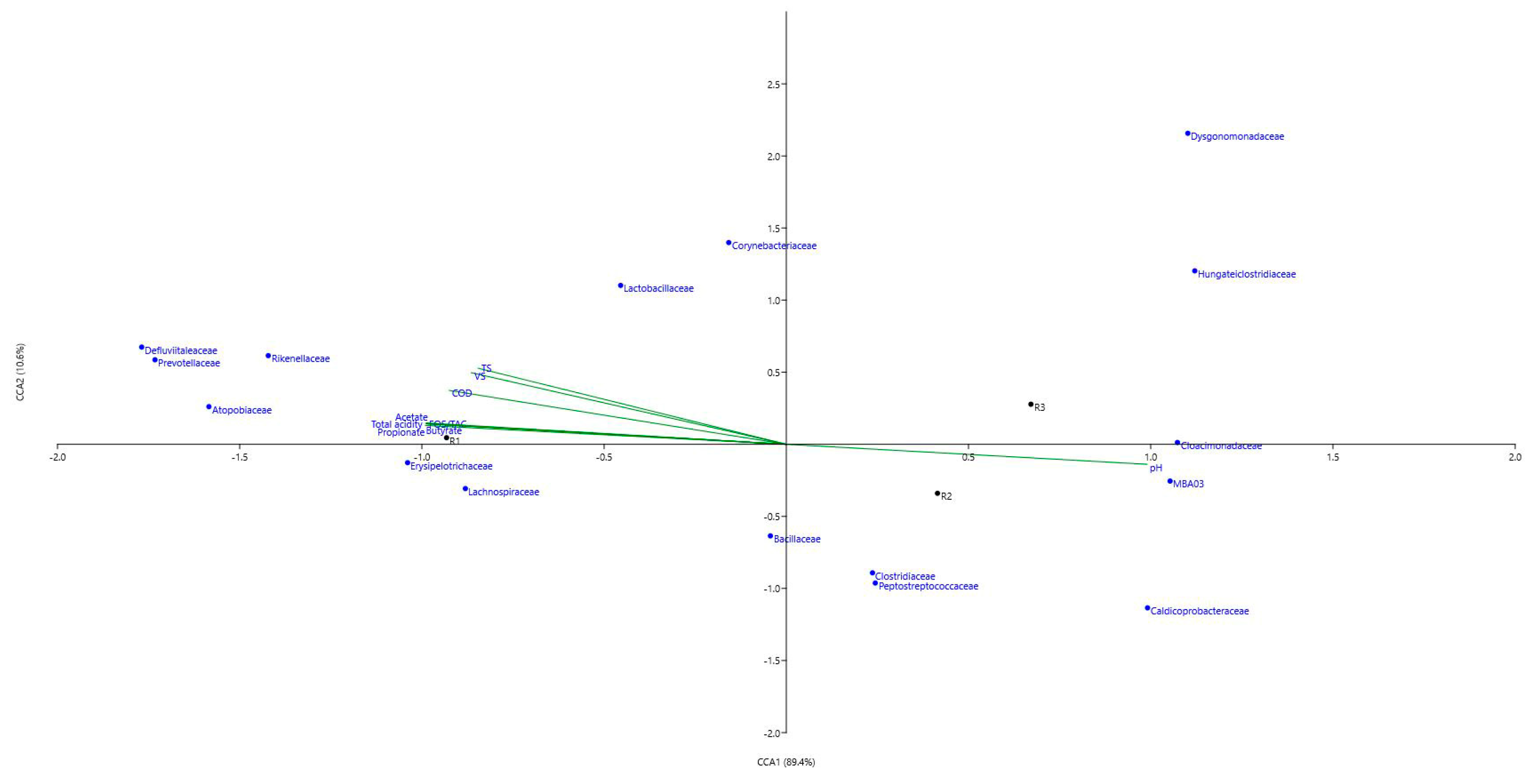
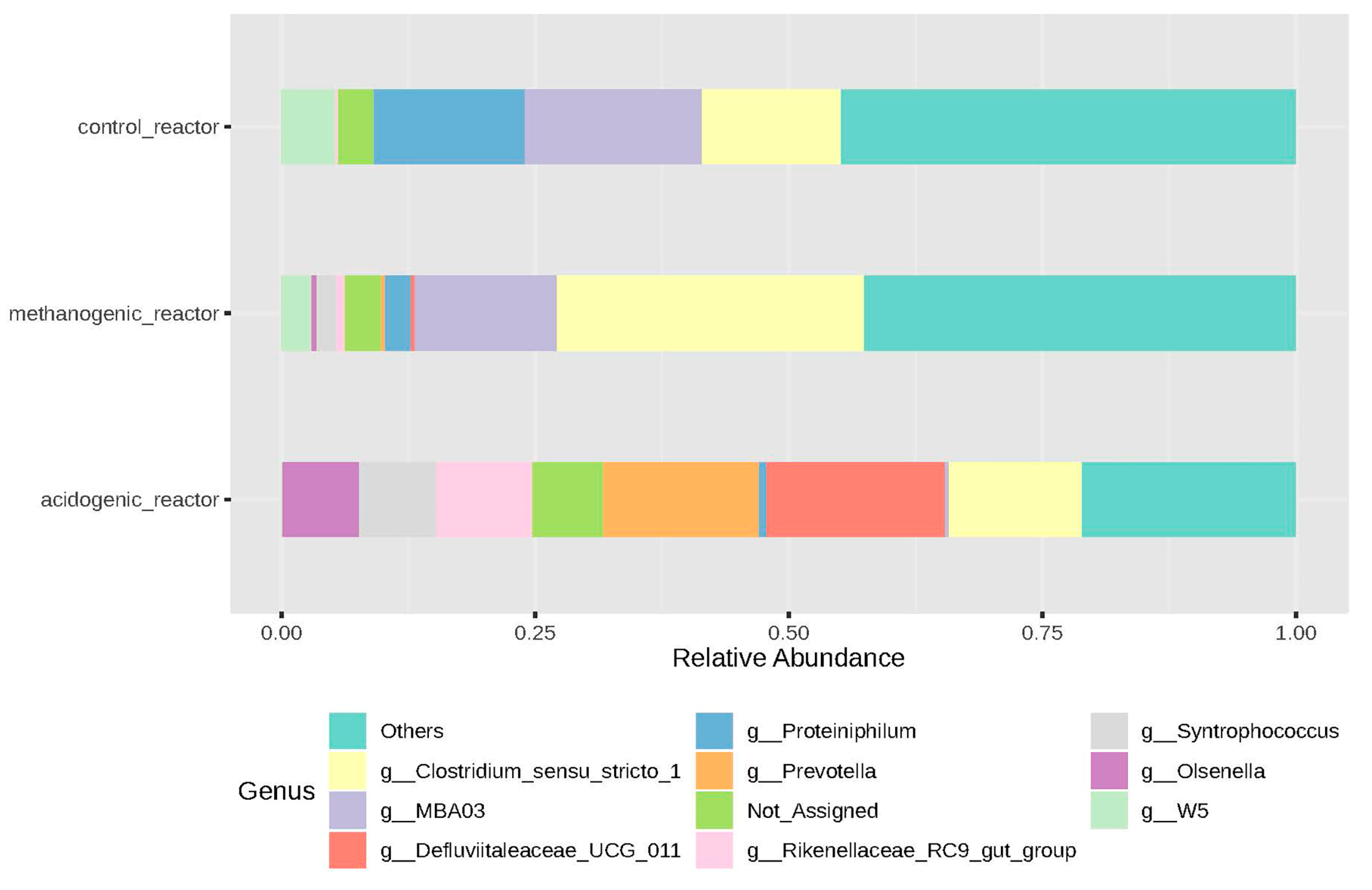
3.2.3. Microbial Genera and LCFAs Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yaashikaa, P.R.; Senthil Kumar, P.; Varjani, S. Valorization of Agro-Industrial Wastes for Biorefinery Process and Circular Bioeconomy: A Critical Review. Bioresour. Technol. 2022, 343, 126126. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, N.; Gurunathan, B.; Han, J.; Krishna, S.; Ananth, A.; Venugopal, K.; Sherly Priyanka, R.B. Recent Advances in Valorization of Organic Municipal Waste into Energy Using Biorefinery Approach, Environment and Economic Analysis. Bioresour. Technol. 2021, 337, 125498. [Google Scholar] [CrossRef] [PubMed]
- Adekunle, K.F.; Okolie, J.A. A Review of Biochemical Process of Anaerobic Digestion. Adv. Biosci. Biotechnol. 2015, 6, 205–212. [Google Scholar] [CrossRef]
- Cysneiros, D.; Banks, C.J.; Heaven, S.; Karatzas, K.-A.G. The Effect of pH Control and ‘Hydraulic Flush’ on Hydrolysis and Volatile Fatty Acids (VFA) Production and Profile in Anaerobic Leach Bed Reactors Digesting a High Solids Content Substrate. Bioresour. Technol. 2012, 123, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Dareioti, M.A.; Kornaros, M. Effect of Hydraulic Retention Time (HRT) on the Anaerobic Co-Digestion of Agro-Industrial Wastes in a Two-Stage CSTR System. Bioresour. Technol. 2014, 167, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Schievano, A.; Tenca, A.; Lonati, S.; Manzini, E.; Adani, F. Can Two-Stage Instead of One-Stage Anaerobic Digestion Really Increase Energy Recovery from Biomass? Appl. Energy 2014, 124, 335–342. [Google Scholar] [CrossRef]
- Demirel, B.; Yenigün, O. Two-phase Anaerobic Digestion Processes: A Review. J. Chem. Technol. Biotechnol. 2002, 77, 743–755. Available online: https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/full/10.1002/jctb.630 (accessed on 23 April 2024). [CrossRef]
- Kumar, G.; Shobana, S.; Nagarajan, D.; Lee, D.-J.; Lee, K.-S.; Lin, C.-Y.; Chen, C.-Y.; Chang, J.-S. Biomass Based Hydrogen Production by Dark Fermentation-Recent Trends and Opportunities for Greener Processes. Curr. Opin. Biotechnol. 2018, 50, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.; Jones, R.J.; Oram, L.; Massanet-Nicolau, J.; Guwy, A. Intensification of Acidogenic Fermentation for the Production of Biohydrogen and Volatile Fatty Acids—A Perspective. Fermentation 2022, 8, 325. [Google Scholar] [CrossRef]
- Elsamadony, M.; Mostafa, A.; Fujii, M.; Tawfik, A.; Pant, D. Advances towards Understanding Long Chain Fatty Acids-Induced Inhibition and Overcoming Strategies for Efficient Anaerobic Digestion Process. Water Res. 2021, 190, 116732. [Google Scholar] [CrossRef]
- Ziels, R.M.; Karlsson, A.; Beck, D.A.C.; Ejlertsson, J.; Yekta, S.S.; Bjorn, A.; Stensel, H.D.; Svensson, B.H. Microbial Community Adaptation Influences Long-Chain Fatty Acid Conversion during Anaerobic Codigestion of Fats, Oils, and Grease with Municipal Sludge. Water Res. 2016, 103, 372–382. [Google Scholar] [CrossRef]
- Diamantis, V.; Eftaxias, A.; Stamatelatou, K.; Noutsopoulos, C.; Vlachokostas, C.; Aivasidis, A. Bioenergy in the Era of Circular Economy: Anaerobic Digestion Technological Solutions to Produce Biogas from Lipid-Rich Wastes. Renew. Energy 2021, 168, 438–447. [Google Scholar] [CrossRef]
- Neves, L.; Pereira, M.A.; Mota, M.; Alves, M.M. Detection and Quantification of Long Chain Fatty Acids in Liquid and Solid Samples and Its Relevance to Understand Anaerobic Digestion of Lipids. Bioresour Technol. 2009, 100, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Parra-Orobio, B.A.; Cruz-Bournazou, M.N.; Torres-Lozada, P. Single-Stage and Two-Stage Anaerobic Digestion of Food Waste: Effect of the Organic Loading Rate on the Methane Production and Volatile Fatty Acids. Water Air Soil Pollut. 2021, 232, 105. [Google Scholar] [CrossRef]
- Yan, W.; Vadivelu, V.; Maspolim, Y.; Zhou, Y. In-Situ Alkaline Enhanced Two-Stage Anaerobic Digestion System for Waste Cooking Oil and Sewage Sludge Co-Digestion. Waste Manag. 2021, 120, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.V.; Salvador, R.; de Francisco, A.C.; Piekarski, C.M. Mapping of Research Lines on Circular Economy Practices in Agriculture: From Waste to Energy. Renew. Sustain. Energy Rev. 2020, 131, 109958. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association/American Water Works Association/Water Environment Federation: Washington, DC, USA, 2005. [Google Scholar]
- Nordmann, W. Die Überwachung Der Schlammfaulung. KA-Informationen Für Das Betriebspersonal. Beil. Zur Korresp. Abwasser 1977, 3, 77. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Rocchetti, G.; Ghilardelli, F.; Carboni, E.; Atzori, A.S.; Masoero, F.; Gallo, A. Milk Metabolome Reveals Pyrimidine and Its Degradation Products as the Discriminant Markers of Different Corn Silage-Based Nutritional Strategies. J. Dairy Sci. 2022, 105, 8650–8663. [Google Scholar] [CrossRef] [PubMed]
- Baldi, F.; Pecorini, I.; Iannelli, R. Comparison of Single-Stage and Two-Stage Anaerobic Co-Digestion of Food Waste and Activated Sludge for Hydrogen and Methane Production. Renew. Energy 2019, 143, 1755–1765. [Google Scholar] [CrossRef]
- Schievano, A.; Tenca, A.; Scaglia, B.; Merlino, G.; Rizzi, A.; Daffonchio, D.; Oberti, R.; Adani, F. Two-Stage vs Single-Stage Thermophilic Anaerobic Digestion: Comparison of Energy Production and Biodegradation Efficiencies. Environ. Sci. Technol. 2012, 46, 8502–8510. [Google Scholar] [CrossRef]
- Garuti, M.; Sinisgalli, E.; Soldano, M.; Jimenez Rodriguez, A.; Fermoso, F.G. Biochemical Conditions for Anaerobic Digestion of Agricultural Feedstocks: A Full-Scale Study Linking Elements Concentration and Residual Methane Potential. Biomass Bioenergy 2023, 176, 106899. [Google Scholar] [CrossRef]
- Wainaina, S.; Lukitawesa; Kumar Awasthi, M.; Taherzadeh, M.J. Bioengineering of Anaerobic Digestion for Volatile Fatty Acids, Hydrogen or Methane Production: A Critical Review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef]
- Van Ginkel, S.; Logan, B.E. Inhibition of Biohydrogen Production by Undissociated Acetic and Butyric Acids. Environ. Sci. Technol. 2005, 39, 9351–9356. [Google Scholar] [CrossRef] [PubMed]
- Valentino, F.; Munarin, G.; Biasiolo, M.; Cavinato, C.; Bolzonella, D.; Pavan, P. Enhancing Volatile Fatty Acids (VFA) Production from Food Waste in a Two-Phases Pilot-Scale Anaerobic Digestion Process. J. Environ. Chem. Eng. 2021, 9, 106062. [Google Scholar] [CrossRef]
- Fu, Z.; Sun, L.; Hou, M.; Hao, J.; Lu, Q.; Liu, T.; Ren, X.; Jia, Y.; Wang, Z.; Ge, G. Effects of Different Harvest Frequencies on Microbial Community and Metabolomic Properties of Annual Ryegrass Silage. Front. Microbiol. 2022, 13, 971449. [Google Scholar] [CrossRef]
- Vaccalluzzo, A.; Celano, G.; Pino, A.; Calabrese, F.M.; Foti, P.; Caggia, C.; Randazzo, C. Metagenetic and Volatilomic Approaches to Elucidate the Effect of Lactiplantibacillus Plantarum Starter Cultures on Sicilian Table Olives. Front. Microbiol. 2022, 12, 771636. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Kougias, P.G.; Treu, L.; Kovalovszki, A.; Valle, G.; Cappa, F.; Morelli, L.; Angelidaki, I.; Campanaro, S. Microbial Activity Response to Hydrogen Injection in Thermophilic Anaerobic Digesters Revealed by Genome-Centric Metatranscriptomics. Microbiome 2018, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Khatami, K.; Atasoy, M.; Ludtke, M.; Baresel, C.; Eyice, Ö.; Cetecioglu, Z. Bioconversion of Food Waste to Volatile Fatty Acids: Impact of Microbial Community, pH and Retention Time. Chemosphere 2021, 275, 129981. [Google Scholar] [CrossRef] [PubMed]
- Merlino, G.; Rizzi, A.; Schievano, A.; Tenca, A.; Scaglia, B.; Oberti, R.; Adani, F.; Daffonchio, D. Microbial Community Structure and Dynamics in Two-Stage vs Single-Stage Thermophilic Anaerobic Digestion of Mixed Swine Slurry and Market Bio-Waste. Water Res. 2013, 47, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, X.; Yellezuome, D.; Liu, R.; Liu, X.; Sun, C.; Abd-Alla, M.; Rasmey, A.-H. Effects of Adding Thermoanaerobacterium Thermosaccharolyticum in the Hydrogen Production Stage of a Two-Stage Anaerobic Digestion System on Hydrogen-Methane Production and Microbial Communities. Fuel 2023, 342, 127831. [Google Scholar] [CrossRef]
- Perman, E.; Schnürer, A.; Björn, A.; Moestedt, J. Serial Anaerobic Digestion Improves Protein Degradation and Biogas Production from Mixed Food Waste. Biomass Bioenergy 2022, 161, 106478. [Google Scholar] [CrossRef]
- Owusu-Agyeman, I.; Plaza, E.; Cetecioglu, Z. Long-Term Alkaline Volatile Fatty Acids Production from Waste Streams: Impact of pH and Dominance of Dysgonomonadaceae. Bioresour. Technol. 2022, 346, 126621. [Google Scholar] [CrossRef]
- Suksong, W.; Kongjan, P.; Prasertsan, P.; O-Thong, S. Thermotolerant Cellulolytic Clostridiaceae and Lachnospiraceae Rich Consortium Enhanced Biogas Production from Oil Palm Empty Fruit Bunches by Solid-State Anaerobic Digestion. Bioresour Technol. 2019, 291, 121851. [Google Scholar] [CrossRef]
- Li, Y.; Xu, H.; Hua, D.; Zhao, B.; Mu, H.; Jin, F.; Meng, G.; Fang, X. Two-Phase Anaerobic Digestion of Lignocellulosic Hydrolysate: Focusing on the Acidification with Different Inoculum to Substrate Ratios and Inoculum Sources. Sci. Total Environ. 2020, 699, 134226. [Google Scholar] [CrossRef]
- Wang, C.; Zuo, J.; Chen, X.; Xing, W.; Xing, L.; Li, P.; Lu, X.; Li, C. Microbial Community Structures in an Integrated Two-Phase Anaerobic Bioreactor Fed by Fruit Vegetable Wastes and Wheat Straw. J. Environ. Sci. 2014, 26, 2484–2492. [Google Scholar] [CrossRef]
- Poszytek, K.; Pyzik, A.; Sobczak, A.; Lipinski, L.; Sklodowska, A.; Drewniak, L. The Effect of the Source of Microorganisms on Adaptation of Hydrolytic Consortia Dedicated to Anaerobic Digestion of Maize Silage. Anaerobe 2017, 46, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Micolucci, F.; Gottardo, M.; Pavan, P.; Cavinato, C.; Bolzonella, D. Pilot Scale Comparison of Single and Double-Stage Thermophilic Anaerobic Digestion of Food Waste. J. Clean. Prod. 2017, 171, 1376–1385. [Google Scholar] [CrossRef]
- Silvestre, G.; Illa, J.; Fernández, B.; Bonmatí, A. Thermophilic Anaerobic Co-Digestion of Sewage Sludge with Grease Waste: Effect of Long Chain Fatty Acids in the Methane Yield and Its Dewatering Properties. Appl. Energy 2014, 117, 87–94. [Google Scholar] [CrossRef]
- Usman, M.; Zhao, S.; Jeon, B.-H.; Salama, E.-S.; Li, X. Microbial β-Oxidation of Synthetic Long-Chain Fatty Acids to Improve Lipid Biomethanation. Water Res. 2022, 213, 118164. [Google Scholar] [CrossRef] [PubMed]
- Lalman, J.A.; Bagley, D.M. Anaerobic Degradation and Methanogenic Inhibitory Effects of Oleic and Stearic Acids. Water Res. 2001, 35, 2975–2983. [Google Scholar] [CrossRef]
- Deng, Z.; Muñoz Sierra, J.; Ferreira, A.L.M.; Cerqueda-Garcia, D.; Spanjers, H.; van Lier, J.B. Effect of Operational Parameters on the Performance of an Anaerobic Sequencing Batch Reactor (AnSBR) Treating Protein-Rich Wastewater. Environ. Sci. Ecotechnol. 2024, 17, 100296. [Google Scholar] [CrossRef] [PubMed]
- DeCola, A.C.; Toppen, L.C.; Brown, K.P.; Dadkhah, A.; Rizzo, D.M.; Ziels, R.M.; Scarborough, M.J. Microbiome Assembly and Stability during Start-up of a Full-Scale, Two-Phase Anaerobic Digester Fed Cow Manure and Mixed Organic Feedstocks. Bioresour. Technol. 2024, 394, 130247. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, J.; Liu, K.; Li, Q.; Li, W.; Fan, J.; Wang, S.; Shi, F.; Zuo, X.; Li, P. Relationship between the Effects of Heat Pre-Treatment on Anaerobic Performance of Pig Manure and the Microbial Variation within Reactors. Chem. Eng. J. 2023, 461, 141991. [Google Scholar] [CrossRef]
- Braga Nan, L.; Trably, E.; Santa-Catalina, G.; Bernet, N.; Delgenès, J.-P.; Escudié, R. Biomethanation Processes: New Insights on the Effect of a High H2 Partial Pressure on Microbial Communities. Biotechnol. Biofuels 2020, 13, 141. [Google Scholar] [CrossRef]
- Zhuravleva, E.A.; Shekhurdina, S.V.; Kotova, I.B.; Loiko, N.G.; Popova, N.M.; Kryukov, E.; Kovalev, A.A.; Kovalev, D.A.; Litti, Y.V. Effects of Various Materials Used to Promote the Direct Interspecies Electron Transfer on Anaerobic Digestion of Low-Concentration Swine Manure. Sci. Total Environ. 2022, 839, 156073. [Google Scholar] [CrossRef]


| Parameter | Inoculum | Steady State | ||||||
|---|---|---|---|---|---|---|---|---|
| T0 | T1 | T2 | ||||||
| R1 | R2/R3 | R1 | R2 | R3 | R1 | R2 | R3 | |
| TS (g/kg) | 139.8 ± 10.1 | 48.1 ± 3.2 | 102.4 ± 9.9 | 58.7 ± 4.2 | 78.3 ± 4.1 | 127.0 ± 10.2 | 61.0 ± 3.6 | 79.3 ± 6.2 |
| VS (g/kg) | 123.1 ± 9.2 | 38.0 ± 2.2 | 88.6 ± 9.2 | 45.5 ± 3.2 | 62.7 ± 3.5 | 111.5 ± 10.2 | 48.9 ± 2.6 | 65.0 ± 5.8 |
| COD (mg/kg) | 247,513.0 ± 13,336.6 | 55,075.0 ± 1423.6 | 105,821.3 ± 19,472.0 | 46,872.0 ± 13,186.8 | 56,694.3 ± 19,175.5 | 117,311.3 ± 9333.1 | 47,288.7 ± 4230.7 | 49,235.0 ± 2138.0 |
| FOS/TAC | 5.6 ± 0.7 | 0.3 ± 0.0 | 6.1 ± 0.7 | 0.2 ± 0.0 | 0.3 ± 0.0 | 5.0 ± 0.4 | 0.3 ± 0.0 | 0.3 ± 0.0 |
| pH | 5.5 ± 0.2 | 8.2 ± 0.0 | 5.4 ± 0.2 | 7.8 ± 0.0 | 7.8 ± 0.1 | 5.6 ± 0.0 | 7.7 ± 0.1 | 7.7 ± 0.1 |
| Acetate (mg/kg) | 5617.0 ± 1492.1 | 413.0 ± 223.6 | 5201.7 ± 1199.9 | 59.1 ± 51.1 | 73.7 ± 69.5 | 5080.0 ± 471.1 | 101.3 ± 92.4 | 135.0 ± 123.4 |
| Propionate (mg/kg) | 3483.0 ± 39.7 | n.d. 1 | 2904.3 ± 68.9 | n.d. | n.d. | 3140.0 ± 351.6 | n.d. | n.d. |
| Butyrate (mg/kg) | 5395.0 ± 447.7 | n.d. | 2698.3 ± 552.4 | n.d. | n.d. | 2463.0 ± 647.1 | n.d. | n.d |
| LCFA | R1 (mg/kg) | R2 (mg/kg) | R3 (mg/kg) |
|---|---|---|---|
| Ottanoic Acid (C8) | 208.5 ± 27.1 | 30.3 ± 1.5 | 27.6 ± 1.1 |
| Palmitic Acid (C16) | 2764.2 ± 420.9 | 445.7 ± 187.5 | 543.4 ± 165.1 |
| Stearic Acid (C18) | 1751.5 ± 154.1 | 153.1 ± 10.0 | 21.9 ± 4.5 |
| Oleic Acid (C18:1) | 1369.5 ± 258.3 | 252.4 ± 141.3 | 391.0 ± 118.9 |
| Linoleic Acid (C18:2) | 1794.7 ± 389.4 | 450.6 ± 300.1 | 620.2 ± 201.7 |
| Linolenic Acid (C18:3) | 545.5 ± 140.1 | 213.7 ± 145.7 | 223.5 ± 86.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanfoni, E.; Sinisgalli, E.; Fontana, A.; Soldano, M.; Garuti, M.; Morelli, L. Microbial Characterisation of a Two-Stage Anaerobic Digestion Process for Conversion of Agri-Based Feedstock in Biogas and Long-Chain Fatty Acids in a Circular Economy Framework. Fermentation 2024, 10, 293. https://doi.org/10.3390/fermentation10060293
Fanfoni E, Sinisgalli E, Fontana A, Soldano M, Garuti M, Morelli L. Microbial Characterisation of a Two-Stage Anaerobic Digestion Process for Conversion of Agri-Based Feedstock in Biogas and Long-Chain Fatty Acids in a Circular Economy Framework. Fermentation. 2024; 10(6):293. https://doi.org/10.3390/fermentation10060293
Chicago/Turabian StyleFanfoni, Elisabetta, Erika Sinisgalli, Alessandra Fontana, Mariangela Soldano, Mirco Garuti, and Lorenzo Morelli. 2024. "Microbial Characterisation of a Two-Stage Anaerobic Digestion Process for Conversion of Agri-Based Feedstock in Biogas and Long-Chain Fatty Acids in a Circular Economy Framework" Fermentation 10, no. 6: 293. https://doi.org/10.3390/fermentation10060293






