Abstract
Poly-γ-glutamic acid (γ-PGA) is an attractive biopolymer for medical, agri-food, and environmental applications. Although microbial synthesis by Bacilli fed on waste streams has been widely adopted, the obtainment of efficient sustainable production processes is still under investigation by bioprocess and metabolic engineering approaches. The abundant glycerol-rich waste generated in the biodiesel industry can be used as a carbon source for γ-PGA production. Here, we studied fermentation performance in different engineered Bacillus subtilis strains in glycerol-based media, considering a swrA+ degU32Hy mutant as the initial producer strain and glucose-based media for comparison. Modifications included engineering the biosynthetic pgs operon regulation (replacing its native promoter with Physpank), precursor accumulation (sucCD or odhAB deletion), and enhanced glutamate racemization (racE overexpression), predicted as crucial reactions by genome-scale model simulations. All interventions increased productivity in glucose-based media, with Physpank-pgs ∆sucCD showing the highest γ-PGA titer (52 g/L). Weaker effects were observed in glycerol-based media: ∆sucCD and Physpank-pgs led to slight improvements under low- and high-glutamate conditions, respectively, reaching ~22 g/L γ-PGA (26% increase). No performance decrease was detected by replacing pure glycerol with crude glycerol waste from a biodiesel plant, and by a 30-fold scale-up. These results may be relevant for improving industrial γ-PGA production efficiency and process sustainability using waste feedstock. The performance differences observed between glucose and glycerol media also motivate additional computational and experimental studies to design metabolically optimized strains.
1. Introduction
Poly-γ-glutamic acid (γ-PGA) is a biopolymer composed of D- and/or L-glutamic acid monomers connected by amide linkages between α-amino and γ-carboxyl groups. It has emerged as a multifaceted compound with a wide range of applications across various industries. γ-PGA’s attractiveness is due to its biodegradability, biocompatibility, water solubility, and high hygroscopicity, with the ability to absorb moisture up to 5000 times its own weight [1]. These remarkable features make this polymer a relevant tool for applications in the health, agriculture, cosmetic, and food fields [2,3]. Products based on γ-PGA such as thickening gels, dietary supplements, cream humectants, soil additives, metal flocculants, bioplastics, drug carriers, and biomaterials for medical devices have been developed, with many of them already available as commercial products [4,5,6,7].
The main strategy for γ-PGA bioproduction is microbial fermentation using strains from different Bacillus species, e.g., B. subtilis, B. amyloliquefaciens, B. licheniformis, B. velezensis, and B. methylotrophicus [5,8,9]. Many Bacillus strains are considered safe because of their long track record of safe use in human foods, e.g., to produce enzymes used for food processing or as microbial strains used for the production of fermented foods [10]. This safe status of different natural γ-PGA producer strains facilitates their industrial use in terms of downstream processing for the purification of the polymer [11]. Although naturally occurring strains have been successfully adopted for high-level polymer production, the cost-effectiveness of the process is still a challenge, mainly due to the trade-off between productivity and the cost of fermentation medium substrates [12,13,14].
Improvements in bioprocess were reported, optimizing fermentation parameters such as pH and aeration, and using diverse strains [14,15,16]. Several non-canonical substrates were also reported to support γ-PGA biosynthesis, in an effort to lower production costs [17,18]. For example, agricultural residues, such as rice bran, can serve as cost-effective and sustainable substrates for γ-PGA production [19]. This approach not only reduces the environmental impact associated with waste disposal but also transforms by-products into value-added biocommodities. Additionally, the use of renewable feedstock aligns with the principles of a circular economy, promoting a closed-loop system in which resources are reused and recycled to minimize waste. Similarly, industrial waste streams can enhance the sustainability of fermentation processes [14]. Crude glycerol is a major by-product of biodiesel production plants, representing about 10% w/w of the total product [20,21]. Despite impurities that may pose challenges in its cost-effective utilization, it has been considered a convenient carbon source for polymer production [14,22]. In fact, purified glycerol, a known substrate for the growth of microbes such as B. subtilis and B. amyloliquefaciens, has been previously adopted in γ-PGA fermentation processes as an additive to glucose-based media to enhance polymer production [23,24] or as the main carbon source [12,25,26]; however, glycerol utilization is less efficient than that of glucose and biopolymer production needs to be improved.
Previous works indicated that metabolic engineering interventions in different hosts were able to enhance γ-PGA yield and increase the sustainability of polymer production. The expression of a completely heterologous γ-PGA synthesis pathway has been shown to provide polymer production capability to Escherichia coli [27] and Corynebacterium glutamicum [28,29], the latter reaching γ-PGA titers of about 50 g/L, which is a competitive production level, comparable with natural producers. However, most reports describe the genetic optimization of natural γ-PGA producers such as B. subtilis and B. amyloliquefaciens. The most common strategies include the deletion of pathways competing with γ-PGA precursor accumulation [30,31,32,33], the removal of polymer-degrading enzymes [34,35,36], the enhancement of oxygen availability by the Vitreoscilla hemoglobin gene [35,37], the improvement of glutamate metabolism [38,39,40], and the tuning of the pgs biosynthetic operon regulatory network by engineering its promoter or by mutating the degU response regulator [30,31,41,42]. Some of these interventions were conceived by rational model-based design, in which modifications were suggested by mathematical models [32]. Metabolic engineering aimed at improving glycerol catabolism to enhance the carbon flux towards γ-PGA in B. licheniformis resulted in up to a 70% improvement in polymer production using purified or crude glycerol [31,43,44]. Other strain design strategies were also developed, aimed at tuning the stereochemical composition and the molecular weight of the produced biopolymer, by swapping glutamate racemase and/or the γ-PGA synthesis genes with variants from different species [40,45,46]. The above strategies led to improvements in polymer production, typically by 2–4-fold by deleting competing pathways, by up to 2-fold by removing degrading enzymes, by up to 1.3-fold by enhancing oxygen availability, and by up to 1.7-fold by improving glutamate metabolism, while modifying pgs regulation allowed γ-PGA production in Bacillus strains that did not exhibit polymer production. However, it is worth mentioning that these strategies were applied to a wide array of species or strains, for which performances were described under diverse growth conditions and fermentation times, making it hard to fairly compare the calculated indexes. Also, the success of the above strategies often turns out to be strain- and/or media-dependent, leading to inconsistent results among different works, as the endogenous regulations leading to γ-PGA production may differ across Bacillus species or even among strains of the same species [45,47].
In summary, the design of low-cost media and the enhancement of microbial productivity of γ-PGA are acknowledged as key factors limiting the industrial-scale production and use of this polymer. This work aims to investigate γ-PGA production in glycerol-based media using engineered B. subtilis strains bearing novel combinations of genomic modifications.
2. Materials and Methods
2.1. Strains
All the strains used in this work are derivatives of the JH642 domesticated B. subtilis strain [48] and are listed in Table 1. Briefly, PB5383 carries the swrA+ degU32Hy double mutation that confers γ-PGA production capability [42]. PB5716 and PB5691 are derivatives of PB5383, into which the sucCD and odhAB knockouts were introduced, respectively, to enhance γ-PGA flux by blocking the TCA cycle at the succinyl-CoA to succinate reaction and the 2-oxoglutarate to succinyl-CoA reaction steps, to promote γ-PGA precursor accumulation (Figure 1). These deletion strains were previously constructed based on the predictions of a genome-scale model and tested in glucose-based media [32]. Conversely, PB5741 does not have the degU32Hy mutation, but the pgs operon expression depends on a transcriptional fusion of the IPTG-inducible Physpank promoter with the pgsB gene. This strain was previously constructed to study the regulation of pgs [47], and here, the modification was leveraged to test γ-PGA production, not characterized before, without relying on the endogenous transcriptional regulations occurring in the native Ppgs promoter. The new strains constructed in this work are derivatives of PB5741 and contain either the sucCD knockout or the racE overexpression (encoding a glutamate racemase), or both modifications.

Table 1.
Strains used in this work.
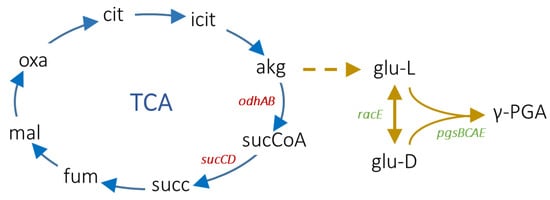
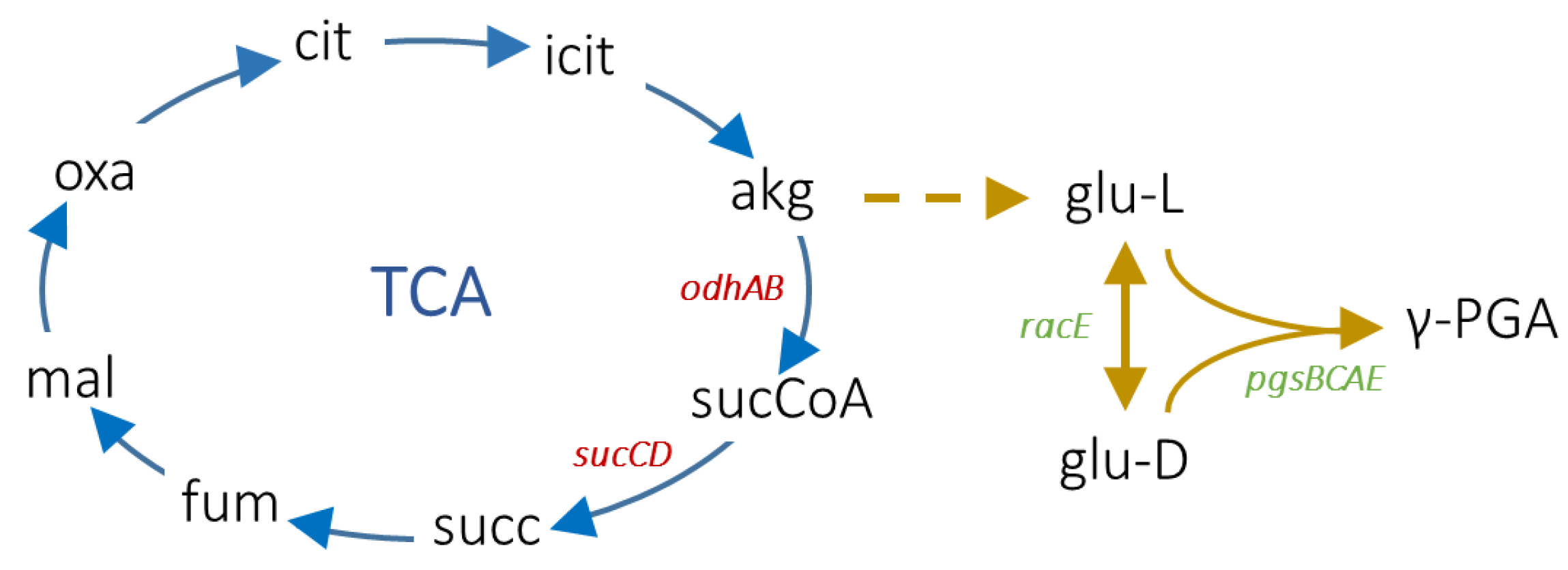
Figure 1.
Main metabolic reactions leading to γ-PGA production in B. subtilis. The tricarboxylic acid (TCA) cycle is shown with blue arrows, and the γ-PGA biosynthesis reactions are shown with yellow arrows. The dashed arrow indicates a set of reactions leading to L-glutamate production from 2-oxoglutarate (reported in [32]). Abbreviations: citrate (cit), isocitrate (icit), 2-oxoglutarate (akg), succinyl-CoA (sucCoA), succinate (succ), fumarate (fum), malate (mal), oxalacetate (oxa), L-glutamate (glu-L), and D-glutamate (glu-D). The relevant genes investigated in this work are also reported with red (deleted genes) and green (overexpressed genes) text. Reactions and molecules have been retrieved from the B. subtilis iYO844_ec genome-scale metabolic model [32].
2.2. Media and Reagents
The strain construction steps were carried out in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, 1.5% agar). Pre-cultures for fermentation experiments were grown in Penassay broth (Difco Antibiotic Medium 3) with 0.5% glucose. Fermentations were carried out in E medium [49]: 1.2% citric acid, 0.7% NH4Cl, 0.05% K2HPO4, 0.05% MgSO4·7H2O, 0.004% FeCl3·6H2O, 0.015% CaCl2·2H2O, and 0.0104% MnSO4·H2O. In this medium, tryptophan and phenylalanine (both at 50 μg/mL) were always added to complement the auxotrophies of the strains. L-glutamic acid was added at the indicated concentrations, in the 0–4% w/v range. Finally, glucose or glycerol were added as main carbon sources at the 8% w/v concentration (glucose) or in the 2–8% w/v range (glycerol). The 8% glycerol concentration was chosen according to the original E medium formulation [49], and the 2% and 4% concentrations were selected as 4- and 2-fold dilutions to test lower glycerol levels. When indicated, laboratory-grade glycerol was replaced by crude glycerol from a biodiesel plant (Oxem S.p.A., Mezzana Bigli, PV, Italy). Crude glycerol was the by-product of an industrial process that used animal fats and accounted for 83% w/v of glycerol with no detectable heavy metals or methanol, based on the analyses carried out by Oxem. Crude glycerol was autoclaved and then added to the fermentation medium at the concentrations indicated in Section 3. To facilitate comparisons between crude and laboratory-grade glycerol as fermentation feedstock, the crude glycerol concentration reported for the media refers to the sole glycerol content.
Enzymes and molecular biology kits were from New England Biolabs and were used according to the manufacturer’s instructions. Antibiotics were added during the cloning steps and for debugging purposes to assess the stability of engineered strains: erythromycin (Em, 50 μg/mL) was used to select PB5741 and its derivatives; tetracycline (Tet, 20 μg/mL) was used to select for sucCD knockout; chloramphenicol (Cat, 5 μg/mL) was used to select for odhAB knockout, or the racE overexpression; and spectinomycin (Sm, 60 μg/mL) was used to select the strains with the degU32Hy mutation.
When required, isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added to fermentation medium at 1 mM final concentration to trigger the expression of the pgs operon; antifoam (#A5757, Sigma Aldrich, St. Louis, MI, USA) was used in bioreactor experiments by injecting a 4-fold-diluted emulsion, prepared with deionized water.
2.3. Strain Construction
The pHCMC05 expression vector [50], containing the IPTG-inducible Pspac promoter, was digested with XbaI. The racE gene was amplified from B. subtilis 168 genomic DNA with primers ForRacE-05 (5′-agcttaaggaggtgaggatcctatgttggaacaaccaataggagtc-3′) and RevRacE-05 (5′-ctgccccggggacgtcgactctatcttttaatcggttcttgcag-3′). Finally, pHCMRacE was obtained by assembling digested pHCMC05 and racE via Gibson Assembly [51] and amplifying the resulting plasmid using competent DH5α Escherichia coli cells. The pHCMRacE was then digested with BglII in order to remove the replication origin of B. subtilis to allow for chromosomal integration, obtaining the pRACE plasmid.
The PB5763 strain was constructed by transforming the pRACE plasmid into PB5741 and selecting with chloramphenicol. The PB5764 and PB5765 strains were obtained by using the genomic DNA of strain GP791 (ΔsucC-sucD::Tet) (Jörg Stülke’s Lab, Gottingen, Germany) for PB5763 and PB5741 transformation, respectively, and selecting for tetracycline resistance. B. subtilis transformation was carried out using the protocol described in [52]. The correct integration of pRACE and sucCD deletion were confirmed via colony PCR with the primers pairs PspacFOR (5′-ttgactttatctacaaggtgtggc-3′)—PraceREV (5′-aggttttcaaccttcaaatcaggg-3′) and SucCDForCheck (5′-gattttgcatcgaactgtagac-3′)—TetRRevCheck (5′-gtcgtaaattcgattgtgaa-3′), respectively.
2.4. Fermentation
Strains from a streaked LB agar plate were grown for about 7 h in Penassay broth at 37 °C, at 250 rpm, to an optical density at 600 nm (OD600) of at least 2. For shake flask experiments, cultures were diluted to an OD600 of 0.1 in 20 mL of E medium and incubated as above for about 74 h. For bioreactor experiments, strains were diluted to an OD600 of 0.1 in 600 mL of E medium and incubated in a glass thermo-jacketed 3 L vessel (Applikon, Ghaziabad, Uttar Pradesh, India), with temperature control at 37 °C, agitation at 480 rpm, and 1.25 L/min aeration. At the beginning of fermentation, 600 µL antifoam was added. When required, antifoam was added again during the experiment, to a maximum of 960 µL.
2.5. Quantification of Cell Density and γ-PGA
Culture samples were withdrawn at the indicated time points during fermentations, to measure OD600. Samples were also collected to quantify γ-PGA: 500 µL culture broth was centrifuged (16,000 rpm, 4 °C, 20 min) and three volumes of cold methanol were added to the supernatant to precipitate γ-PGA. Samples were kept at −20 °C for at least 12 h. γ-PGA was recovered by centrifugation (14,000 rpm, 4 °C, 15 min), and the supernatant was discarded. A vacuum concentrator (Concentrator 5301, Eppendorf, Hamburg, Germany) was used to dry the γ-PGA pellet, which was finally dissolved in deionized water. Polymer quantification was carried out spectrophotometrically at 216 nm [53] in quartz cuvettes (NanoPhotometer UV/Vis spectrophotometer, Implen, Schatzbogen, München, Germany). Deionized water was used as a blank, and purified γ-PGA (#G1049, Sigma Aldrich) dissolved in deionized water was used to construct a calibration curve.
For the 600 mL bioreactor experiments, γ-PGA concentration was corrected to consider water loss over time in the culture due to evaporation, by multiplying the polymer quantification by Volume(measured, in mL)/600.
γ-PGA titer was calculated as the maximum polymer concentration reached in the time course, expressed as g/L. γ-PGA production rate was calculated as the maximum numeric time derivative value of the polymer concentration time series, expressed as g/L/h [54].
2.6. Genome-Scale Metabolic Modeling
To further improve the production performance of γ-PGA, the enzyme-constrained model of B. subtilis (ec_iYO844 [32]), with the addition of the synthesis reaction reported in the genome-scale metabolic model of B. licheniformis (0.77 glu-D + 0.23 glu-L → γ-pga [55]), was analyzed to find the candidate genes to be overexpressed, using the concept of Flux Scanning based on Enforced Objective Flux (FSEOF [56]). The FSEOF method identifies the reaction fluxes that show an increase for increasing flux towards target production. In particular, the fluxes of selected reactions obtained from wild-type configuration and under enforced γ-PGA production flux were normalized with respect to growth rate. The fold changes were computed (Equation (1)) for each i-th selected reaction and the average values through the j-th enforced steps and were used as final scores to support the selection of overexpression targets.
Flux Variability Analysis (FVA, [57]) was finally applied to assess the robustness of results. The algorithm was implemented via MathWorks MATLAB R2012a and run with the COBRA toolbox [58].
2.7. Statistical Analysis and Visualization
Differences in γ-PGA titers were evaluated between the swrA+ degU32Hy and Physpank-pgs strains via two-sided t-tests for independent samples. Two-way ANOVA was used to evaluate the effect of genetic modifications (knockouts and/or racE overexpression) in the swrA+ degU32Hy or the Physpank-pgs strains. The Dunnett method was used to perform multiple comparisons. A p-value (p) cutoff of 0.05 was adopted to evaluate statistical significance. GraphPad Prism 8.3.0 was used for statistical analysis and data visualization.
3. Results
3.1. Evaluation of γ-PGA Production from Glycerol Using Strains with Engineered Accumulation of Polymer Precursors in the swrA+ degU32Hy Background
We previously constructed a γ-PGA hyper-producing strain by joining the swrA+ and degU32Hy mutations [42], herein used as a reference strain, and its derivatives with the ΔsucCD and ΔodhAB deletions [32]. Both deletions affect the TCA cycle, promoting γ-PGA precursor accumulation by removing competing pathways. In shake flask fermentations in glucose-based medium, both deletions positively affected γ-PGA production [32]. Here, we aimed to assess whether their productivity could be maintained using glycerol as a carbon source; in addition, we explored the possibility of improving process sustainability by deriving the carbon source from glycerol-rich industrial waste.
3.1.1. Selection of Glycerol Concentration in Fermentation Media
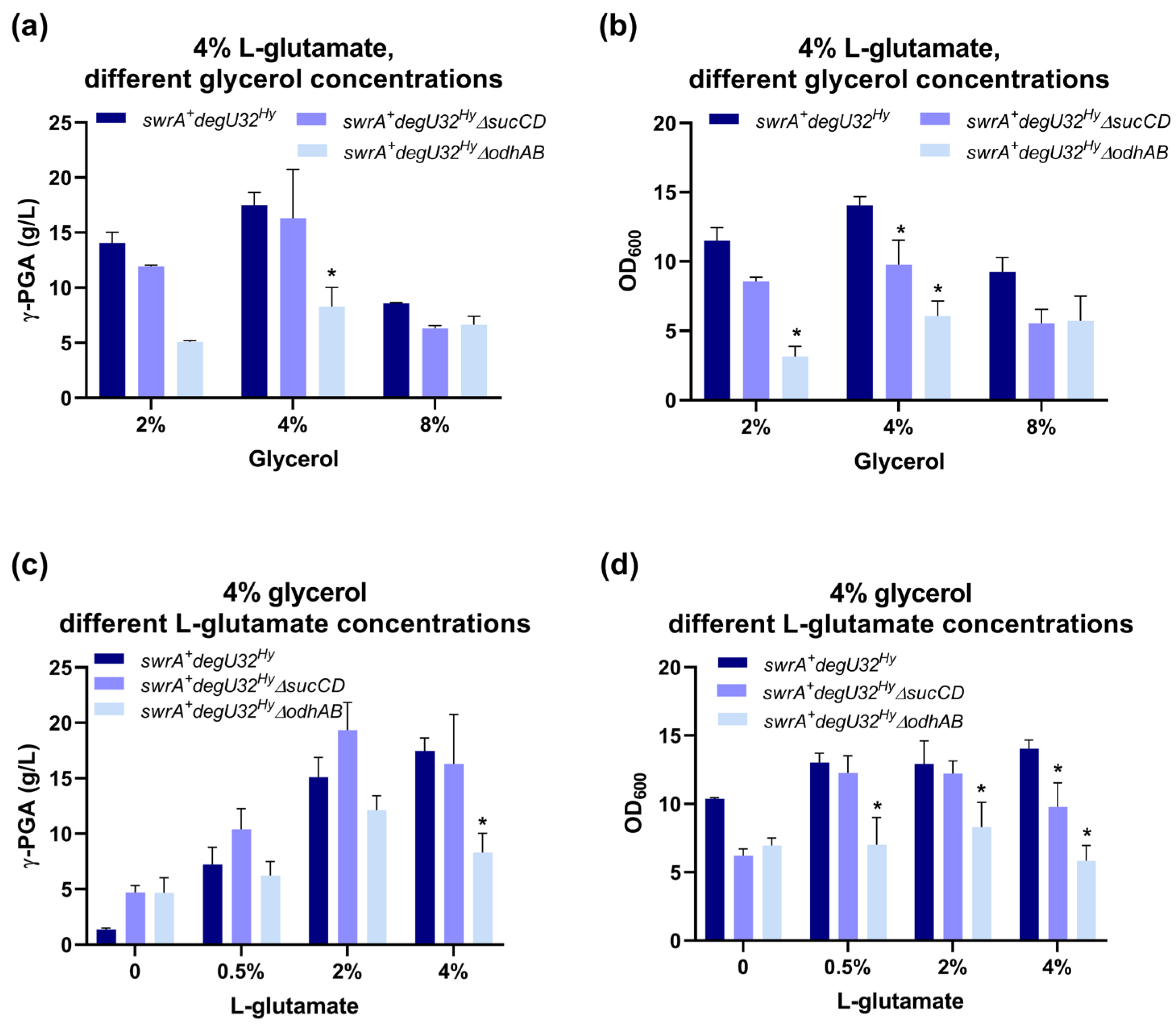
To screen the optimal glycerol concentration for fermentation, different concentrations (2, 4, and 8% w/v) were used in the γ-PGA production medium (E medium) with 4% L-glutamic acid in flask fermentations. For all the above strains, γ-PGA titer followed a similar trend as a function of the glycerol level in the fermentation medium (Figure 2a). However, the highest γ-PGA titer did not correspond to the highest glycerol concentration. In each case, the optimal glycerol level was 4%, for which the parent and the ΔsucCD strains reached significantly higher polymer levels than the ΔodhAB strain (p < 0.05; ANOVA). The 2% glycerol concentration showed a slightly lower γ-PGA titer than the 4% concentration for the three strains. Finally, the 8% glycerol condition showed a drop in γ-PGA titer for all the strains compared with the other glycerol levels tested, although the parent strain reached a slightly higher polymer titer than the deleted strains. Considering the optimal glycerol concentration (4%), the parent and ΔsucCD strains performed similarly in terms of polymer titer. However, the cell growth performances, herein measured in terms of maximum cell density attained (Figure 2b), indicated that both the ΔsucCD and ΔodhAB strains reached a significantly lower OD600 than the parent strain (p < 0.05, ANOVA). This growth defect, more pronounced for the ΔodhAB strain, was probably due to the perturbation of the TCA cycle due to the two knockouts, even though no growth decrease was previously detected in E medium with glucose as the main carbon source [32].

Figure 2.
Characterization of the swrA+ degU32Hy strain (PB5383) and its two derivatives (PB5691 and PB5716) in glycerol media. (a) Maximum γ-PGA concentrations reached in shake flask fermentations for the indicated strains as a function of glycerol concentration. (b) Maximum OD600 reached in the experiments of panel (a). (c) Maximum γ-PGA concentrations reached in shake flask fermentations in 4% glycerol at different L-glutamate levels. (d) Maximum OD600 reached in the experiments of panel (c). Bars represent the mean of at least two independent replicates, and error bars indicate the standard error of the mean. For each glycerol (panels (a,b)) or glutamate (panels (c,d)) condition, asterisks indicate statistically significant differences from the swrA+ degU32Hy strain.
These data demonstrated that variations in the carbon source had dramatic effects on growth and γ-PGA production, resulting in non-monotonic trends; in addition, the ranking of the strains in terms of γ-PGA titer differed when fermentation was carried out in glycerol (herein tested) or glucose (previously characterized [32]). The 8% glycerol concentration was excluded from the subsequent experiments due to poor growth and low polymer production. Most of the further experiments were conducted in E medium containing 4% glycerol as optimal concentration.
3.1.2. Fermentation of Crude Glycerol
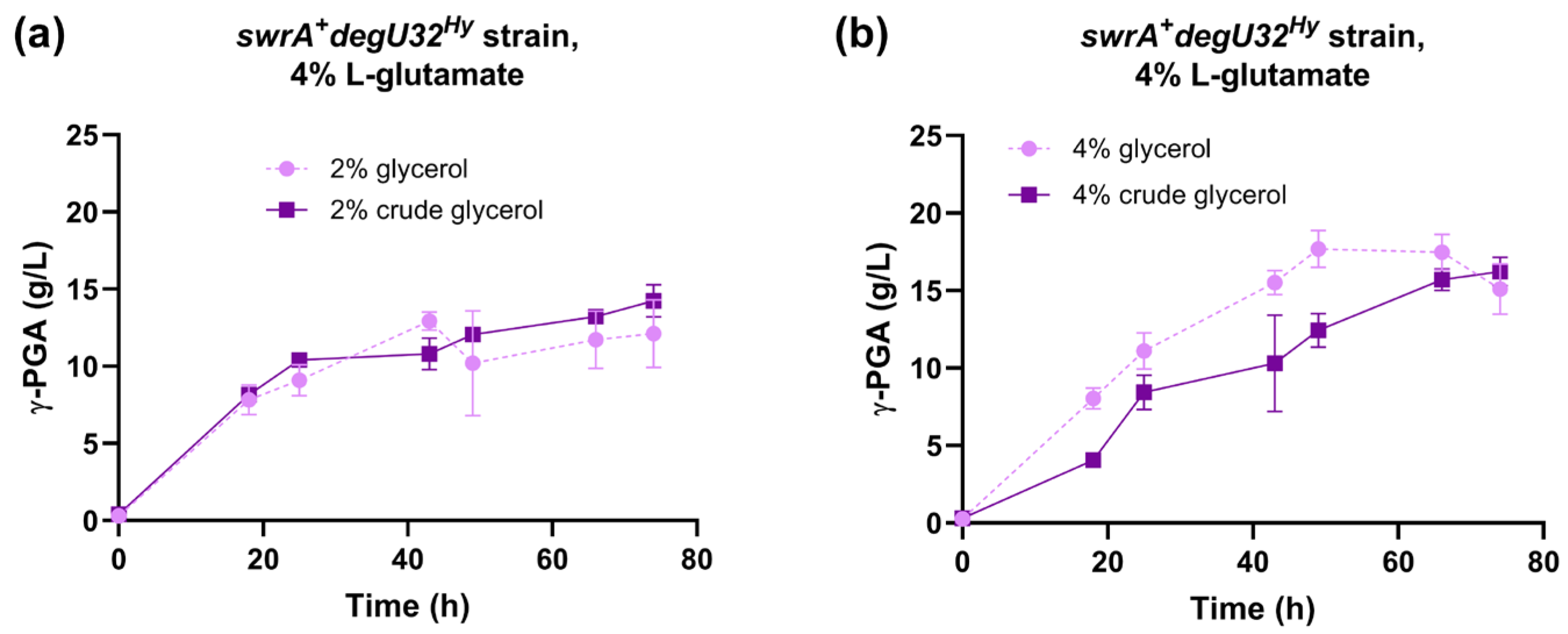
The fermentation capabilities of the reference strain (swrA+ degU32Hy) were tested in E medium containing 4% L-glutamic acid and 2% or 4% crude glycerol derived from a waste stock of an industrial biodiesel plant. The time course of γ-PGA production was similar in growth media containing laboratory-grade purified glycerol or crude glycerol (Figure 3). In fact, both glycerol concentrations reached the same γ-PGA titer at the end of the experiment. In the 2% glycerol condition, the profile of γ-PGA production was also highly similar between purified and crude glycerol (Figure 3a). In fermentations with 4% glycerol, only a slight delay was observed in the crude glycerol condition compared with the purified glycerol (Figure 3b). Overall, these data demonstrate that no relevant inhibitors of fermentation are present in the crude glycerol used in this work, which thus represents a convenient feedstock to replace purified glycerol.
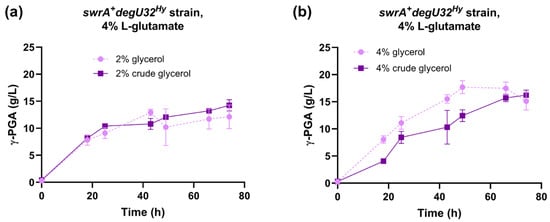
Figure 3.
Characterization of the swrA+ degU32Hy strain (PB5383) in medium with crude glycerol. Time course of γ-PGA concentration in shake flask fermentations for the swrA+ degU32Hy strain, with 2% (a) or 4% (b) glycerol content. The time course obtained with laboratory-grade glycerol is shown for comparison. Data points represent the mean of at least two independent replicates, and error bars indicate the standard error of the mean.
3.1.3. Fermentations in Glycerol-Based Media with Different Glutamate Concentrations
Previous characterization of the three strains in glucose media showed that lowering the L-glutamate concentration in the fermentation medium resulted in lower γ-PGA production. This outcome was expected since glutamic acid is a precursor of γ-PGA. However, in low-glutamate conditions, the ΔsucCD and ΔodhAB strains reached higher polymer titers than the parent strain [32]. Here, we tested different glutamic acid levels in E medium with 4% glycerol as the carbon source to investigate if the knockout strains could outperform the parent strain in such precursor-poor conditions (Figure 2c).
Consistent with expectation, when no glutamic acid was added, both ΔsucCD and ΔodhAB strains showed a higher γ-PGA than the parent strain. γ-PGA titer increased, as expected, with increasing L-glutamate concentrations. However, the ΔodhAB strain produced significantly less polymer than the other two strains for L-glutamate concentrations greater than zero (statistically significant for the 4% glutamate concentration; p < 0.05, ANOVA). This behavior was not expected, but it was consistent with the low performance of the ΔodhAB strain grown in 4% glycerol, even in the presence of L-glutamate (Section 3.1.1). As expected, the parent strain showed a monotonic increase in γ-PGA production as a function of L-glutamate. The ΔsucCD strain showed a slightly better production performance than the parent strain at intermediate glutamate concentrations (although the difference was not statistically significant—p > 0.05; ANOVA). In other words, data suggest that the sucCD deletion decreased L-glutamate requirement, consistent with previous data obtained with glucose [32].
An analysis of cell growth (Figure 2d) showed that the OD600 in the ΔsucCD strain was always lower than or equal to the cell density of the parent strain (statistically significant difference for the 4% glutamate concentration; p < 0.05, ANOVA). The poor growth in the ΔsucCD strain was consistent with the data described in Section 3.1.1 and indicates that this deletion strain was able to reach γ-PGA titers equal to (4% glutamate condition) or greater than (0 to 2% glutamate) the parent strain, even with a lower amount of cells.
3.2. Engineering the γ-PGA Biosynthetic Operon Regulation Using a Synthetic Promoter
To investigate the impact of different carbon sources on γ-PGA production excluding confounding factors linked to their effects on the pgs promoter modulation, we replaced the native Ppgs promoter with a new one. Promoter replacement is an alternative strategy to guarantee the expression of the pgs operon genes without modifying the transcriptional regulation pathway of Ppgs (e.g., occurring with the swrA+ degU32Hy mutations). A reporter strain, PB5741, was recently constructed in our lab to study the regulation of the pgs promoter [47]. PB5741 contains an IPTG-inducible Physpank promoter [59] upstream of the pgs operon genes, enabling user-defined tuning of operon expression and removing all the existing regulations. PB5741 has a strong IPTG-inducible mucoid phenotype, qualitatively indicating γ-PGA production [60]. This behavior motivated us to quantitatively characterize γ-PGA production in this strain under different conditions. Conflicting results have been reported about the replacement of the pgs operon promoter with synthetic promoters, producing either strong or mild effects compared with other metabolic engineering strategies, and even being detrimental to γ-PGA production performance [30,33,61,62]. This lack of consensus is likely due to the different strains, promoters, and fermentation conditions used in the previous works.
3.2.1. γ-PGA Production in Glucose-Based Media
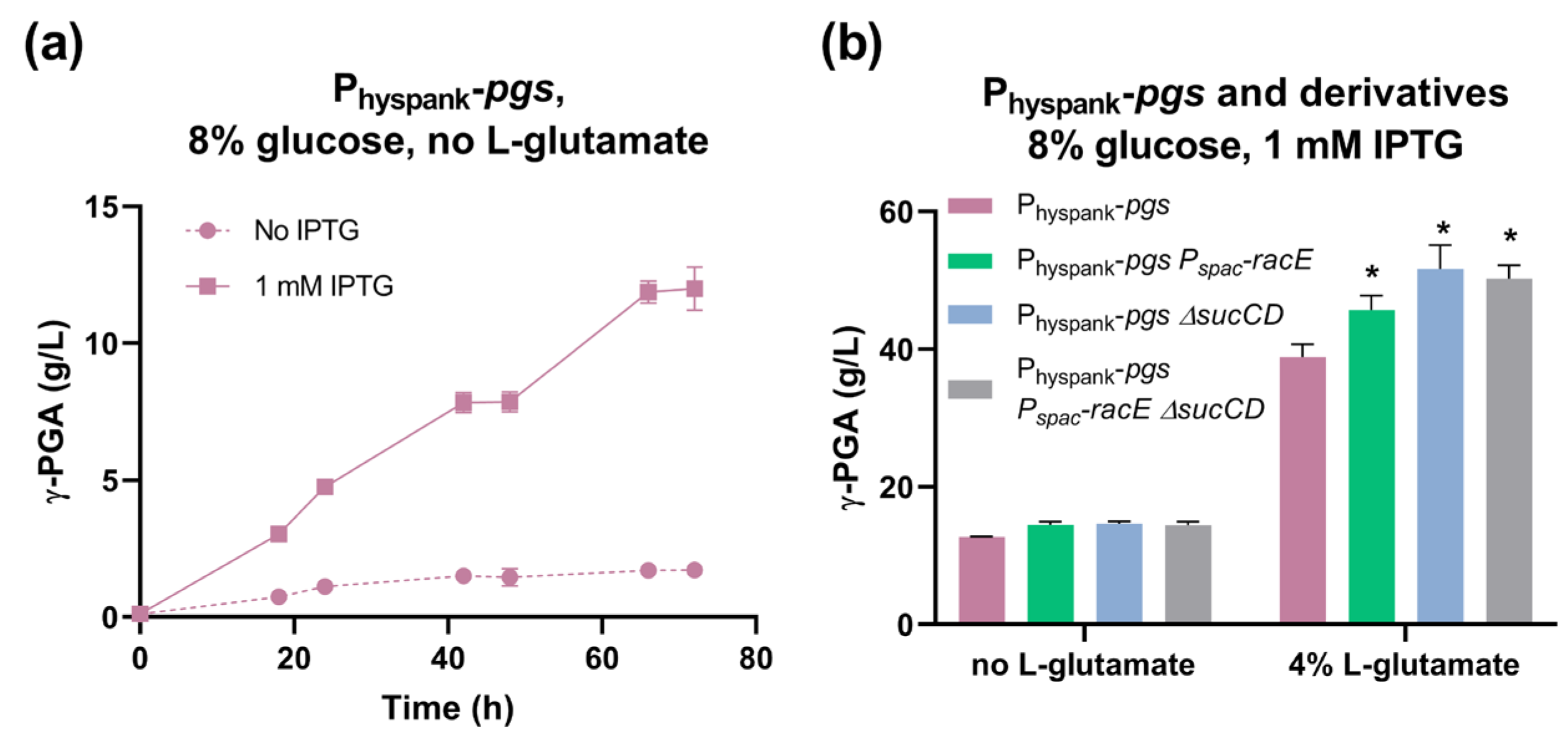
As expected, γ-PGA production in the Physpank-pgs strain was IPTG-inducible (Figure 4a): in the absence of L-glutamate and the presence of glucose and IPTG, the strain performed significantly better than the previously characterized swrA+ degU32Hy derivatives. PB5741 almost reached 13 g/L γ-PGA, while under the same condition, the swrA+ degU32Hy strain and its derivatives reached 3 to 7 g/L [32]. In the presence of L-glutamate, the Physpank-pgs strain showed a performance (39 g/L γ-PGA, Figure 4b) similar to that of the swrA+ degU32Hy strain and its derivatives (32 to 43 g/L γ-PGA [32]). Due to the strong positive effect on γ-PGA production, the Physpank-pgs strain was cultured in the presence of IPTG for the subsequent experiments.

Figure 4.
Characterization of the Physpank-pgs strain (PB5741) and its derivatives (PB5763, PB5765, and PB5764) in glucose media. (a) γ-PGA concentration over time for Physpank-pgs strain in glutamate-free media without IPTG and with induction by IPTG in shake flask fermentations. (b) Maximum γ-PGA titer reached for the indicated strains in shake flask fermentations, with 1 mM IPTG. Bars and data points represent the mean of at least three independent replicates, and error bars indicate the standard error of the mean. For each glutamate concentration in the bar plot, asterisks indicate statistically significant differences from the Physpank-pgs strain.
3.2.2. γ-PGA Production in Glycerol-Based Media
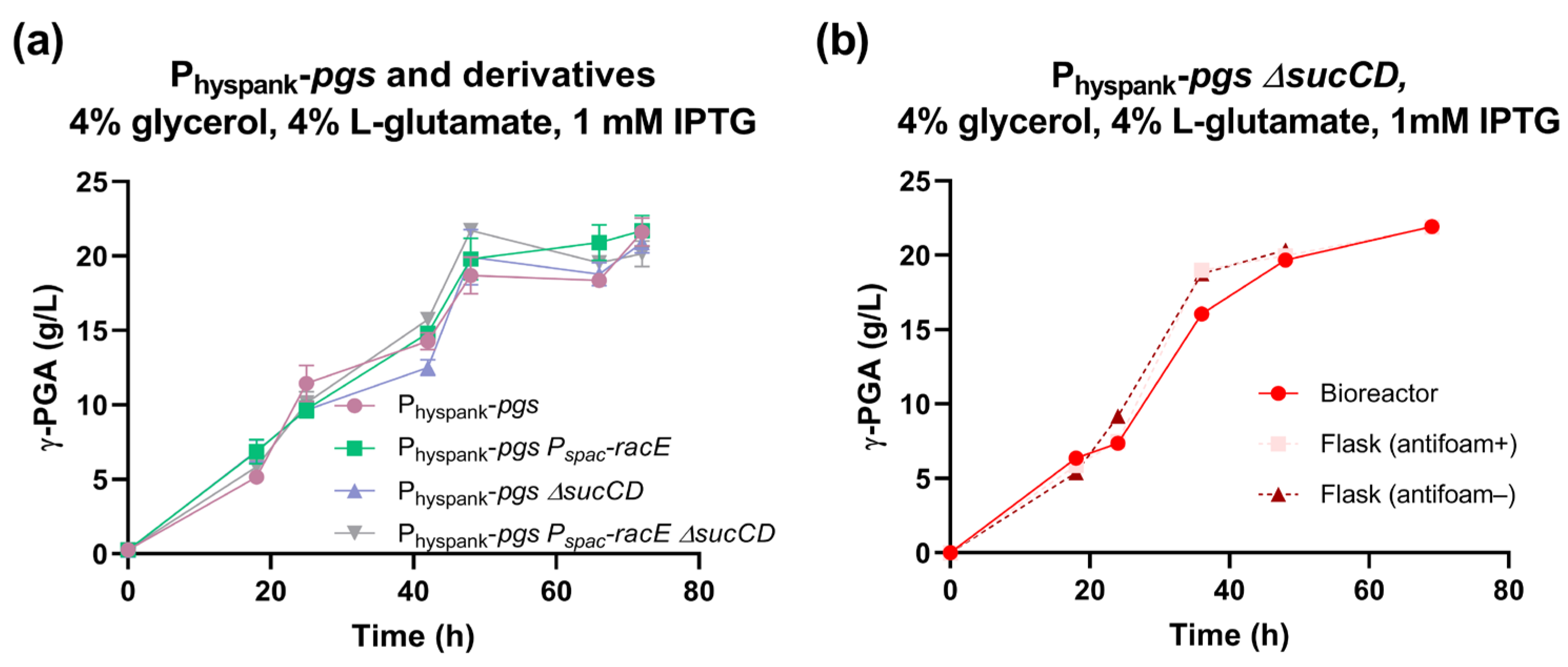
The Physpank-pgs strain was then tested in a medium with 4% glycerol and 4% L-glutamate and showed a maximum γ-PGA titer of 22 g/L with a production rate of 0.94 g/L/h (Figure 5a). These data represent a statistically significant 26% and 68% increase in γ-PGA titer and production rate, respectively, compared with those of the swrA+ degU32Hy strain (p < 0.05, t-test). This suggests that, under these conditions, the Physpank promoter replacement strategy outperformed the degU(Hy)-based strategy for γ-PGA biosynthesis in both glucose and glycerol fermentations.
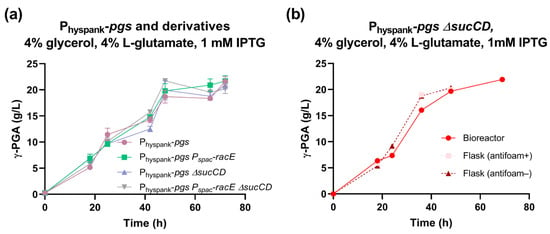
Figure 5.
Characterization of the Physpank-pgs strain and its derivatives in glycerol media. (a) γ-PGA concentration over time for the indicated strains (PB5741, PB5763, PB5765, and PB5764) in shake flask fermentations with 4% glutamate and 1 mM IPTG. Data points represent the mean of at least three independent replicates, and error bars indicate the standard error of the mean. (b) γ-PGA concentration over time for the Physpank-pgs ΔsucCD strain (PB5765) with 4% glutamate and 1 mM IPTG, in flask or bioreactor conditions as specified in the graph. Single data points are shown from a parallel experiment, run on the same day.
3.3. Metabolic Engineering of the Inducible γ-PGA Production Strain to Remove Competing Pathways for Precursor Accumulation and Enhance Glutamate Metabolism
To remove competing pathways and enhance glutamate metabolism, new derivatives of the Physpank-pgs strain were constructed by integrating previous and new recommendations by the ec_iYO844 genome-scale metabolic model [32].
3.3.1. In Silico Identification of Overexpression Targets
New simulations were carried out to search for overexpression targets via the FSEOF method (see Section 2.6) using the ec_iYO844 model. This model was previously parametrized using omics data from glucose minimal media, for which extensive data of B. subtilis are available, and was used for identifying optimal gene knockouts [32]. For this reason, model recommendations may be less accurate when dealing with media containing different carbon sources. Since no extensive knowledge of B. subtilis physiology and proteomics (both required for model parametrization) in glycerol-based media is available, we decided to use the available model to gain insight into the recommended overexpression targets.
The racE gene, encoding for a glutamate racemase [63], was selected as the best target for upregulation. This gene product affects the L- to D-glutamate racemization, theoretically contributing to the supply of both forms of glutamate as precursors for γ-PGA biosynthesis. Previous experimental works focused on racE overexpression, including its effects on the γ-PGA stereochemistry, reported variable success rates for the improvement in polymer yield [38,40].
3.3.2. Introducing a sucCD Knockout and/or racE Overexpression in the Physpank-pgs Strain Background
Our first racE overexpression attempt exploited the pHCMRacE replicative plasmid, in which the racE gene was inserted under the control of the IPTG-inducible Pspac promoter. When this construct was inserted in a γ-PGA producer strain, a significant decrease in the γ-PGA titer was observed and the same decrease was detected even when inserting the pHCMC05 empty plasmid, used as a control. This behavior was probably due to the burden imposed by the presence of pHCMC05, which resulted in lower fermentation performances, as observed previously in other species engineered for bioproduction [54,64]. For this reason, we decided to integrate Pspac-racE into the chromosome of the Physpank-pgs strain, as described in Section 2.3. In addition, we chose the ΔsucCD mutation, described in Section 3.1, as a further candidate to try to enhance γ-PGA production in the Physpank-pgs strain. Three strains were thus constructed in the Physpank-pgs background (Pspac-racE, ΔsucCD, and Pspac-racE ΔsucCD).
In glucose media and the absence of L-glutamate, these modifications did not affect γ-PGA production compared with Physpank-pgs (Figure 4b). In the presence of 4% L-glutamate, we observed a statistically significant increase (p < 0.05, ANOVA) in γ-PGA production from 39 g/L (Physpank-pgs) to 46 g/L by overexpressing racE (+17%) and to 52 g/L by deleting sucCD (+33%) (Figure 4b). The Pspac-racE ΔsucCD double mutant showed a γ-PGA titer of 50 g/L (+29%), very similar to the ΔsucCD strain, demonstrating that the two mutations did not bring additive benefits.
In glycerol media, no relevant improvement in polymer production was observed (Figure 5a), except for a slight and not statistically significant increase in γ-PGA production rate to 1.2 g/L/h in the Physpank-pgs ΔsucCD strain, corresponding to a 31% improvement in this index.
3.3.3. Scale-Up in Bioreactor Experiments
Fermentation in glycerol-based media was carried out in a benchtop bioreactor with agitation and aeration in a 0.6 L volume, corresponding to a 30-fold scale-up with respect to the shake flask experiments (Figure 5b). We obtained γ-PGA production profiles very similar to those obtained by flask fermentations, reaching ~22 g/L of polymer. Although no improvement over flask experiments was detected, data showed that the process could be easily scaled up.
4. Discussion
Addressing the challenges of sustainable development requires a change from the currently dominant linear economy model to a circular economy. Due to the increasing popularity of the circular economy, the growing production of organic waste, and the need to manage it, the concept of biorefinery is becoming more and more popular. In this concept, waste and by-products constitute the basis for the production of compounds with added value. One of the compounds that can be biologically produced is γ-PGA, widely used in the food, pharmaceutical, polymer, and cosmetic industries. Various Bacillus species can produce this compound as a metabolite, but B. subtilis stands out among them due to its safe characteristics. Attempts are being made to increase the efficiency of γ-PGA bioproduction using various genetic modifications. We addressed this task by using a domesticated B. subtilis strain, for which a compendium of physiological and genomic information is available, making it amenable to several rational metabolic engineering interventions.
In this work, we showed that purified glycerol and glycerol-rich industrial waste can be used as feedstock for γ-PGA production with engineered B. subtilis, reaching γ-PGA titer and synthesis rate of up to 22 g/L and 1.2 g/L/h, respectively.
Among the strain engineering interventions we tested in glycerol fermentation in shake flasks, the expression of the pgs operon genes by the Physpank synthetic promoter led to a 26% improvement in γ-PGA titer, compared with the initial producer (swrA+ degU32Hy strain). Also, sucCD deletion led to a slight (though not statistically significant) improvement in γ-PGA titer in the presence of 0.5% and 2% L-glutamate in the swrA+ degU32Hy strain background, and in the γ-PGA production rate with 4% L-glutamate in the Physpank-pgs strain background. A strain bearing these two genetic modifications was analyzed in a bioreactor test, resulting in performances identical to the shake flask experiments. Overall, the tested modifications pave the way to lowering L-glutamate demand and increasing the efficiency of γ-PGA production.
The metabolic engineering interventions herein tested include novel combinations of deletion and overexpression targets that had never been tested in glycerol-based fermentation media. This is especially relevant for the integration of Physpank, used to express the pgs operon genes, which highlighted the strong impact of the pgs operon transcription on γ-PGA production compared with the activated original promoter (by the swrA+ degU32Hy mutation). The overexpression of the pgs operon genes through different promoters produced conflicting results in terms of polymer production performance [28,31,58,59]. The use of different promoters in different strains and the use of different carbon sources is likely to provide non-comparable outcomes.
In this context, the data herein presented highlight differences between glucose and glycerol media in terms of strain performance. In fact, some of the benefits of genomic modifications that have been analyzed here and reported in a previous work [32] in glucose media were not detected in glycerol media (e.g., the improved γ-PGA production in odhAB deletion strains detected in glucose-based media). Indeed, it has already been acknowledged that glycerol fermentation in Bacillus strains is more challenging than glucose fermentation [24,43]. Moreover, it is worth mentioning that the genome-scale mathematical model used to identify some of the deletion and overexpression targets was parametrized using physiological, proteomic, and metabolomic data of wild-type and mutant strains grown in glucose minimal media; for this reason, it is not surprising that some metabolic engineering interventions analyzed in this work, namely sucCD or odhAB deletion and racE overexpression, are more effective in glucose-based media than in glycerol-based media. The set of physiological and omics data available for B. subtilis in glycerol media is less extensive than those in glucose media; therefore, our genome-scale model could not be used to accurately simulate the growth and the reaction fluxes occurring in fermentations with glycerol as a carbon source. These aspects stress the importance of carrying out metabolic engineering strategies tailored for each carbon source, and the need to develop more accurate computational tools.
5. Conclusions
Thanks to the novel combinations of genetic interventions herein analyzed in glycerol media, this work significantly increments our knowledge toward the exploitation of a domesticated strain of B. subtilis for the sustainable production of γ-PGA by model-guided metabolic engineering. Using the metabolically engineered strains obtained, we anticipate that the scale-up of fermentation in bioreactors and the replacement of purified glycerol with crude glycerol, both successfully tested in this work without a decrease in fermentation performance, will provide useful insights into the design of efficient and sustainable microbial processes for the valorization of waste feedstock. Industrial-scale γ-PGA production processes may benefit from the findings of this work, due to the successfully demonstrated increase in polymer production titer, use of a waste material as a carbon source, and scale-up fermentation under bioreactor conditions.
Author Contributions
Conceptualization, L.P., P.M. and C.C.; methodology, L.P., I.M. and C.C.; validation, I.M. and C.C.; formal analysis, L.P., P.M. and C.C.; investigation, L.P., I.M., P.M. and C.C.; data curation, L.P. and C.C.; writing—original draft preparation, L.P.; writing—review and editing, P.M., I.M. and C.C.; visualization, L.P.; supervision, P.M. and C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fondazione Cariplo (grant number 2015-0397), Innovation HUB Regione Lombardia (grant number 1139857), MIUR (Dipartimenti di Eccellenza Program 2018–2022, Department of Biology and Biotechnology, University of Pavia), and the NODES project which received funding from the MUR—M4C2 1.5 of PNRR with grant agreement no. ECS00000036.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The main contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors.
Acknowledgments
The authors want to thank Massimo Bellato, Michela Casanova, and Erlinda Rama for their help during fermentation experiments. The authors also thank Jörg Stülke for the generous gift of the GP791 strain.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bajaj, I.; Singhal, R. Poly (glutamic acid)—An emerging biopolymer of commercial interest. Bioresour. Technol. 2011, 102, 5551–5561. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.H.; Huang, K.Y.; Kunene, S.C.; Lee, T.Y. Poly-γ-glutamic acid synthesis, gene regulation, phylogenetic relationships, and role in fermentation. Int. J. Mol. Sci. 2017, 18, 2644. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.L.; Van, Y.T. The production of poly-(gamma-glutamic acid) from microorganisms and its various applications. Bioresour. Technol. 2001, 79, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Han, Y.; Zheng, X.; Xue, B.; Zhang, X.; Mahmut, Z.; Wang, Y.; Dong, B.; Zhang, C.; Gao, D.; et al. Synthesis of poly-γ-glutamic acid and its application in biomedical materials. Materials 2024, 17, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hou, L.; Gao, Y.; Tian, Z.; Fan, B.; Wang, F.; Li, S. Recent advances in microbial synthesis of poly-gamma-glutamic acid: A review. Foods 2022, 11, 739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Guo, Y.; Ding, S.; Chen, G.; Liang, Z.; Zeng, W. An integrated strategy for recovery and purification of poly-γ-glutamic acid from fermentation broth and its techno-economic analysis. Sep. Purif. Technol. 2022, 278, 119575. [Google Scholar] [CrossRef]
- Rodriguez-Carmona, E.; Villaverde, A. Nanostructured bacterial materials for innovative medicines. Trends Microbiol. 2010, 18, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yan, Q.; Wang, Y.; Li, Y.; Jiang, Z. Efficient production of poly-γ-glutamic acid by Bacillus velezensis via solid-state fermentation and its application. Food Biosci. 2022, 46, 101575. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Jiang, B.; Zhang, T.; Mu, W.M.; Miao, M.; Hua, Y.F. High-level production of poly(γ-glutamic acid) by a newly isolated glutamate-independent strain, Bacillus methylotrophicus. Process Biochem. 2015, 50, 329–335. [Google Scholar] [CrossRef]
- Sirisansaneeyakul, S.; Cao, M.; Kongklom, N.; Chuensangjun, C.; Shi, Z.; Chisti, Y. Microbial production of poly-γ-glutamic acid. World J. Microbiol. Biotechnol. 2017, 33, 173. [Google Scholar] [CrossRef]
- McNulty, M.J.; Gleba, Y.; Tuse, D.; Hahn-Lobmann, S.; Giritch, A.; Nandi, S.; McDonald, K.A. Techno-economic analysis of a plant-based platform for manufacturing antimicrobial proteins for food safety. Biotechnol. Prog. 2020, 36, e2896. [Google Scholar] [CrossRef] [PubMed]
- Elbanna, K.; Alsulami, F.S.; Neyaz, L.A.; Abulreesh, H.H. Poly (γ) glutamic acid: A unique microbial biopolymer with diverse commercial applicability. Front. Microbiol. 2024, 15, 1348411. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.G.; Navale, G.R.; Dharne, M.S. Poly-gamma-glutamic acid biopolymer: A sleeping giant with diverse applications and unique opportunities for commercialization. Biomass Convers. Biorefin. 2023, 13, 4555–4573. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Guo, Y.; Liu, J.; Zhao, M.; Zou, W.; Li, S. Microbial synthesis of poly-γ-glutamic acid: Current progress, challenges, and future perspectives. Biotechnol. Biofuels 2016, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.L.; Van, Y.T.; Chang, Y.N. Application of statistical experimental methods to optimize production of poly(γ-glutamic acid) by Bacillus licheniformis CCRC 12826. Enzyme Microb Technol. 2002, 31, 213–220. [Google Scholar] [CrossRef]
- Cromwick, A.M.; Birrer, G.A.; Gross, R.A. Effects of pH and aeration on γ-poly(glutamic acid) formation by Bacillus licheniformis in controlled batch fermentor cultures. Biotechnol. Bioeng. 1996, 50, 222–227. [Google Scholar]
- Nair, P.G.; Dharne, M.S. Sustainable and cleaner production of poly-gamma-glutamic acid (γ-PGA) biopolymer using floral waste and its anti-staling properties. J. Clean. Prod. 2023, 425, 138709. [Google Scholar] [CrossRef]
- Song, D.Y.; Reddy, L.V.; Charalampopoulos, D.; Wee, Y.J. Poly-(γ-glutamic acid) production and optimization from agro-industrial bioresources as renewable substrates by Bacillus sp. FBL-2 through response surface methodology. Biomolecules 2019, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Liu, Y.; Huan, C.; Xu, L.; Ji, G.; Yan, Z. Comparison of poly-γ-glutamic acid production between sterilized and non-sterilized solid-state fermentation using agricultural waste as substrates. J. Clean. Prod. 2020, 255, 120248. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, B.; Lawal, A. Recovery and utilization of crude glycerol, a biodiesel byproduct. RSC Adv. 2022, 12, 27997–28008. [Google Scholar] [CrossRef]
- Chilakamarry, C.R.; Sakinah, A.M.M.; Zularisam, A.W.; Pandey, A. Glycerol waste to value added products and its potential applications. Syst. Microbiol. Biomanufacturing 2021, 1, 378–396. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hanna, M.A.; Sun, R. Value-added uses for crude glycerol--a byproduct of biodiesel production. Biotechnol. Biofuels 2012, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, H.; Liang, J.; Yao, J. Contribution of glycerol on production of poly(gamma-Glutamic Acid) in Bacillus subtilis NX-2. Appl. Biochem. Biotechnol. 2010, 160, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.H.; Gross, R.A. Effects of glucose and glycerol on gamma-poly(glutamic acid) formation by Bacillus licheniformis ATCC 9945a. Biotechnol Bioeng. 1998, 57, 430–437. [Google Scholar] [CrossRef]
- Tork, S.E.; Aly, M.M.; Alakilli, S.Y.; Al-Seeni, M.N. Purification and characterization of gamma poly glutamic acid from newly Bacillus licheniformis NRC20. Int. J. Biol. Macromol. 2015, 74, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Birrer, G.A.; Cromwick, A.M.; Gross, R.A. Gamma-poly(glutamic acid) formation by Bacillus licheniformis 9945a: Physiological and biochemical studies. Int. J. Biol. Macromol. 1994, 16, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Geng, W.; Zhang, W.; Sun, J.; Wang, S.; Feng, J.; Zheng, P.; Jiang, A.; Song, C. Engineering of recombinant Escherichia coli cells co-expressing poly-γ-glutamic acid (γ-PGA) synthetase and glutamate racemase for differential yielding of γ-PGA. Microb. Biotechnol. 2013, 6, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, J.; Shen, J.; Zhu, Y.; Liu, W.; Chen, Y.; Zha, J.; Zhang, X.; Zhang, X.; Shi, J.; et al. Enhanced poly-γ-glutamic acid synthesis in Corynebacterium glutamicum by reconstituting PgsBCA complex and fermentation optimization. Metab. Eng. 2024, 81, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zha, J.; Cheng, H.; Ibrahim, M.H.A.; Yang, F.; Dalton, H.; Cao, R.; Zhu, Y.; Fang, J.; Chi, K.; et al. Engineering Corynebacterium glutamicum for the de novo biosynthesis of tailored poly-γ-glutamic acid. Metab. Eng. 2019, 56, 39–49. [Google Scholar] [CrossRef]
- Halmschlag, B.; Volker, F.; Hanke, R.; Putri, S.P.; Fukusaki, E.; Buchs, J.; Blank, L.M. Metabolic engineering of B. subtilis 168 for increased precursor supply and poly-γ-glutamic acid production. Front. Food Sci. Technol. 2023, 3, 1111571. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Du, S.S.; Yan, Y.F.; Pan, F.; Wang, R.; Li, S.; Xu, H.; Luo, Z.S. Systematic engineering of Bacillus amyloliquefaciens for efficient production of poly-γ-glutamic acid from crude glycerol. Bioresour. Technol. 2022, 359, 127382. [Google Scholar] [CrossRef] [PubMed]
- Massaiu, I.; Pasotti, L.; Sonnenschein, N.; Rama, E.; Cavaletti, M.; Magni, P.; Calvio, C.; Herrgard, M.J. Integration of enzymatic data in Bacillus subtilis genome-scale metabolic model improves phenotype predictions and enables in silico design of poly-gamma-glutamic acid production strains. Microb. Cell Fact. 2019, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Gu, Y.; Quan, Y.; Cao, M.; Gao, W.; Zhang, W.; Wang, S.; Yang, C.; Song, C. Improved poly-γ-glutamic acid production in Bacillus amyloliquefaciens by modular pathway engineering. Metab. Eng. 2015, 32, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhang, Z.; Feng, J.; Dang, Y.; Quan, Y.; Gu, Y.; Wang, S.; Song, C. Effects of MreB paralogs on poly-γ-glutamic acid synthesis and cell morphology in Bacillus amyloliquefaciens. FEMS Microbiol. Lett. 2016, 363, fnw187. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Gu, Y.; Sun, Y.; Han, L.; Yang, C.; Zhang, W.; Cao, M.; Song, C.; Gao, W.; Wang, S. Metabolic engineering of Bacillus amyloliquefaciens for poly-gamma-glutamic acid (γ-PGA) overproduction. Microb. Biotechnol. 2014, 7, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Scoffone, V.; Dondi, D.; Biino, G.; Borghese, G.; Pasini, D.; Galizzi, A.; Calvio, C. Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis. Biotechnol. Bioeng. 2013, 110, 2006–2012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xie, H.; He, Y.; Feng, J.; Gao, W.; Gu, Y.; Wang, S.; Song, C. Chromosome integration of the Vitreoscilla hemoglobin gene (vgb) mediated by temperature-sensitive plasmid enhances γ-PGA production in Bacillus amyloliquefaciens. FEMS Microbiol. Lett. 2013, 343, 127–134. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sha, Y.; Sun, T.; Qiu, Y.; Zhu, Y.; Zhan, Y.; Zhang, Y.; Xu, Z.; Li, S.; Feng, X.; Xu, H. Investigation of Glutamate Dependence Mechanism for Poly-gamma-glutamic Acid Production in Bacillus subtilis on the Basis of Transcriptome Analysis. J. Agric. Food Chem. 2019, 67, 6263–6274. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Quan, Y.; Gu, Y.; Liu, F.; Huang, X.; Shen, H.; Dang, Y.; Cao, M.; Gao, W.; Lu, X.; et al. Enhancing poly-gamma-glutamic acid production in Bacillus amyloliquefaciens by introducing the glutamate synthesis features from Corynebacterium glutamicum. Microb. Cell Fact. 2017, 16, 88. [Google Scholar] [CrossRef]
- Jiang, F.; Qi, G.; Ji, Z.; Zhang, S.; Liu, J.; Ma, X.; Chen, S. Expression of glr gene encoding glutamate racemase in Bacillus licheniformis WX-02 and its regulatory effects on synthesis of poly-γ-glutamic acid. Biotechnol. Lett. 2011, 33, 1837–1840. [Google Scholar] [CrossRef]
- Hu, L.X.; Zhao, M.; Hu, W.S.; Zhou, M.J.; Huang, J.B.; Huang, X.L.; Gao, X.L.; Luo, Y.N.; Li, C.; Liu, K.; et al. Poly-gamma-Glutamic Acid Production by Engineering a DegU Quorum-Sensing Circuit in Bacillus subtilis. ACS Synth. Biol. 2022, 11, 4156–4170. [Google Scholar] [CrossRef] [PubMed]
- Osera, C.; Amati, G.; Calvio, C.; Galizzi, A. SwrAA activates poly-gamma-glutamate synthesis in addition to swarming in Bacillus subtilis. Microbiology 2009, 155, 2282–2287. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Sheng, B.; Wang, H.; Shi, J.; Cai, D.; Yi, L.; Yang, S.; Wen, Z.; Ma, X.; Chen, S. Rewiring glycerol metabolism for enhanced production of poly-γ-glutamic acid in Bacillus licheniformis. Biotechnol. Biofuels. 2018, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhu, C.; Sheng, B.; Cai, D.; Wang, Q.; Zhiyou Wen, Z.; Chen, S. Improvement of glycerol catabolism in Bacillus licheniformis for production of poly-γ-glutamic acid. Appl. Microbiol. Biotechnol. 2017, 101, 7155–7164. [Google Scholar] [CrossRef] [PubMed]
- Halmschlag, B.; Steurer, X.; Putri, S.P.; Fukusaki, E.; Blank, L.M. Tailor-made poly-γ-glutamic acid production. Metab. Eng. 2019, 55, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Fu, J.; Yu, B.; Wang, L. Development of a conjugation-based genome editing system in an undomesticated Bacillus subtilis strain for poly-γ-glutamic acid production with diverse molecular masses. J. Agric. Food Chem. 2023, 71, 7734–7743. [Google Scholar] [CrossRef] [PubMed]
- Ermoli, F.; Bonta, V.; Vitali, G.; Calvio, C. SwrA as global modulator of the two-component system DegSU in Bacillus subtilis. Res. Microbiol. 2021, 172, 103877. [Google Scholar] [CrossRef]
- Smith, J.L.; Goldberg, J.M.; Grossman, A.D. Complete genome sequences of Bacillus subtilis subsp. subtilis Laboratory Strains 518 JH642 (AG174) and AG1839. Genome Announc. 2014, 2, e00663-14. [Google Scholar] [CrossRef]
- Leonard, C.G.; Housewright, R.D.; Thorne, C.B. Effects of some metallic ions on glutamyl polypeptide synthesis by Bacillus subtilis. J. Bacteriol. 1958, 76, 499–503. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Nguyen, Q.A.; Ferreira, R.C.; Ferreira, L.C.; Tran, L.T.; Schumann, W. Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid 2005, 54, 241–248. [Google Scholar] [CrossRef]
- Gibson, D.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Benda, M.; Schulz, L.M.; Stulke, J.; Rismondo, J. Influence of the ABC transporter YtrBCDEF of Bacillus subtilis on competence, biofilm formation and cell wall thickness. Front. Microbiol. 2021, 12, 587035. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Chen, G.; Zhang, Y.; Wu, K.; Liang, Z. Studies on the UV spectrum of poly (γ-glutamic acid) based on development of a simple quantitative method. Int. J. Biol. Macromol. 2012, 51, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Pasotti, L.; De Marchi, D.; Casanova, M.; Massaiu, I.; Bellato, M.; Cusella De Angelis, M.G.; Calvio, C.; Magni, P. Engineering endogenous fermentative routes in ethanologenic Escherichia coli W for bioethanol production from concentrated whey permeate. New Biotechnol. 2020, 57, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, H.; Wang, C.; Chang, J.W.; Chen, L.L. Construction and analysis of a genome-scale metabolic network for Bacillus licheniformis WX-02. Res. Microbiol. 2016, 167, 282–289. [Google Scholar] [CrossRef]
- Choi, H.S.; Lee, S.Y.; Kim, T.Y.; Woo, H.M. In silico identification of gene amplification targets for improvement of lycopene production. Appl. Environ. Microbiol. 2010, 76, 3097–3105. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef]
- Schellenberger, J.; Que, R.; Fleming, R.; Thiele, I.; Orth, J.D.; Feist, A.M.; Zielinski, D.C.; Bordbar, A.; Lewis, N.E.; Rahmanian, S.; et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 2011, 6, 1290–1307. [Google Scholar] [CrossRef] [PubMed]
- Guiziou, S.; Sauveplane, V.; Chang, H.J.; Clerte, C.; Declerck, N.; Jules, M.; Bonnet, J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016, 44, 7495–7508. [Google Scholar] [CrossRef]
- Usai, F.; Loi, G.; Scocozza, F.; Bellato, M.; Castagliuolo, I.; Conti, M.; Pasotti, L. Design and biofabrication of bacterial living materials with robust and multiplexed biosensing capabilities. Mater. Today Bio. 2022, 18, 100526. [Google Scholar] [CrossRef]
- Gao, W.; He, Y.; Zhang, F.; Zhao, F.; Huang, C.; Zhang, Y.; Zhao, Q.; Wang, S.; Yang, C. Metabolic engineering of Bacillus amyloliquefaciens LL3 for enhanced poly-γ-glutamic acid synthesis. Microb. Biotechnol. 2019, 12, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Shimanouchi, K.; Horiuchi, T.; Kamei, T.; Misono, H. Genetically engineered poly-gamma-glutamate producer from Bacillus subtilis ISW1214. Biosci. Biotechnol. Biochem. 2006, 70, 1794–1797. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Tran, L.P.; Itoh, Y. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 2004, 150, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- Pasotti, L.; Bellato, M.; Politi, N.; Casanova, M.; Zucca, S.; Cusella De Angelis, M.G.; Magni, P. A synthetic close-loop controller circuit for the regulation of an extracellular molecule by engineered bacteria. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 248–258. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).