Efficient Biosynthesis of Ectoine in Recombinant Escherichia coli by Biobrick Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
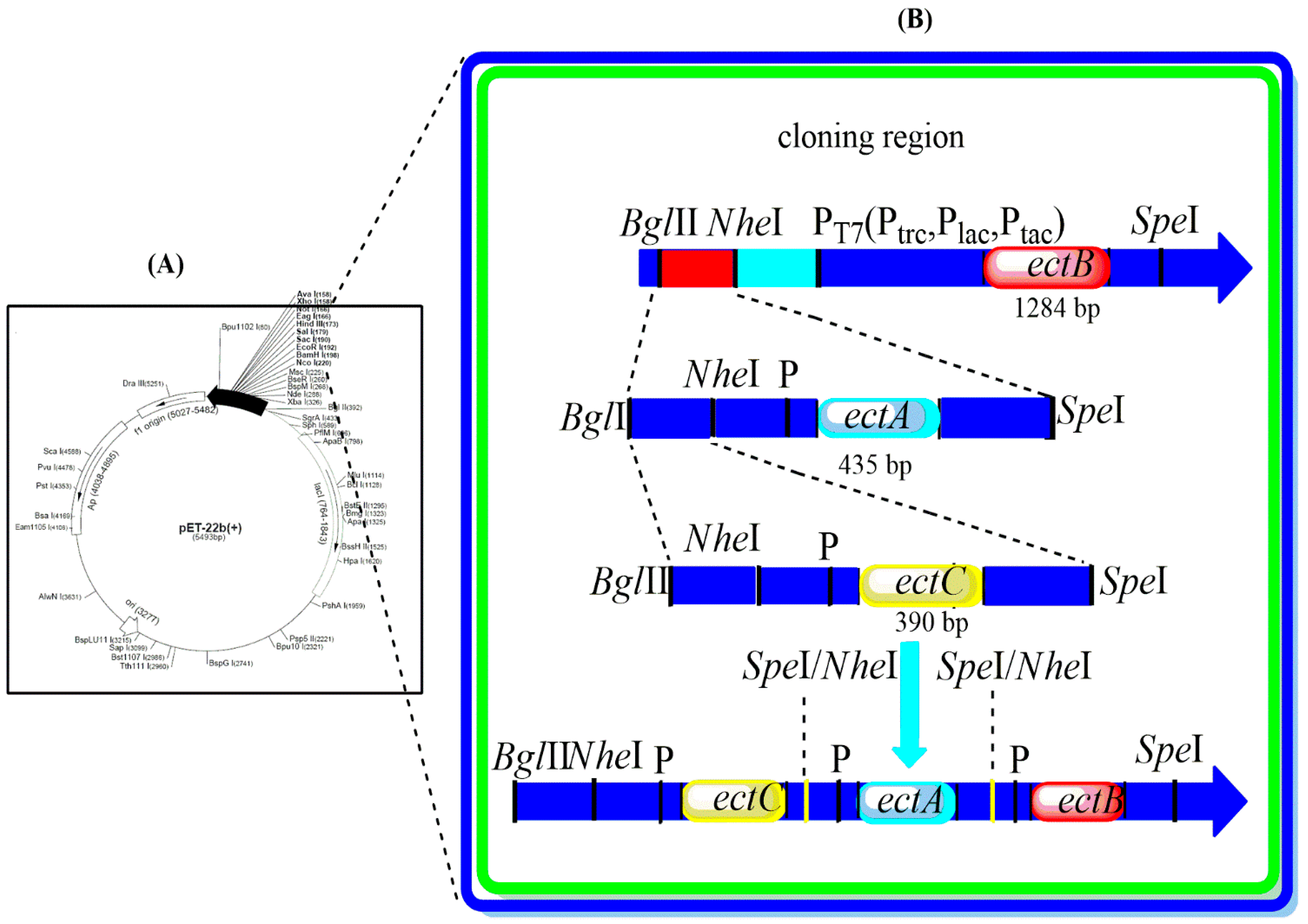
2.2. Construction of the Expression Plasmids
2.3. Heterogeneous Protein Purification
2.4. Protein Expression Analysis
2.5. Ectoine Detection
2.6. Optimizing Conditions for Ectoine Biosynthesis
2.6.1. Optimization of pH
2.6.2. Reaction Temperature
2.6.3. L-aspartic Acid Concentration
2.6.4. Optimization of KCl
2.6.5. Mass-to-Volume Ratio of Cell Pellets
2.6.6. Reaction Time
2.6.7. Protein Induction
2.6.8. Effect of Promoter
2.6.9. Bacterial Reusability
2.7. Stress Resistance Effects
2.7.1. Salt Tolerance
2.7.2. pH Tolerance
2.8. Sequencing Analysis
3. Results
3.1. Construction of Co-Expression Vectors
3.2. Purification Analysis of EctA, EctB, and EctC Recombinant Proteins
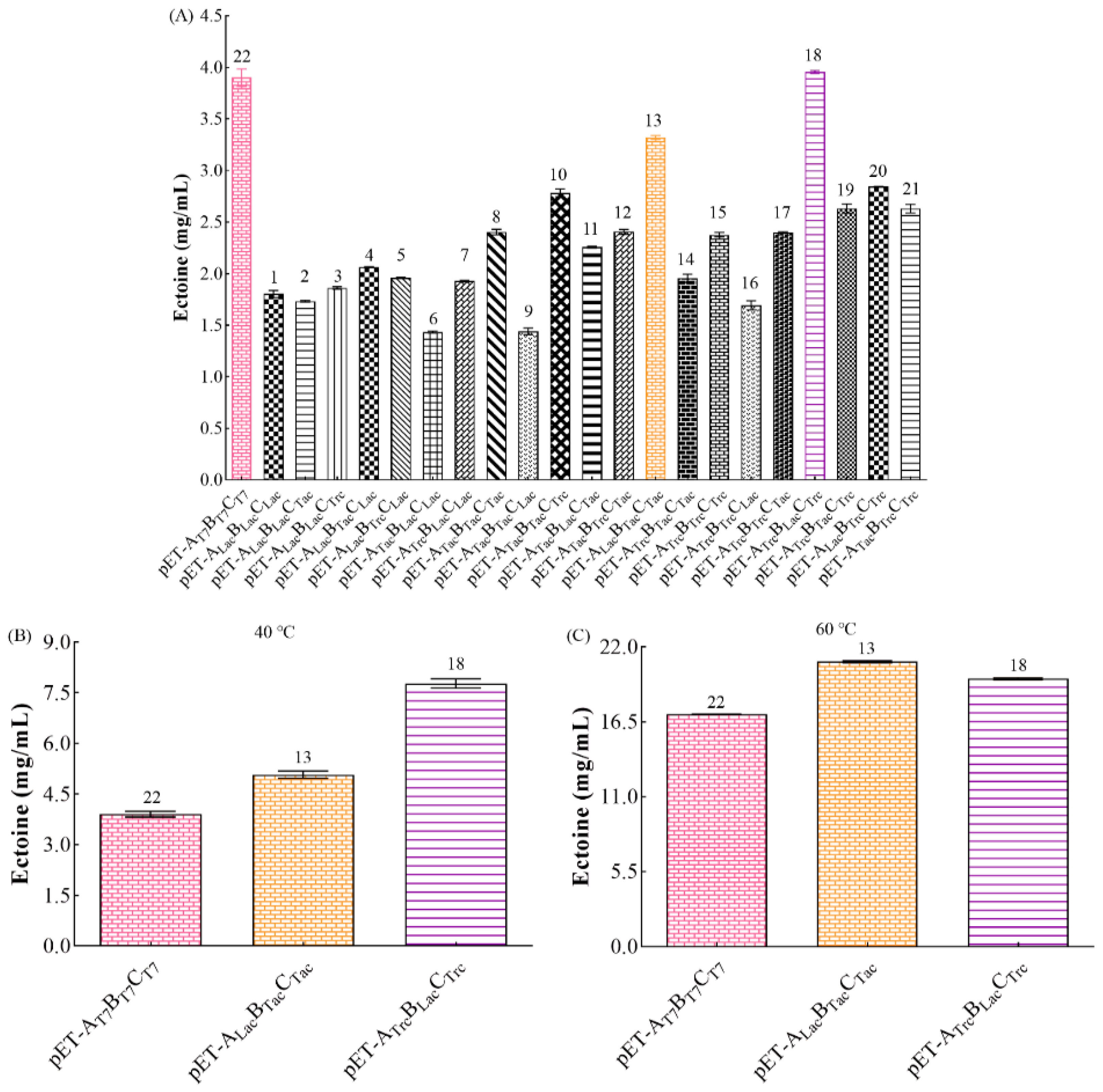
3.3. Optimizing Conditions for First Screening with T7 Promoter
3.4. Effect of Promoter
3.4.1. Optimization Conditions for Second Screening under Different Promoters
3.4.2. Comparison of Ectoine Production among Different Recombinant Strains
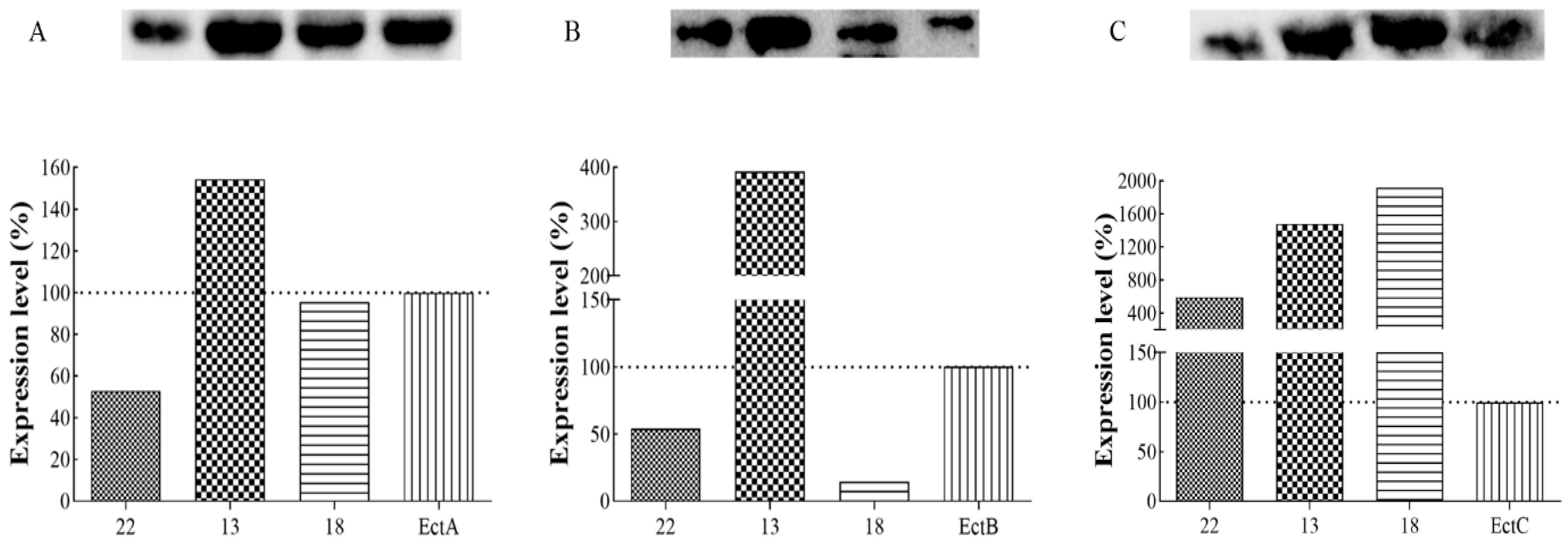
3.5. Protein Expression Analysis
3.6. Saline Alkali Tolerance Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Liu, K.; Kong, F.; Ye, T.; Wang, T. Multiple Functions of Compatible Solute Ectoine and Strategies for Constructing Overproducers for Biobased Production. Mol. Biotechnol. 2023, 66, 1772–1785. [Google Scholar] [CrossRef]
- Czech, L.; Poehl, S.; Hub, P.; Stöveken, N.; Bremer, E. Tinkering with osmotically controlled transcription allows enhanced production and excretion of ectoine and hydroxyectoine from a microbial cell factory. Appl. Environ. Microbiol. 2018, 84, e01772-17. [Google Scholar] [CrossRef]
- Wedeking, A.; Hagen-Euteneuer, N.; Gurgui, M.; Broere, R.; Lentzen, G.; Tolba, R.; Galinski, E.; van Echten-Deckert, G. A lipid anchor improves the protective effect of ectoine in inflammation. Curr. Med. Chem. 2014, 21, 2565–2572. [Google Scholar] [CrossRef]
- Unfried, K.; Krämer, U.; Sydlik, U.; Autengruber, A.; Bilstein, A.; Stolz, S.; Marini, A.; Schikowski, T.; Keymel, S.; Krutmann, J. Reduction of neutrophilic lung inflammation by inhalation of the compatible solute ectoine: A randomized trial with elderly individuals. Int. J. Chronic Obstr. Pulm. Dis. 2016, 11, 2573–2583. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L. Ectoine improves yield of biodiesel catalyzed by immobilized lipase. J. Mol. Catal. B Enzym. 2010, 62, 90–95. [Google Scholar] [CrossRef]
- Ma, Z.; Wu, C.; Zhu, L.; Chang, R.; Ma, W.; Deng, Y.; Chen, X. Bioactivity profiling of the extremolyte ectoine as apromising protectant and its heterologous production. 3 Biotech 2022, 12, 331. [Google Scholar] [CrossRef] [PubMed]
- Galinski, E.A.; Pfeiffer, H.P.; Trüper, H.G. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 1985, 149, 135–139. [Google Scholar] [CrossRef]
- Hobmeier, K.; Oppermann, M.; Stasinski, N.; Kremling, A.; Pflüger-Grau, K.; Kunte, H.J.; Marin-Sanguino, A. Metabolic engineering of Halomonas elongata: Ectoine secretion is increased by demand and supply driven approaches. Front. Microbiol. 2022, 13, 968983. [Google Scholar] [CrossRef]
- Kang, J.Y.; Lee, B.; Kim, J.A.; Kim, M.-S.; Kim, C.H. Identification and characterization of an ectoine biosynthesis gene cluster from Aestuariispira ectoiniformans sp. nov., isolated from seawater. Microbiol. Res. 2022, 254, 126898. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, H.; Shi, M.; Jiang, M.; Li, L.; Zheng, Y. Microbial production of ectoine and hydroxyectoine as high-value chemicals. Microb. Cell Factories 2021, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Peng, W.; Wang, T.; Li, Y.; Zhao, L.; Wang, X.; Li, Y.; Lin, L. Ectoine Production Using Novel Heterologous EctABC S. salarius from Marine Bacterium Salinicola salarius. Appl. Sci. 2021, 11, 6873. [Google Scholar] [CrossRef]
- Niknam, K.; Mirzaee, S. Silica Sulfuric Acid, an Efficient and Recyclable Solid Acid Catalyst for the Synthesis of 4,4′-(Arylmethylene) bis (1 H-pyrazol-5-ols). Synth. Commun. 2011, 41, 2403–2413. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, Z.; Zhao, M.; Ma, Y.; Luo, Z.; Li, S.; Xu, H. Metabolic engineering of Escherichia coli for ectoine production with a fermentation strategy of supplementing the amino donor. Front. Bioeng. Biotechnol. 2022, 10, 824859. [Google Scholar] [CrossRef] [PubMed]
- Sauer, T.; Galinski, E.A. Bacterial milking: A novel bioprocess for production of compatible solutes. Biotechnol. Bioeng. 1998, 57, 306–313. [Google Scholar] [CrossRef]
- Chen, W.-C.; Hsu, C.-C.; Lan, J.C.-W.; Chang, Y.-K.; Wang, L.-F.; Wei, Y.-H. Production and characterization of ectoine using a moderately halophilic strain Halomonas salina BCRC17875. J. Biosci. Bioeng. 2018, 125, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Parwata, I.P.; Wahyuningrum, D.; Suhandono, S.; Hertadi, R. Heterologous ectoine production in Escherichia coli: Optimization using response surface methodology. Int. J. Microbiol. 2019, 2019, 5745361. [Google Scholar] [CrossRef]
- Ng, H.S.; Wan, P.-K.; Kondo, A.; Chang, J.-S.; Lan, J.C.-W. Production and recovery of ectoine: A review of current state and future prospects. Processes 2023, 11, 339. [Google Scholar] [CrossRef]
- Matsumura, I. Methylase-assisted subcloning for high throughput BioBrick assembly. PeerJ 2020, 8, e9841. [Google Scholar] [CrossRef]
- Yamazaki, K.-I.; de Mora, K.; Saitoh, K. Biobrick-based ‘quick gene assembly’ in vitro. Synth. Biol. 2017, 2, ysx003. [Google Scholar] [CrossRef]
- Chaudhari, V.R.; Hanson, M.R. GoldBricks: An improved cloning strategy that combines features of Golden Gate and BioBricks for better efficiency and usability. Synth. Biol. 2021, 6, ysab032. [Google Scholar] [CrossRef]
- Shetty, R.P.; Endy, D.; Knight, T.F. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, N.; Li, J.-M.; Liu, H.; Abu-Romman, A.; Yaman, E.; Bian, F.; de Paiva, C.S.; Pflugfelder, S.C.; Li, D.-Q. Ectoine, from a Natural Bacteria Protectant to a New Treatment of Dry Eye Disease. Pharmaceutics 2024, 16, 236. [Google Scholar] [CrossRef]
- Shu, Z.; Zhang, X.; Wang, R.; Xing, J.; Li, Y.; Zhu, D.; Shen, G. Metabolic engineering of Halomonas campaniensis strain XH26 to remove competing pathways to enhance ectoine production. Sci. Rep. 2023, 13, 9732. [Google Scholar] [CrossRef]
- He, Y.-Z.; Gong, J.; Yu, H.-Y.; Tao, Y.; Zhang, S.; Dong, Z.-Y. High production of ectoine from aspartate and glycerol by use of whole-cell biocatalysis in recombinant Escherichia coli. Microb. Cell Factories 2015, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, P.; Chu, X.; Chen, J.; Zhang, H.; Rowley, D.C.; Wang, H. Metabolic pathway construction and optimization of Escherichia coli for high-level ectoine production. Curr. Microbiol. 2020, 77, 1412–1418. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Wu, X.; Zhang, C.; Xu, Q.; Chen, N.; Xie, X. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab. Eng. 2016, 36, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, J.; Wang, Y.; Du, G.; Kang, Z. Engineering Escherichia coli for high-yield production of ectoine. Green Chem. Eng. 2021, 4, 217–223. [Google Scholar] [CrossRef]
- Braman, J.C.; Sheffield, P.J. Seamless assembly of DNA parts into functional devices and higher order multi-device systems. PLoS ONE 2019, 14, 0199653. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, S.; Yu, K.; Xu, S.; Liu, M.; Zheng, J.; Yuan, W. Metabolic Engineering of Escherichia coli for Production of Ectoine. J. Food Nutr. Res. 2021, 9, 626–632. [Google Scholar] [CrossRef]
- Lang, Y.-J.; Bai, L.; Ren, Y.-N.; Zhang, L.-H.; Nagata, S. Production of ectoine through a combined process that uses both growing and resting cells of Halomonas salina DSM 5928 T. Extremophiles 2011, 15, 303–310. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martinez, R.A.; Martinez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the expression system on recombinant protein production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, B.; Chen, W.; Ye, K.; Shen, J.; Liu, P.; Wu, J.; Wang, H.; Chu, X. Highly efficient production of ectoine via an optimized combination of precursor metabolic modules in Escherichia coli BL21. Bioresour. Technol. 2023, 390, 129803. [Google Scholar] [CrossRef]
- Kadam, P.; Khisti, M.; Ravishankar, V.; Barvkar, V.; Dhotre, D.; Sharma, A.; Shouche, Y.; Zinjarde, S. Recent advances in production and applications of ectoine, a compatible solute of industrial relevance. Bioresour. Technol. 2023, 393, 130016. [Google Scholar] [CrossRef]
- Nie, Y.; Yao, M.; Jiang, G.; Yang, Y.; Wang, S.; Xu, H.; Liang, J.; Ren, X.; Tian, Y. Systems engineering of Escherichia coli for high-level l-alanine production. Food Biosci. 2024, 59, 103894. [Google Scholar] [CrossRef]
- Orhan, F.; Ceyran, E.; Akincioğlu, A. Optimization of ectoine production from Nesterenkonia xinjiangensis and one-step ectoine purification. Bioresour. Technol. 2023, 371, 128646. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Liu, H.; Liu, Y.; Mohsin, A.; Liu, Z.; Zheng, Y.; Xing, J.; Han, J.; Zhuang, Y. Temporal dynamics of stress response in Halomonas elongata to NaCl shock: Physiological, metabolomic, and transcriptomic insights. Microb. Cell Factories 2024, 23, 88. [Google Scholar] [CrossRef]
- Chen, J.; Qiao, D.; Yuan, T.; Feng, Y.; Zhang, P.; Wang, X.; Zhang, L. Biotechnological production of ectoine: Current status and prospects. Folia Microbiol. 2024, 69, 247–258. [Google Scholar] [CrossRef]
- Zou, Z.; Kaothien-Nakayama, P.; Ogawa-Iwamura, J.; Nakayama, H. Metabolic engineering of high-salinity-induced biosynthesis of γ-aminobutyric acid improves salt-stress tolerance in a glutamic acid-overproducing mutant of an ectoine-deficient Halomonas elongata. Appl. Environ. Microbiol. 2024, 90, e01905–e01923. [Google Scholar] [CrossRef]
- Rawat, M.; Chauhan, M.; Pandey, A. Extremophiles and their expanding biotechnological applications. Arch. Microbiol. 2024, 206, 247. [Google Scholar] [CrossRef]




| Primers. | Target Sequence (5′→3′) | Description |
|---|---|---|
| EctA F01 EctA R01 EctB-F01 EctB-R01 EctC-F01 EctC-R01 | GCATATGTGGGAATTAGTTAATC CCTCGAGCCTTAATGGTCCAATTC GCATATGAA ACA AACTGA TATG AGAGCTCGTTAGCAACAGGCTCAG GCATATGAA AGT AGTAGCTCT ACTCGAGTTCGTCATCAACTACTG | ectA gene cloning ectB gene cloning ectC gene cloning |
| Strain Type | Ectoine Amount (mg/mL/d) | Disadvantages or Features | Approach/Method | Reference |
|---|---|---|---|---|
| H. elongate DSM142 | 7.4 | salinity issues, low yield | bacterial milking | [14] |
| H. salina BCRC17875 | 7.61 | salinity issues, low yield | bacterial milking | [15] |
| E. coli BW25113 | 25.1 | salinity issues, low yield | bacterial milking | [24] |
| E. coli MG1655 | 12.7 | complicated, low yield | whole cell catalysis | [25] |
| E. coli W3110 | 31.37 | complicated operational design | fed-batch process | [26] |
| H. salina DSM 5928T | 14.86 | low yield, complicated design | fed-batch process | [30] |
| E. coli BL21(DE3) | 167.2 | high yield, efficient design | biobrick approach | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naeem, M.; Yuan, H.; Luo, S.; Zhang, S.; Wei, X.; He, G.; Zhao, B.; Ju, J. Efficient Biosynthesis of Ectoine in Recombinant Escherichia coli by Biobrick Method. Fermentation 2024, 10, 450. https://doi.org/10.3390/fermentation10090450
Naeem M, Yuan H, Luo S, Zhang S, Wei X, He G, Zhao B, Ju J. Efficient Biosynthesis of Ectoine in Recombinant Escherichia coli by Biobrick Method. Fermentation. 2024; 10(9):450. https://doi.org/10.3390/fermentation10090450
Chicago/Turabian StyleNaeem, Muhammad, Huiling Yuan, Suya Luo, Simei Zhang, Xinyue Wei, Guangzheng He, Baohua Zhao, and Jiansong Ju. 2024. "Efficient Biosynthesis of Ectoine in Recombinant Escherichia coli by Biobrick Method" Fermentation 10, no. 9: 450. https://doi.org/10.3390/fermentation10090450
APA StyleNaeem, M., Yuan, H., Luo, S., Zhang, S., Wei, X., He, G., Zhao, B., & Ju, J. (2024). Efficient Biosynthesis of Ectoine in Recombinant Escherichia coli by Biobrick Method. Fermentation, 10(9), 450. https://doi.org/10.3390/fermentation10090450







