Cloning, Expression, Enzymatic Characterization and Mechanistic Studies of M13 Mutant Acetohydroxyacid Synthase That Rescues Valine Feedback Inhibition
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Oligonucleotide Primers, Media, and Growth Conditions
2.2. Acquisition of the Target Gene and Construction of the Expression Vector
2.3. Protein Expression and Purification
2.4. Determination of AHAS Activity
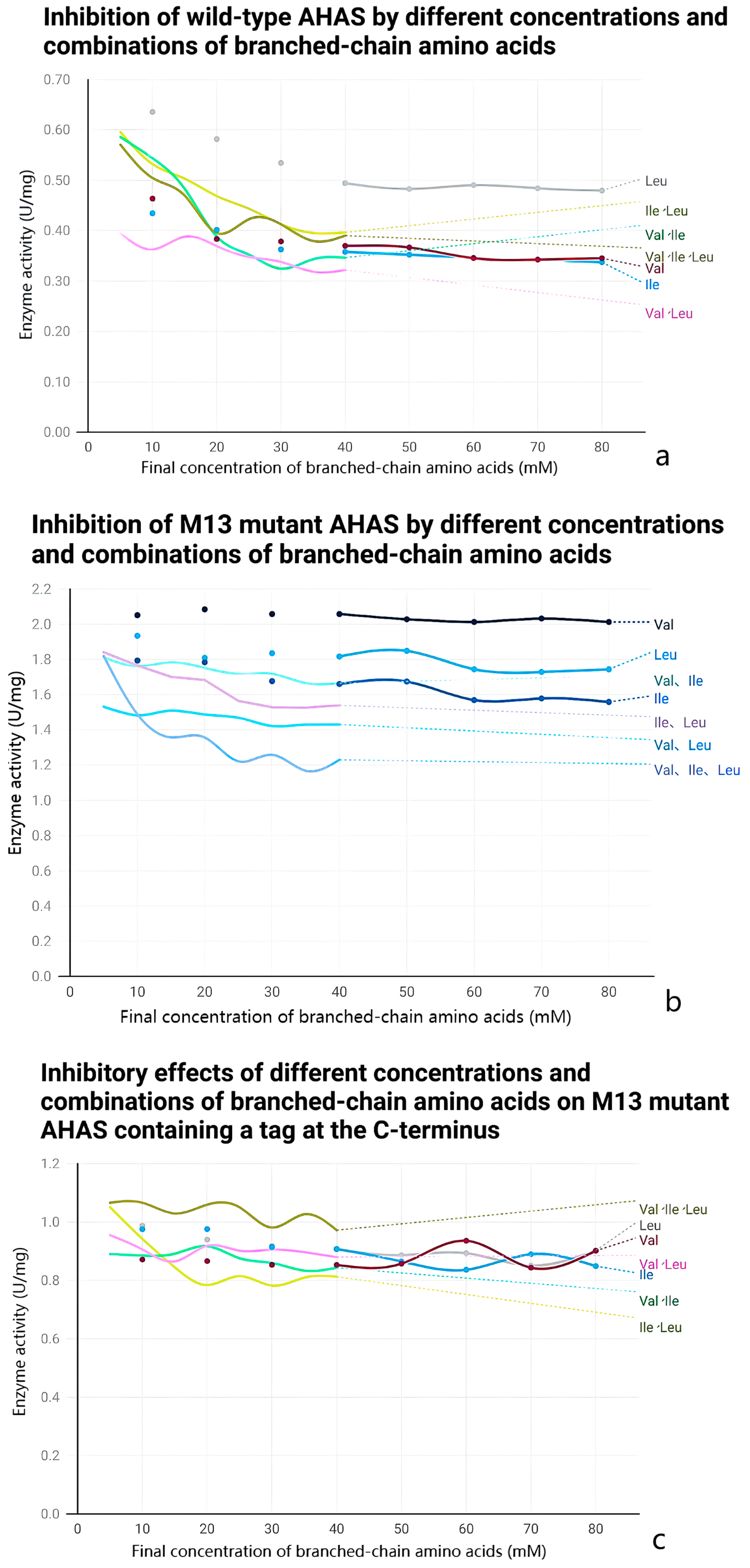
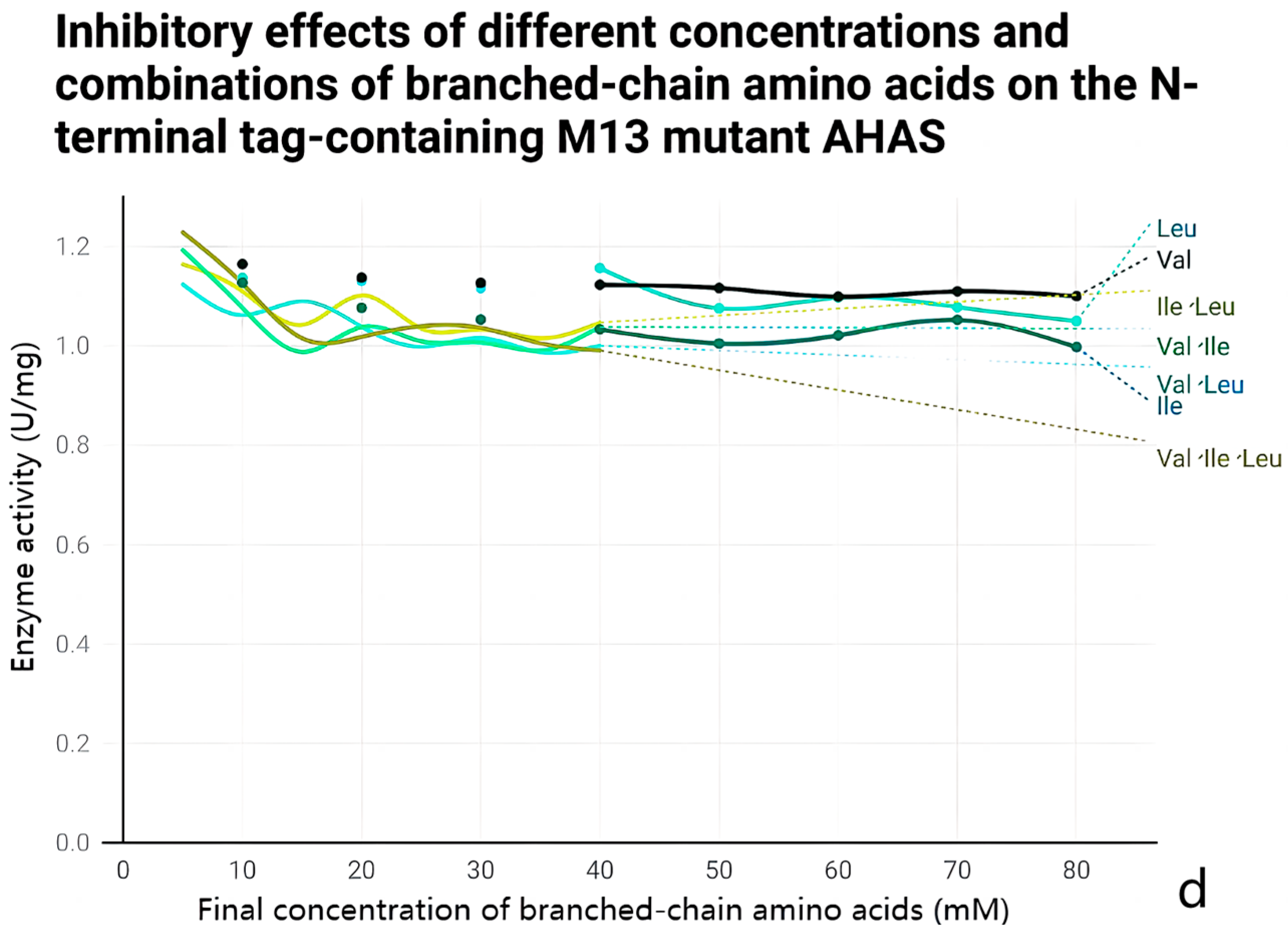
2.5. Anti-Feedback Inhibition Analysis
2.6. Wild-Type and M13 Mutant AHAS
2.7. Molecular Modeling
3. Results and Discussion
3.1. Construction of AHAS Mutants Resistant to Feedback Inhibition
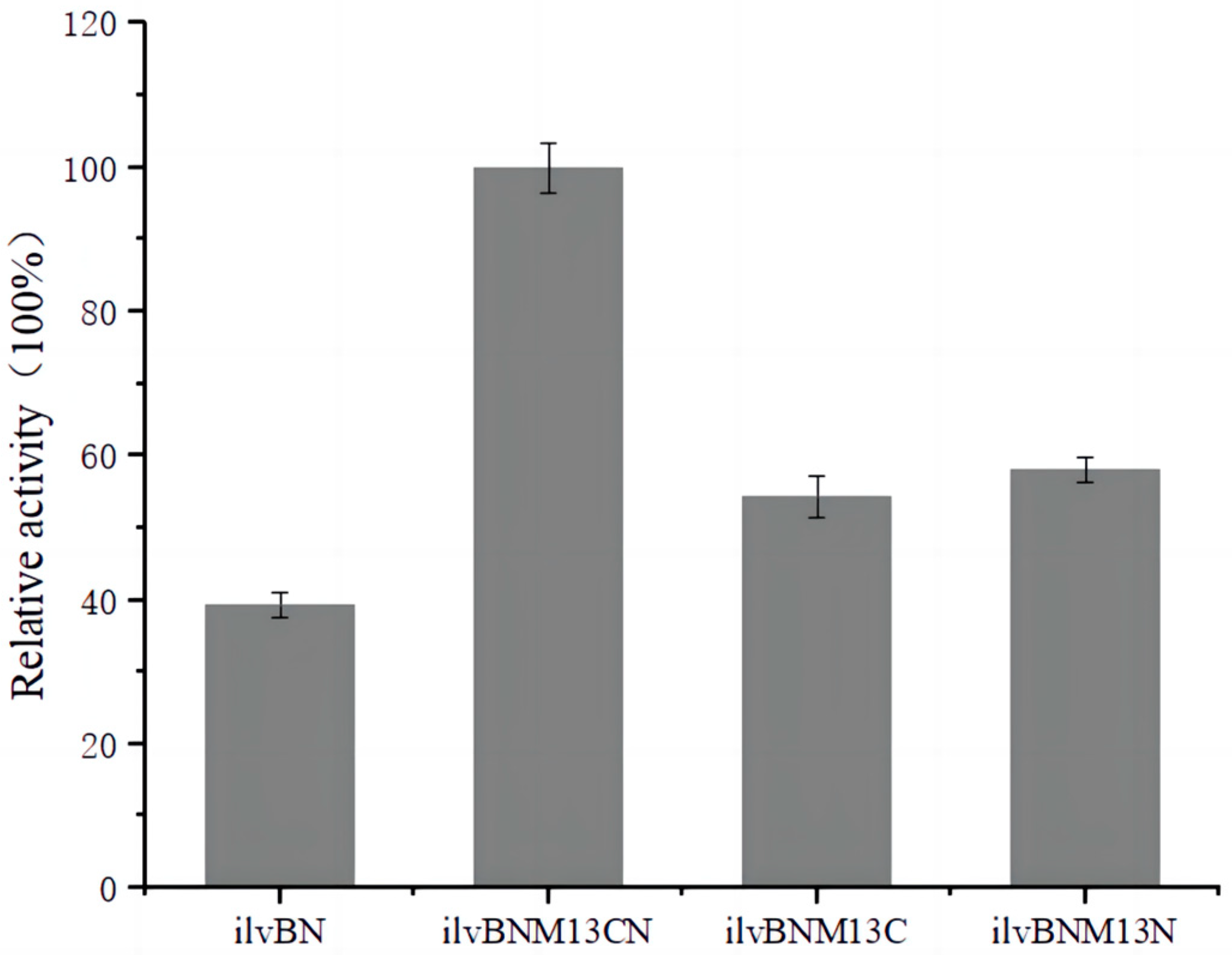
3.2. Determination of AHAS Activity
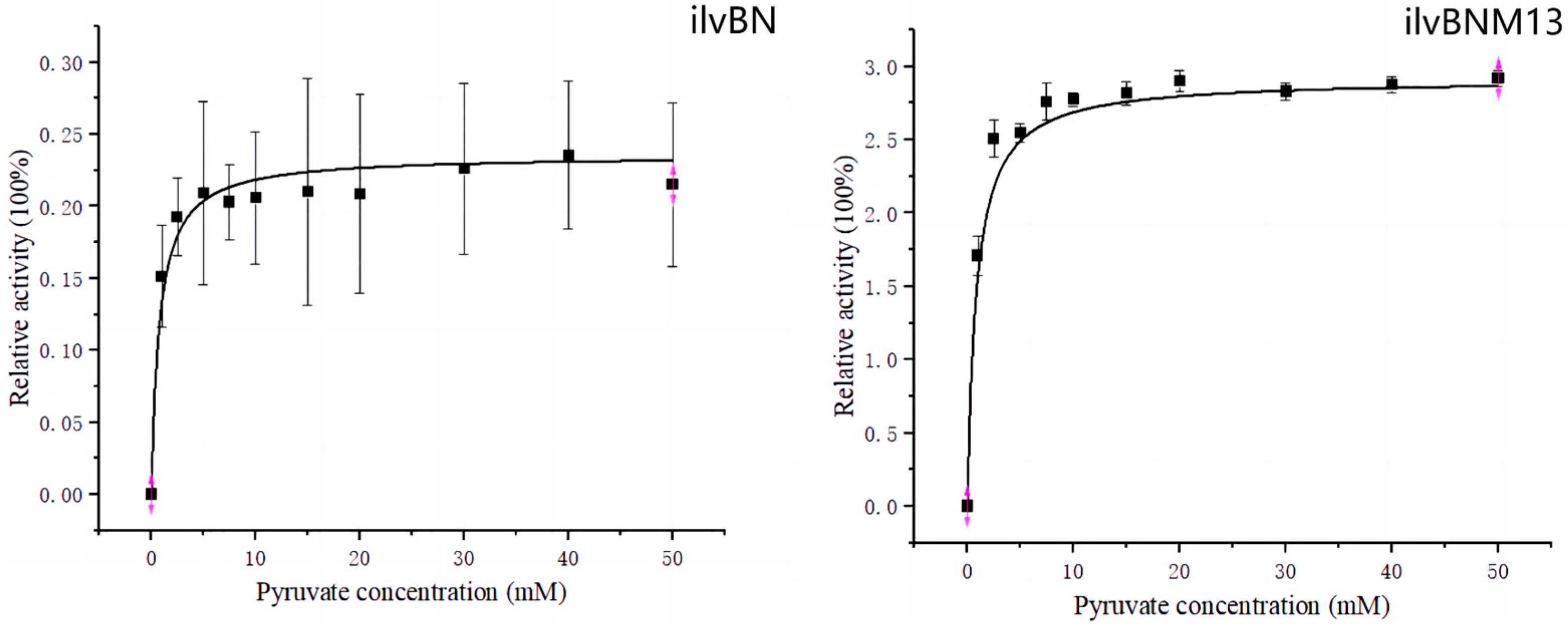
3.3. Enzyme Kinetic Analysis
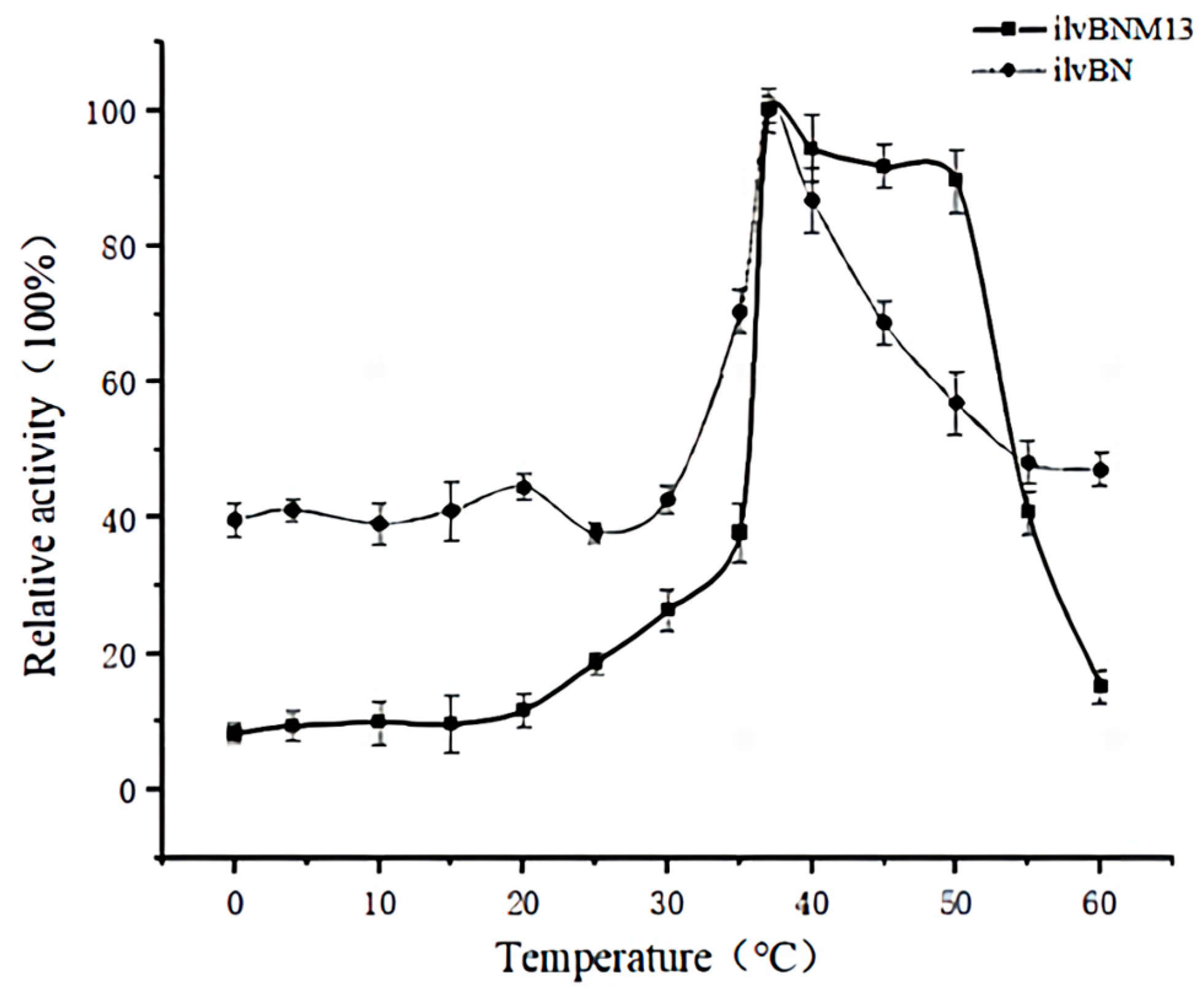
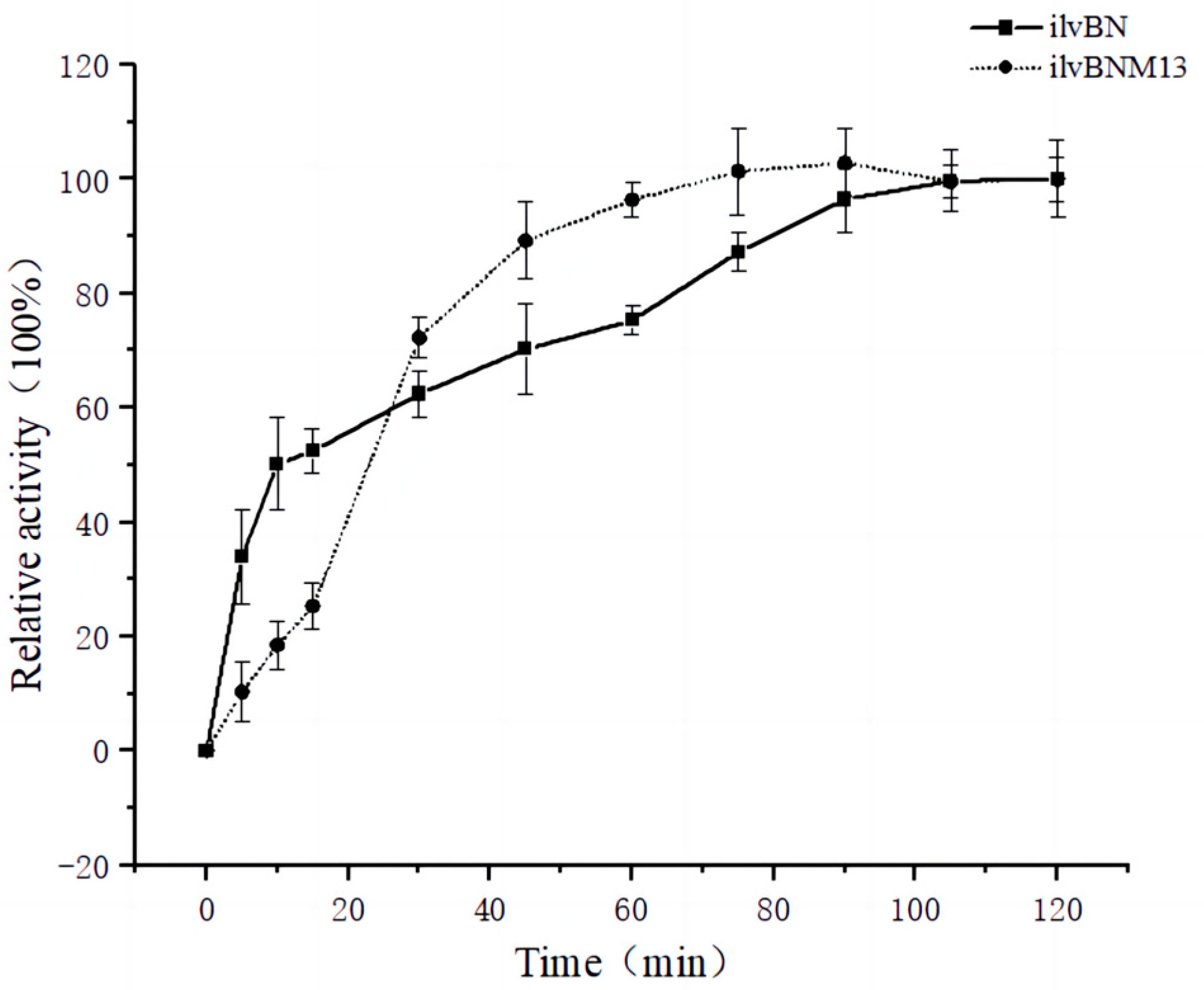
3.4. Effect of Mutations on the Enzymatic Properties of AHAS
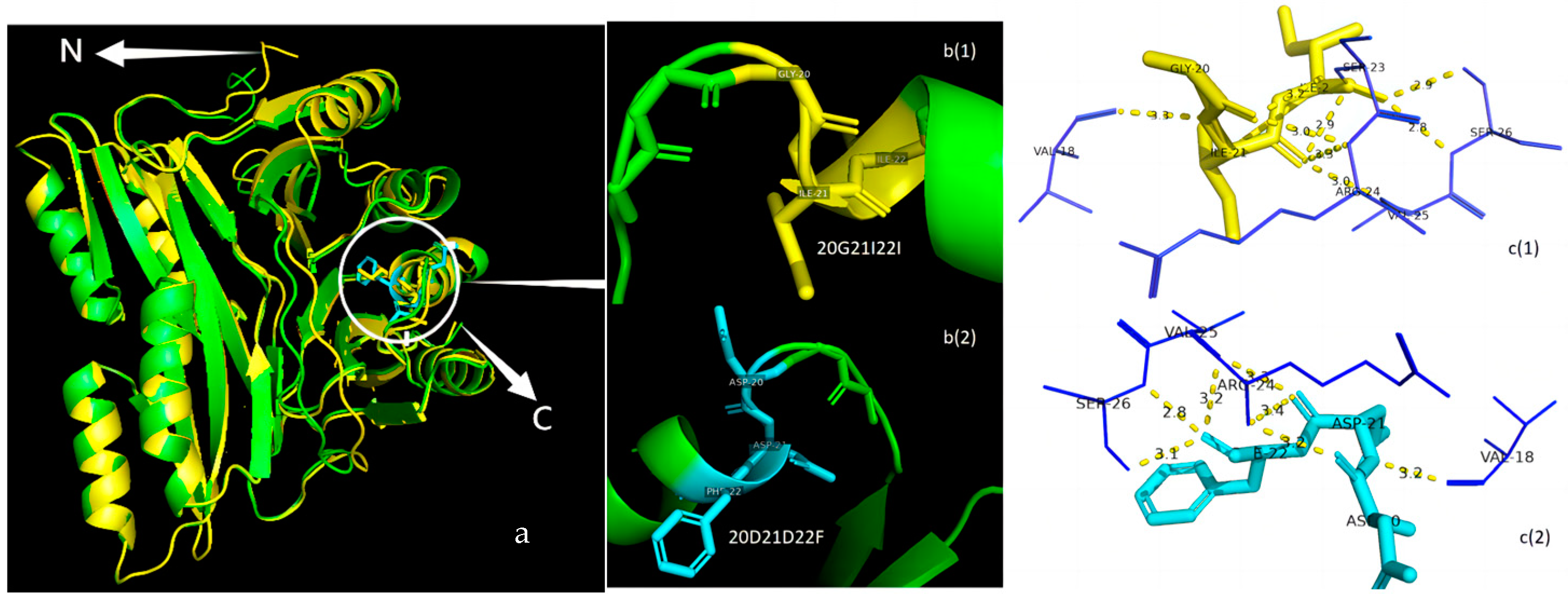
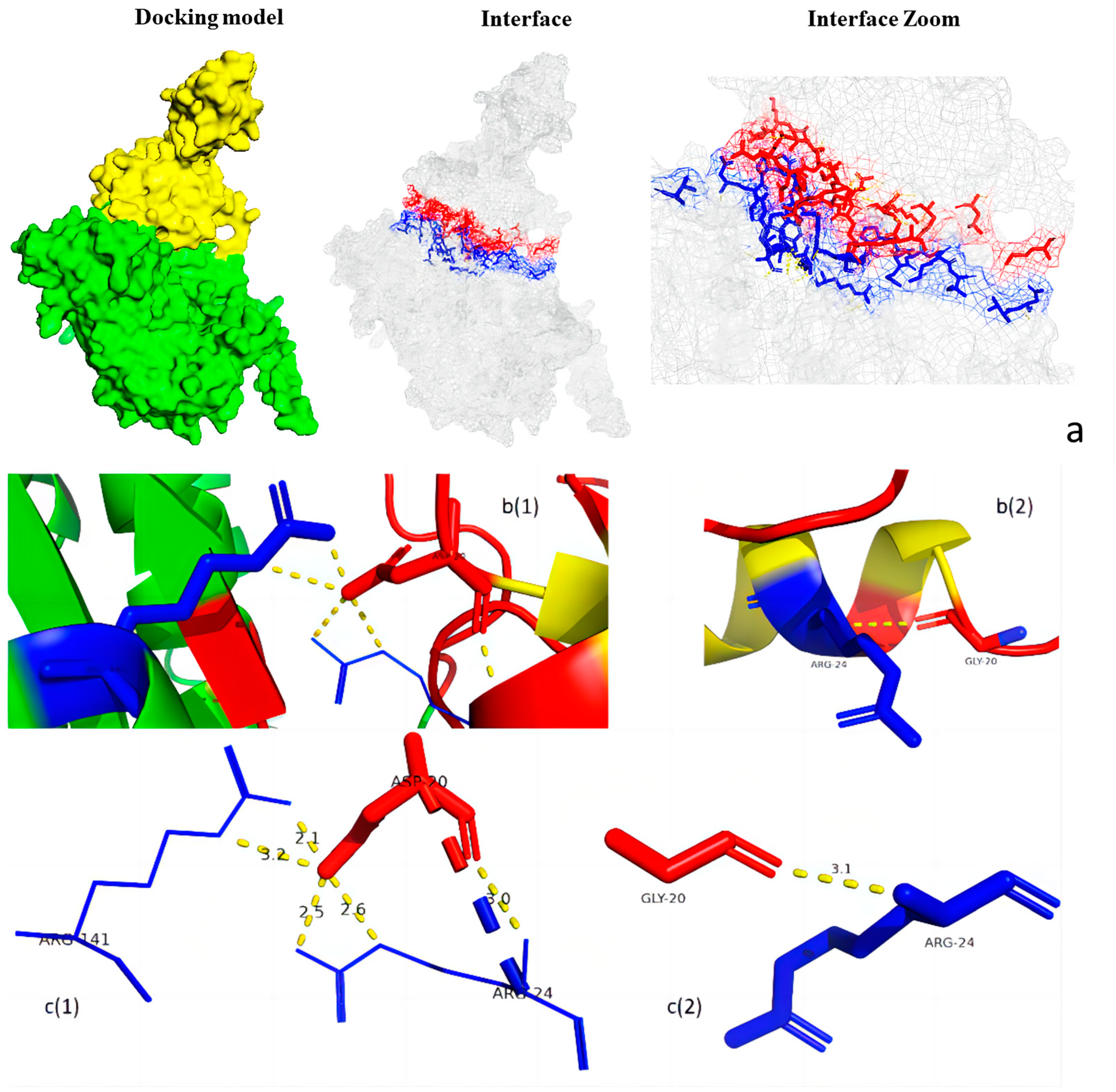
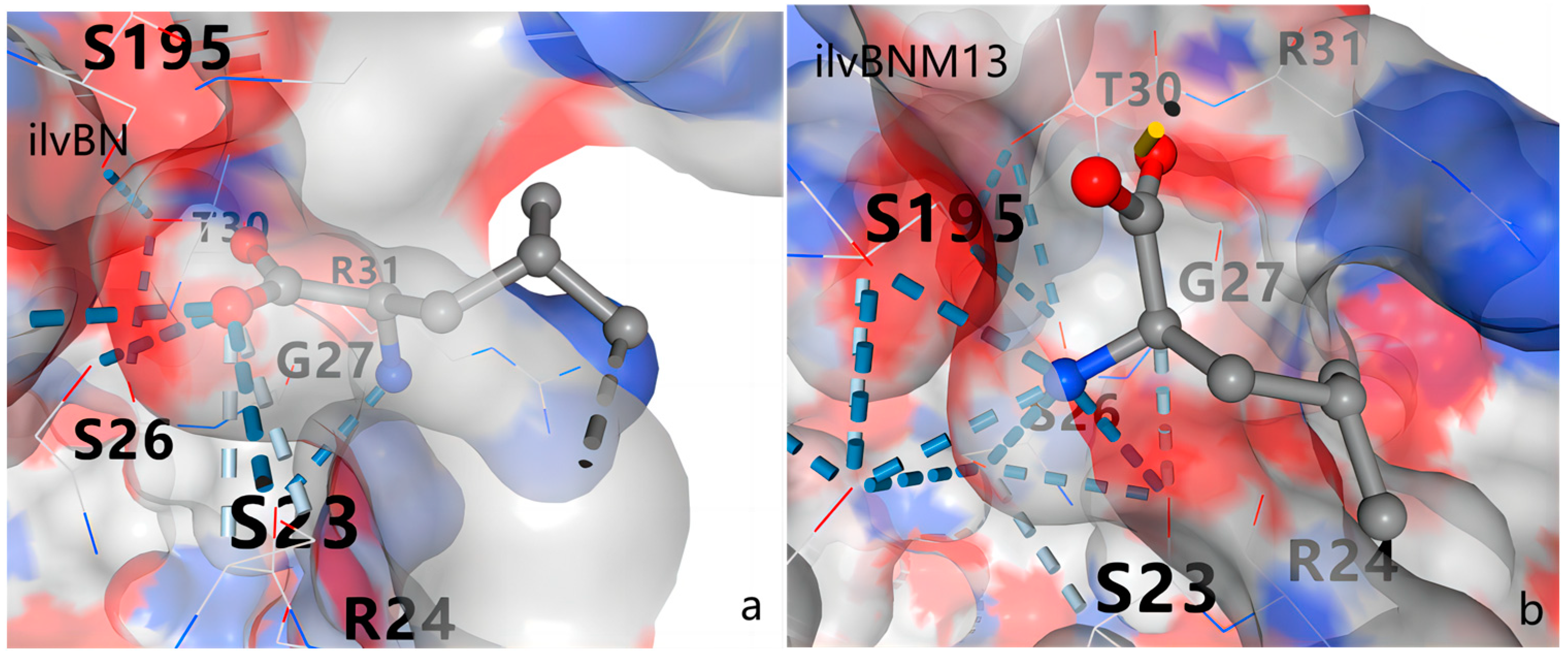
3.5. Protein Structure Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duggleby, R.G. Domain relationships in thiamine diphosphate-dependent enzymes. Acc. Chem. Res. 2006, 39, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Schloss, J.V.; Ciskanik, L.M.; Dyk, D.E.V. Origin of the herbicide binding site of acetolactate synthase. Nature 1988, 331, 360–362. [Google Scholar] [CrossRef]
- Lonhienne, T.; Garcia, M.D.; Guddat, L.W. The Role of a FAD Cofactor in the Regulation of Acetohydroxyacid Synthase by Redox Signaling Molecules. J. Biol. Chem. 2017, 292, 5101–5109. [Google Scholar] [CrossRef] [PubMed]
- Duggleby, R.; Pang, S.S. Acetohydroxyacid Synthase. J. Biochem. Mol. Biol. 2000, 33, 1–36. [Google Scholar]
- Niu, X.; Liu, X.; Zhou, Y.; Niu, C.; Xi, Z.; Su, X.D. Preliminary X-ray crystallographic studies of the catalytic subunit of Escherichia coli AHAS II with its cofactors. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 659–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Liu, X.; Sun, J.; Li, X.; Lin, J.; Yang, X.; Xi, Z.; Shen, Y. Molecular architecture of the acetohydroxyacid synthase holoenzyme. Biochem. J. 2020, 477, 2439–2449. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Wang, X. Molecular evolution of acetohydroxyacid synthase in bacteria. MicrobiologyOpen 2017, 6, e00524. [Google Scholar] [CrossRef]
- Kaplun, A.; Vyazmensky, M.; Zherdev, Y.; Belenky, I.; Slutzker, A.; Mendel, S.; Barak, Z.; Chipman, D.M.; Shaanan, B. Structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from Escherichia coli. J. Mol. Biol. 2006, 357, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, V.; Vyazmensky, M.; Engel, S.; Belenky, I.; Kaplun, A.; Kryukov, O.; Barak, Z.; Chipman, D.M. Acetohydroxyacid synthase isozyme I from Escherichia coli has unique catalytic and regulatory properties. Biochim. Biophys. Acta 2006, 1760, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Barak, Z.; Chipman, D.M. Allosteric regulation in Acetohydroxyacid Synthases (AHASs)—Different structures and kinetic behavior in isozymes in the same organisms. Arch. Biochem. Biophys. 2012, 519, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Low, Y.S.; Garcia, M.D.; Croll, T.; Gao, Y.; Wang, Q.; Brillault, L.; Williams, C.M.; Fraser, J.A.; McGeary, R.P.; et al. Structures of fungal and plant acetohydroxyacid synthases. Nature 2020, 586, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Westerfeld, W.W. A colorimetric determination of blood acetoin. J. Biol. Chem. 1945, 161, 495–502. [Google Scholar] [CrossRef]
- Eliáková, V.; Pátek, M.; Holátko, J.; Nevera, J.; Leyval, D.; Goergen, J.L.; Delaunay, S. Feedback-Resistant Acetohydroxy Acid Synthase Increases Valine Production in Corynebacterium glutamicum. Am. Soc. Microbiol. (ASM) 2005, 71, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Lawther, R.P.; Calhoun, D.H.; Adams, C.W.; Hauser, C.A.; Gray, J.; Hatfield, G.W. Molecular basis of valine resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 1981, 78, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Eoyang, L.; Silverman, P.M. Role of small subunit (IlvN polypeptide) of acetohydroxyacid synthase I from Escherichia coli K-12 in sensitivity of the enzyme to valine inhibition. J. Bacteriol. 1986, 166, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ilan, A.; Balan, V.; Tittmann, K.; Golbik, R.; Vyazmensky, M.; Hübner, G.; Barak, Z.; Chipman, D.M. Binding and activation of thiamin diphosphate in acetohydroxyacid synthase. Biochemistry 2001, 40, 11946–11954. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Garcia, M.D.; Pierens, G.; Mobli, M.; Nouwens, A.; Guddat, L.W. Structural insights into the mechanism of inhibition of AHAS by herbicides. Proc. Natl. Acad. Sci. USA 2018, 115, E1945–E1954. [Google Scholar] [CrossRef] [PubMed]
- Mccourt, J.A.; Pang, S.S.; King-Scott, J.; Guddat, L.W.; Duggleby, R.G. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc. Natl. Acad. Sci. USA 2006, 103, 569–573. [Google Scholar] [CrossRef]
- Karanth, N.M.; Sarma, S.P. The coiL-to-helix transition in IlvN regulates the allosteric control of Escherichia coli acetohydroxyacid synthase I. Biochemistry 2013, 52, 70–83. [Google Scholar] [CrossRef] [PubMed]






| Strain or Plasmid | Relevant Characteristics | Reference or Source |
|---|---|---|
| C. glutamicum strains ATCC 13032 | Wild type | Laboratory preservation |
| E. coli strains DH5α | Wild type | Laboratory preservation |
| E. coli strains BL21 | Wild type | Laboratory preservation |
| DH5α/pET-28a-ilvBN | DH5αharboring pET-28a-ilvBN | This work |
| DH5α/pET-28a-ilvBNCNM13 | DH5αharboring pET-28a-ilvBNCNM13 | This work |
| DH5α/pET-28a-ilvBNCM13 | DH5αharboring pET-28a-ilvBNCM13 | This work |
| DH5α/pET-28a-ilvBNNM13 | DH5αharboring pET-28a-ilvBNNM13 | This work |
| BL 21/pET-28a-ilvBN | BL 21 harboring pET-28a-ilvBN | This work |
| BL 21/pET-28a-ilvBNCNM13 | BL 21 harboring pET-28a-ilvBNCNM13 | This work |
| BL 21/pET-28a-ilvBNCM13 | BL 21 harboring pET-28a-ilvBNCM13 | This work |
| BL 21/pET-28a-ilvBNNM13 | BL 21 harboring pET-28a-ilvBNNM13 | This work |
| plasmids | ||
| pET-28a | E. coli vector, Kmr | Laboratory preservation |
| pET-28a-ilvBN | pET-28a with the insertion of ilvBN | This work |
| pET-28a-ilvBNCNM13 | pET-28a with the insertion of ilvBNCNM13 | This work |
| pET-28a-ilvBNCM13 | pET-28a with the insertion of ilvBNCM13 | This work |
| pET-28a-ilvBNNM13 | pET-28a with the insertion of ilvBNNM13 | This work |
| Primer | 5′-3′ Sequence a |
|---|---|
| ilvBN-F | CGCGGATCCATGAATGTGGCAGCTTCTCAACAG |
| ilvBN-R | GGGAAGCTTGATCTTGGCCGGAGCCATG |
| ilvBNM13-F | GATGACTTTTCCCGCGTATCAGGTATGTTCACC |
| ilvBNM13-R | AAAGTCATCGTCTACGTCCTGAACGAGTACGGAC |
| Pet-CF | AAAGCCCGAAAGGAAGCTGAGT |
| Pet-CR | GCCCATGGTATATCTCCTTCTTAAAGT |
| Pet-NF | GCTGAGTTGGCTGCTGCCACCG |
| Pet-NR | GGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGGGG |
| ilvBNM13C1 | CGCGACGACGACGACAAGGGATCCATGAATGTGGCAGCTTCTCAACAGCCC |
| ilvBNM13N1 | GGGAAGCTTGTCATCGTCATCCTTGATCTTGGCCGGAGCCATGGTCTTC |
| ilvBNCM13-F1 | CATCATCATCATCATCACAGCAGCGGCC |
| ilvBNCM13-R1 | TCAGATCTTGGCCGGAGCCATGGTC |
| ilvBNNM13-F1 | ATGAATGTGGCAGCTTCTCAACAGC |
| ilvBNNM13-R1 | TTTGTTAGCAGCCGGATCAAGCTTT |
| ilvBNCM13-F2 | gaaggagatataccatgggcCATCATCATCATCATCACAGCAGC |
| ilvBNCM13-R2 | tcagcttcctttcgggctttTCAGATCTTGGCCGGAGCC |
| ilvBNNM13-F2 | ctttaagaaggagatataccATGAATGTGGCAGCTTCTCAACA |
| ilvBNNM13-R2 | gtggcagcagccaactcagcTTTGTTAGCAGCCGGATCAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.; Gao, X.; An, Z.; Wang, N.; Ma, Y.; Zhang, H. Cloning, Expression, Enzymatic Characterization and Mechanistic Studies of M13 Mutant Acetohydroxyacid Synthase That Rescues Valine Feedback Inhibition. Fermentation 2024, 10, 311. https://doi.org/10.3390/fermentation10060311
Tan Y, Gao X, An Z, Wang N, Ma Y, Zhang H. Cloning, Expression, Enzymatic Characterization and Mechanistic Studies of M13 Mutant Acetohydroxyacid Synthase That Rescues Valine Feedback Inhibition. Fermentation. 2024; 10(6):311. https://doi.org/10.3390/fermentation10060311
Chicago/Turabian StyleTan, Yaqing, Xingxing Gao, Zhiqiang An, Nan Wang, Yaqian Ma, and Hailing Zhang. 2024. "Cloning, Expression, Enzymatic Characterization and Mechanistic Studies of M13 Mutant Acetohydroxyacid Synthase That Rescues Valine Feedback Inhibition" Fermentation 10, no. 6: 311. https://doi.org/10.3390/fermentation10060311
APA StyleTan, Y., Gao, X., An, Z., Wang, N., Ma, Y., & Zhang, H. (2024). Cloning, Expression, Enzymatic Characterization and Mechanistic Studies of M13 Mutant Acetohydroxyacid Synthase That Rescues Valine Feedback Inhibition. Fermentation, 10(6), 311. https://doi.org/10.3390/fermentation10060311





