Statistical Optimization and Purification of Cellulase Enzyme Production from Trichosporon insectorum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Cultural Conditions
2.2. Measurement of Cellulase Activities
2.3. Experimental Design
2.4. Statistical Analyzes
2.5. Enzyme Purification
2.5.1. Partial Purification of Enzymes
2.5.2. Enzyme Purification by Gel Filtration Chromatography
2.5.3. Thin Layer Chromatography
2.5.4. Determination of the Activities of Partially and Purified Enzymes
2.5.5. Determination of Protein Concentration
3. Results and Discussion
3.1. Experimental Responses
3.1.1. Production of CMCase
°C) − 0.13957 (pH * [FC]) + 0.05609 (T °C * [FC])
3.1.2. Production of FPase
3.2. Statistical Study
3.2.1. Production of CMCase
3.2.2. Production of FPase
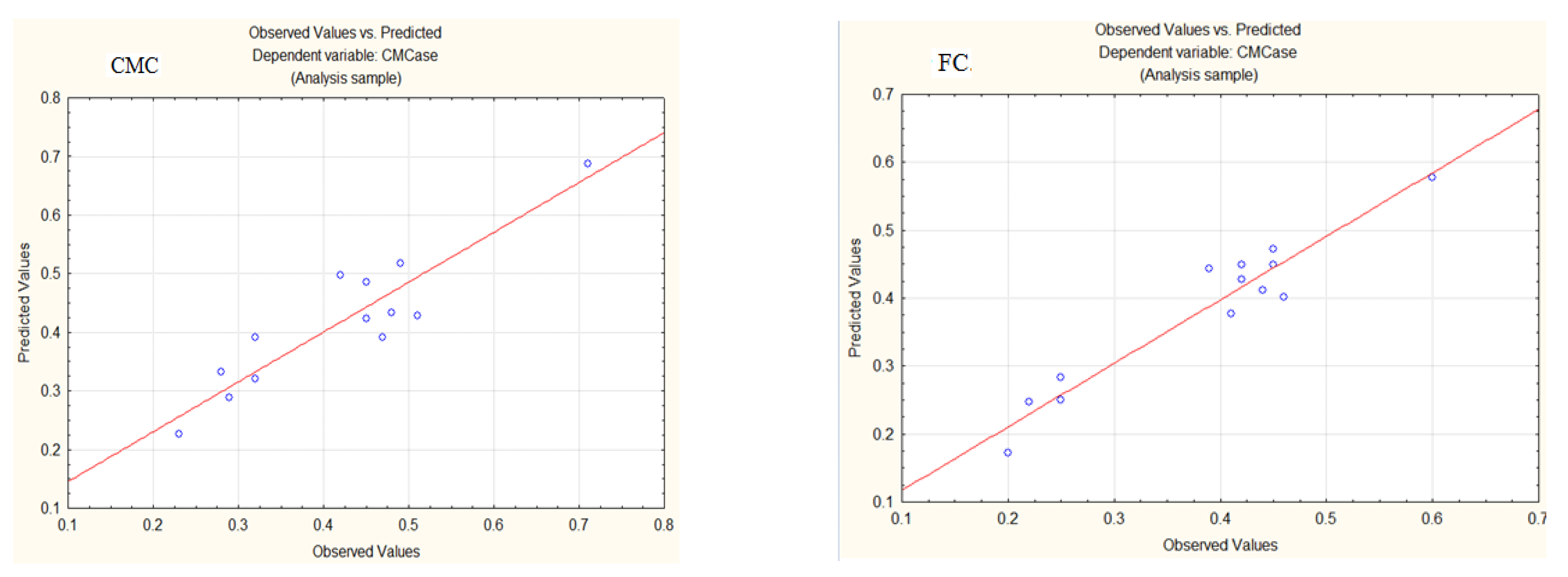
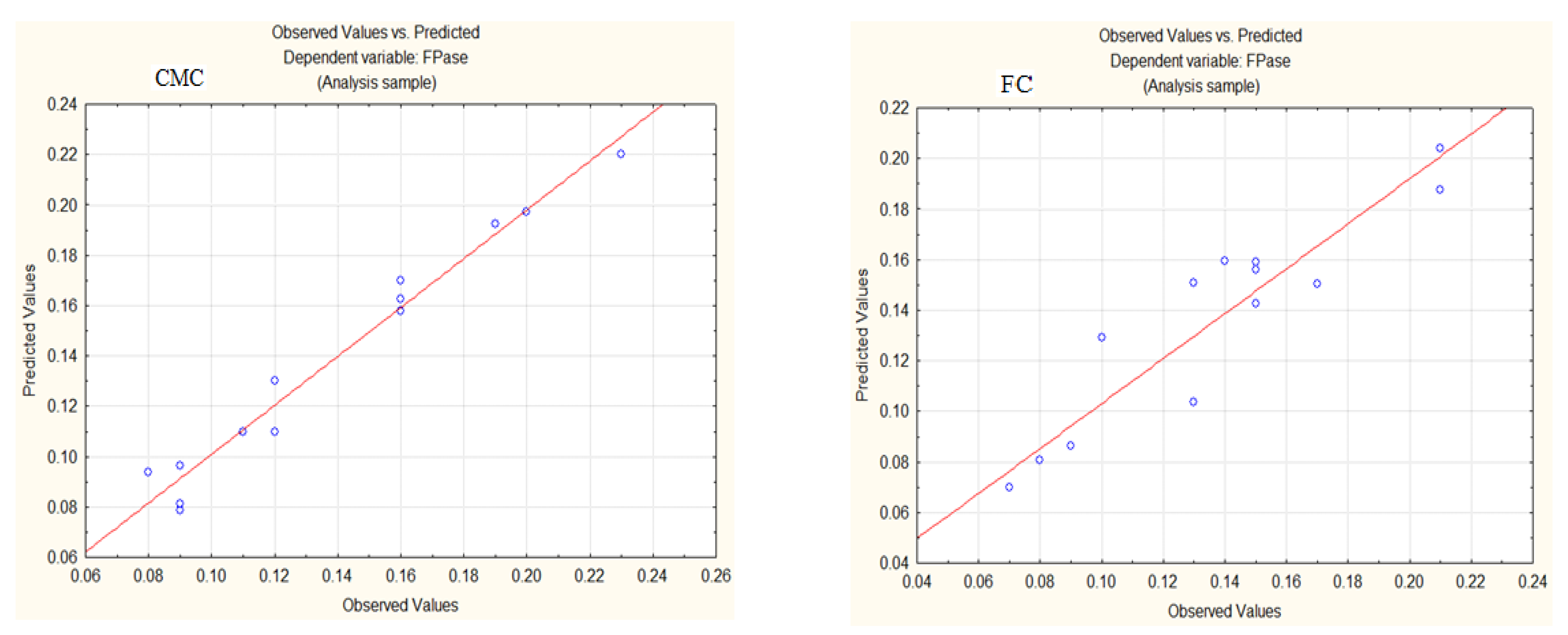
3.3. Model Validation
3.3.1. Production of CMCase
3.3.2. Production of FPase
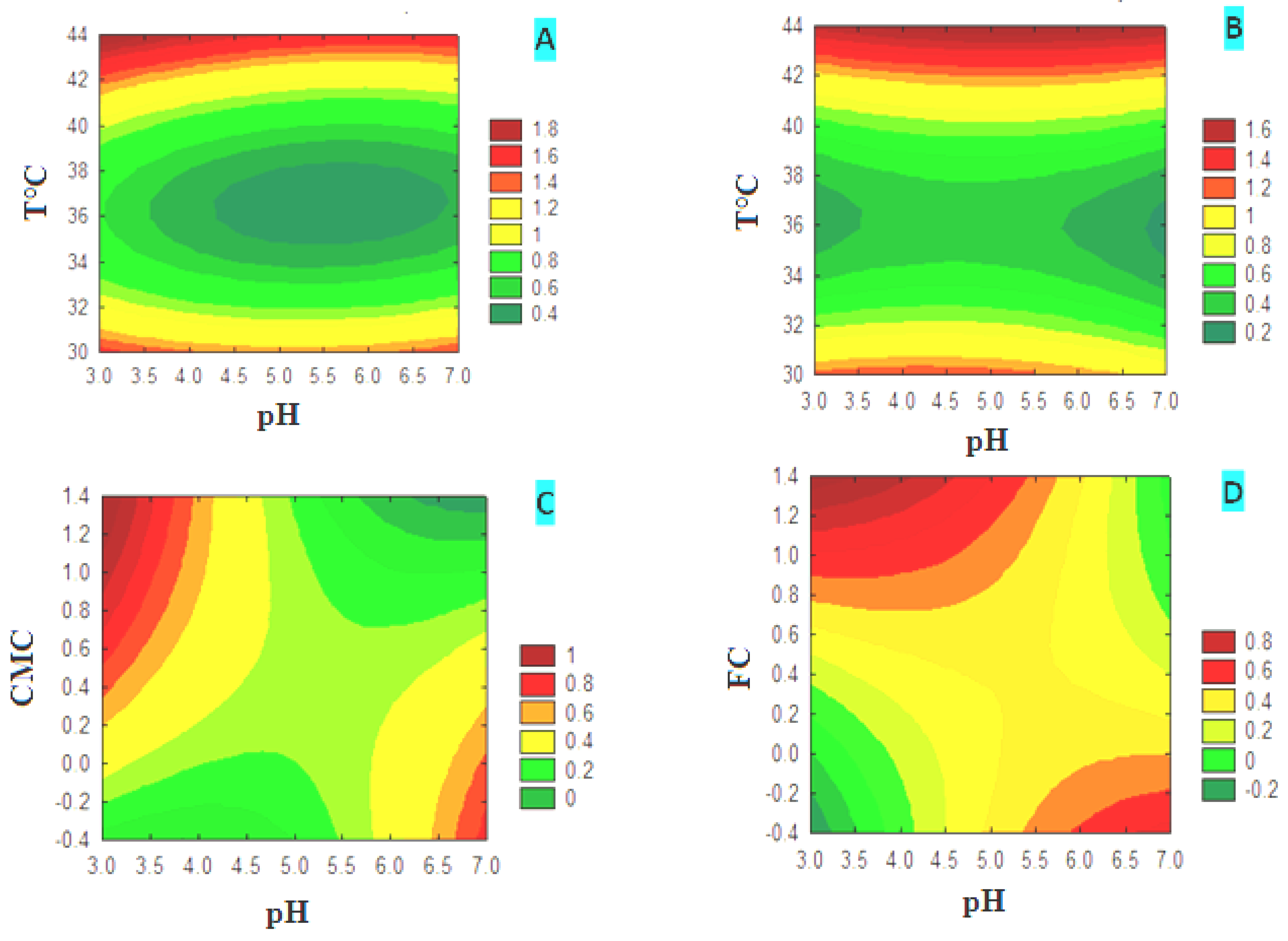
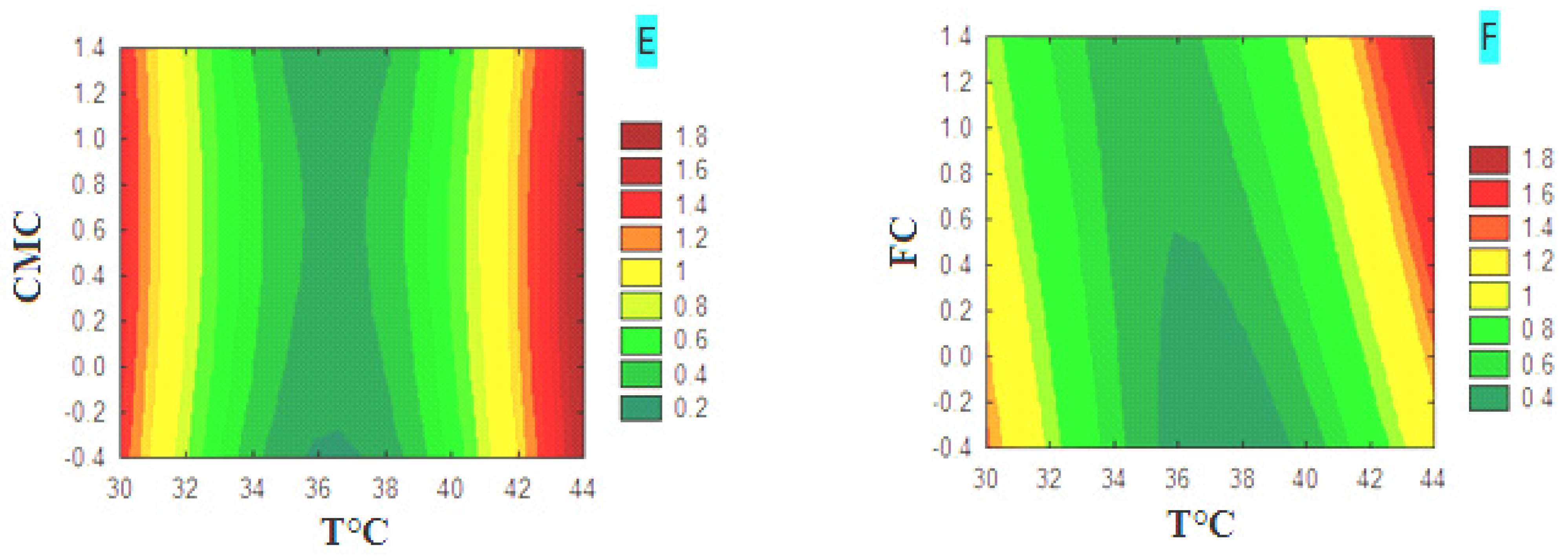
3.4. Optimization of Cellulase Production
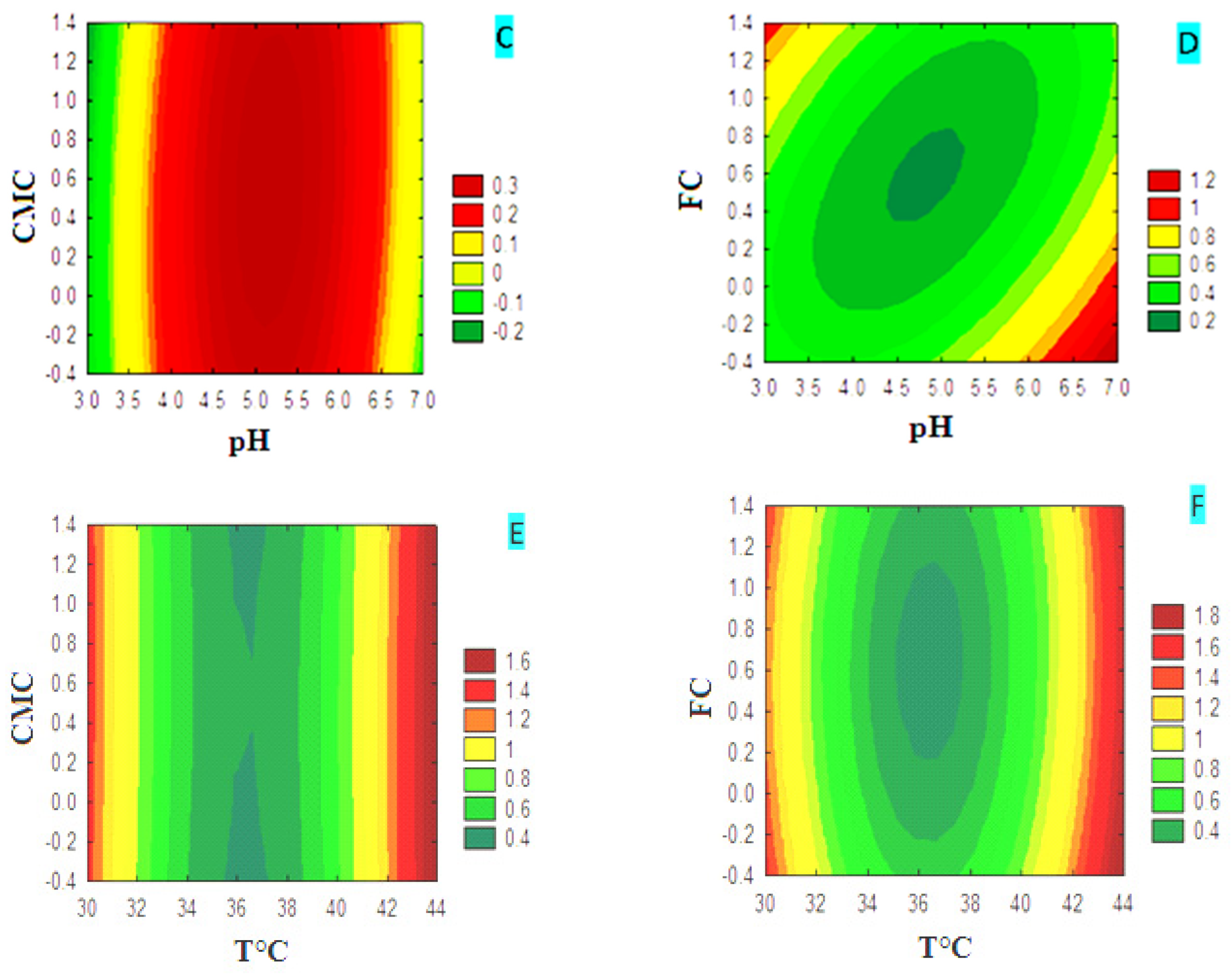
3.4.1. CMCase Contour Diagrams
3.4.2. FPase Contour Diagrams
3.5. Purification of the Enzymes Produced
3.5.1. Ammonium Sulfate Protein Precipitation
3.5.2. Fractionation by Molecular Exclusion Chromatography
Protein density of eluted fractions
Analysis of eluted fractions
3.5.3. Determination of the Enzymatic Activities of the Collected Fractions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahoo, A.; Das, P.K.; Patra, S.; Veeranki, V.D. Engineered Yeasts for the Production of Biofuel and Platform Chemicals; Elsevier: Amsterdam, The Netherlands, 2023; pp. 21–46. [Google Scholar] [CrossRef]
- Carvalho, J.K.; Panatta, A.A.S.; Silveira, M.A.D.; Tav, C.; Johann, S.; Rodrigues, M.L.F.; Martins, C.V.B. Yeasts isolated from a lotic continental environment in Brazil show potential to produce amylase, cellulase and protease. Biotechnol. Rep. 2021, 30, e00630. [Google Scholar] [CrossRef]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.S. Enzyme Immobilization Technologies and Industrial Applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef] [PubMed]
- Sukmawati, D.; Sondana, G.A.; Fikriyyah, N.N.; Afifah, Z.N.; Firhandini, A.; Khumaiya, U.; Komsiatun, D.A.; Asmara, Y.T.; Supiyani, A.; Puspitaningrum, R.; et al. Cellulase-producing yeast isolated from fermented cocoa beans as biocontrol for pathogenic mold chocolate fruit collected from Sentul, Jawa Barat, Indonesia. J. Phys. Conf. Ser. 2021, 1869, 012043. [Google Scholar] [CrossRef]
- Narisetty, V.; Tarafdar, A.; Bachan, N.; Madhavan, A.; Tiwari, A.; Chaturvedi, P.; Sindhu, R. An Overview of Cellulase Immobilization Strategies for Biofuel Production. Bioenergy Res. 2023, 16, 4–15. [Google Scholar] [CrossRef]
- Wanderley Siqueira, J.G.; Rodrigues, C.; de Souza Vandenberghe, S.P.; Woiciechowski, A.L.; Soccol, C.R. Current advances in on-site cellulase production and application on lignocellulosic biomass conversion to biofuels: A review. Biomass Bioenergy 2019, 132, 105419. [Google Scholar] [CrossRef]
- Wu, W.; Li, P.; Huang, L.; Wei, Y.; Li, J.; Zhang, L.; Jin, Y. The Role of Lignin Structure on Cellulase Adsorption and Enzymatic Hydrolysis. Biomass 2023, 3, 96–107. [Google Scholar] [CrossRef]
- Bautista-Cruz, A.; Aquino-Bolaños, T.; Hernández-Canseco, J.; Quiñones-Aguilar, E.E. Cellulolytic Aerobic Bacteria Isolated from Agricultural and Forest Soils: An Overview. Biology 2024, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Bajar, S.; Devi, A.; Pant, D. An overview on the recent developments in fungal cellulase production and their industrial applications. Bioresour. Technol. Rep. 2020, 14, 100652. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Zhang, Y.-H.P. Cellulases: Characteristics, Sources, Production, and Applications. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 131–146. [Google Scholar] [CrossRef]
- Auxenfans, T.; Husson, E.; Sarazin, C. Simultaneous pretreatment and enzymatic saccharification of (ligno) celluloses in aqueous-ionic liquid media: A compromise. Biochem. Eng. J. 2017, 117, 77–86. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Alhazmi, A.; Kausar, T.; Haque, S.; Singh, R.; Ramteke, P.; Mishra, P.K.; Tuohy, M.; Leitgeb, M.; et al. Technological advances for improving fungal cellulase production from fruit wastes for bioenergy application: A review. Environ. Pollut. 2021, 287, 117370. [Google Scholar] [CrossRef]
- Juhász, T.; Szengyel, Z.; Réczey, K.; Siika-Aho, M.; Viikari, L. Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem. 2005, 40, 3519–3525. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.-M.; Wan, C.; Haan, R.-D.; Bai, F.-W.; Zhao, X.-Q. Improved cellulase production in recombinant Saccharomyces cerevisiae by disrupting the cell wall protein-encoding gene CWP2. J. Biosci. Bioeng. 2019, 129, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Adebola, A.B.; Sarafadeen, K.O.; Idowu, A.A.; Monilola, W.S. Optimization of cellulase enzyme from sorghum straw by yeasts isolated from plant feeding—Termite Zonocerus variegatus. Food Appl. Biosci. J. 2019, 7, 81–99. [Google Scholar]
- Otero, D.M.; Cadaval, C.L.; Teixeira, L.M.; Rosa, C.A.; Sanzo, A.V.L.; Kalil, S.J. Screening of yeasts capable of producing cellulase-free xylanase. African J. Biotechnol. 2015, 14, 1961–1969. [Google Scholar] [CrossRef]
- Stevens, B.J.H.; Payne, J. Cellulase and Xylanase Production by Yeasts of the Genus Trichosporon. J. Gen. Microbiol. 1977, 100, 381–393. [Google Scholar] [CrossRef]
- Thongekkaew, J.; Ikeda, H.; Masaki, K.; Iefuji, H. An acidic and thermostable carboxymethyl cellulase from the yeast Cryptococcus sp. S-2: Purification, characterization and improvement of its recombinant enzyme production by high cell-density fermentation of Pichia pastoris. Protein Expr. Purif. 2008, 60, 140–146. [Google Scholar] [CrossRef]
- Rong, Y.; Zhang, L.; Chi, Z.; Wang, X. A carboxymethyl cellulase from a marine yeast (Aureobasidium pullulans 98): Its purification, characterization, gene cloning and carboxymethyl cellulose digestion. J. Ocean Univ. China. 2015, 14, 913–921. [Google Scholar] [CrossRef]
- Vyas, S. and Chhabra, M. Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour. Technol. 2017, 223, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Giesea, E.C.; Dussán, K.J.; Pierozzia, M.; Chandela, A.K.; Pagnoccad, F.C.; da Silvaa, S.S. Cellulase Production by Trichosporon laibachii. Orbital-Electron. J. Chem. 2017, 9, 271–278. [Google Scholar] [CrossRef]
- Ilić, N.; Milić, M.; Beluhan, S.; Dimitrijević-Branković, S. Cellulases: From Lignocellulosic Biomass to Improved Production. Energies 2023, 16, 3598. [Google Scholar] [CrossRef]
- Yang, J.; Yue, H.R.; Pan, L.Y.; Feng, J.X.; Zhao, S.; Suwannarangsee, S.; Chempreda, V.; Liu, C.G.; Zhao, X.Q. Fungal strain improvement for efficient cellulase production and lignocellulosic biorefinery: Current status and future prospects. Bioresour. Technol. 2023, 385, 129449. [Google Scholar] [CrossRef] [PubMed]
- Davison, S.A.; Den Haan, R.; Van Zyl, W.H. Exploiting strain diversity and rational engineering strategies to enhance recombinant cellulase secretion by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2020, 104, 5163–5184. [Google Scholar] [CrossRef]
- Egwim, E.C.; Agboola, A.O.; Saidu, A.N. Immobilization of cellulase and yeast for the hydrolysis and fermentation of pre-treated bagasse for ethanol production. Nig. J. Biotech. 2019, 36, 113–121. [Google Scholar] [CrossRef]
- Boro, M.; Kumar, A.; Chettri, D. Environmental Technology & Innovation Strategies inved in biofuel production from agro-based lignocellulose biomass. Environ. Technol. Innov. 2022, 28, 102679. [Google Scholar] [CrossRef]
- Bhat, M.K. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 2000, 18, 355–383. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Mushtaq, Q.; Tabssum, F.; Shakir, H.A.; Qazi, J.I. Carboxymethyl cellulase production optimization from newly isolated thermophilic Bacillus subtilis K-18 for saccharification using response surface methodology. AMB Express. 2017, 29, 7. [Google Scholar] [CrossRef] [PubMed]
- Touijer, H.; Benchemsi, N.; Ettayebi, M.; Janati Idrissi, A.; Chaouni, B.; Bekkari, H. Thermostable Cellulases from the Yeast Trichosporon sp. Enzyme Res. 2019, 2019, 2790414. [Google Scholar] [CrossRef]
- Rosyida, V.T.; Indrianingsih, A.W.; Maryana, R.; Wahono, S.K. Effect of Temperature and Fermentation Time of Crude Cellulase Production by Trichoderma reesei on Straw Substrate. Energy Procedia. 2015, 65, 368–371. [Google Scholar] [CrossRef]
- Miller, G.L. Use of DinitrosaIicyIic Acid Reagent for Determination of Reducing Sugar. Lancet 1959, 372, 475–488. [Google Scholar] [CrossRef]
- Parkhey, P.; Gupta, P.; Eswari, J.S. Optimization of Cellulase Production From Isolated Cellulolytic Bacterium: Comparison Between Genetic Algorithms, Simulated Annealing and Response Surface Methodology. Chem. Eng. Commun. 2016, 204, 28–38. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Anwar, Z.; Irshad, M.; Javid, A.; Hussain, A.; Ali, S. Biocatalysis and Agricultural Biotechnology Optimization of cellulase production from a novel strain of Aspergillus Tubingensis IMMIS2 through response surface methodology. Biocatal. Agric. Biotechnol. 2017, 12, 191–198. [Google Scholar] [CrossRef]
- Saini, R.; Saini, J.K.; Adsul, M.; Patel, A.K.; Mathur, A.; Tuli, D.; Singhania, R.R. Enhanced cellulase production by Penicillium oxalicum for bio-ethanol application. Bioresour. Technol. 2015, 188, 240–246. [Google Scholar] [CrossRef]
- Datta, R. Enzymatic degradation of cellulose in soil: A review. Heliyon 2024, 10, e24022. [Google Scholar] [CrossRef]
- Askari, H.; Soleimanian-zad, S.; Kadivar, M.; Shahbazi, S. Creating a novel genetic diversity of Trichoderma afroharzianum by γ -radiation for xylanase-cellulase production. Heliyon 2024, 10, e28349. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Raheja, Y.; Basotra, N.; Sharma, G.; Tsang, A.; Chadha, B.S. CRISPR/Cas9 mediated gene editing of transcription factor ACE1 for enhanced cellulase production in thermophilic fungus Rasamsonia emersonii. Fungal Biol. Biotechnol. 2023, 10, 18. [Google Scholar] [CrossRef]
- Boondaeng, A.; Keabpimai, J.; Trakunjae, C.; Vaithanomsat, P.; Srichola, P.; Niyomvong, N. Cellulase production under solid-state fermentation by Aspergillus sp. IN5: Parameter optimization and application. Heliyon 2024, 10, e26601. [Google Scholar] [CrossRef]
- Zhao, Z.; Gu, S.; Liu, D.; Liu, D.; Chen, B.; Li, J.; Tian, C. The putative methyltransferase LaeA regulates mycelium growth and cellulase production in Myceliophthora thermophila. Biotechnol. Biofuels Bioprod. 2023, 16, 58. [Google Scholar] [CrossRef]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef]
- Korsa, G.; Konwarh, R.; Masi, C.; Ayele, A.; Haile, S. Microbial cellulase production and its potential application for textile industries. Ann. Microbiol. 2023, 73, 13. [Google Scholar] [CrossRef]
- Li, H.; Zhang, M.; Zhang, Y.; Xu, X.; Zhao, Y.; Jiang, X.; Zhang, R.; Gui, Z. Characterization of Cellulose-Degrading Bacteria Isolated from Silkworm Excrement and Optimization of Its Cellulase Production. Polymers 2023, 15, 4142. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.; Barzkar, N.; Michaud, P.; Tamadoni Jahromi, S.; Babich, O.; Sukhikh, S.; Das, R.; Nahavandi, R. Cellulolytic and Xylanolytic Enzymes from Yeasts: Properties and Industrial Applications. Molecules 2022, 27, 3783. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Gupta, V.K.; Molina, G.; Rodriguez-Couto, S.; Manikanta, A.; Ramteke, P.W. Applications of fungal cellulases in biofuel production: Advances and limitations. Renew. Sustain. Energy Rev. 2017, 82, 2379–2386. [Google Scholar] [CrossRef]
- Abdel-Fattah, Y.R.; El-Helow, E.R.; Ghanem, K.M.; Lotfy, W.A. Application of factorial designs for optimization of avicelase production by a thermophilic Geobacillus isolate. Res. J. Microbiol. 2007, 2, 13–23. [Google Scholar]
- Deka, D.; Bhargavi, P.; Sharma, A.; Goyal, D.; Jawed, M.; Goyal, A. Enhancement of cellulase activity from a new strain of bacillus subtilis by medium optimization and analysis with various cellulosic substrates. Enzyme Res. 2011, 2011, 151656. [Google Scholar] [CrossRef]
- Korish, M. Production, Purification, Properties and Application of the Cellulases from a Wild type Strain of a Yeast isolate. Ph.D Thesis, Faculty of Biology of the Johannes Gutenberg-University Mainz, Mainz, Germany, 2003. [Google Scholar]



| Code | Levels | |||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| Substrate concentration [CS] (%) | X1 | 0.1 | 0.5 | 1 |
| Temperature (°C) | X2 | 33 | 36.5 | 40 |
| pH | X3 | 4 | 5 | 6 |
| Run No. | Variables (Unit) | Carbon Source CMC | Carbon Source FC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | [CS] (%) | CMCase Activity (IU/mL) | CMCase Activity (IU/mL) | |||||
| Observed | Predicted | Residual Value | Observed | Predicted | Residual Value | ||||
| 1 | 0 | 1 | 1 | 0.3900 | 0.4462 | −0.0562 | 0.7100 | 0.6762 | 0.0337 |
| 2 | −1 | 0 | 1 | 0.4100 | 0.3737 | 0.0362 | 0.4800 | 0.4275 | 0.0525 |
| 3 | 1 | −1 | 0 | 0.4500 | 0.4700 | −0.0200 | 0.4700 | 0.3837 | 0.0862 |
| 4 | −1 | −1 | 0 | 0.4200 | 0.4525 | −0.0325 | 0.4500 | 0.4212 | 0.0287 |
| 5 | 0 | 1 | −1 | 0.4500 | 0.4462 | 0.0037 | 0.5100 | 0.4287 | 0.0812 |
| 6 | 1 | 0 | 1 | 0.2000 | 0.1762 | 0.0237 | 0.2900 | 0.2950 | −0.0050 |
| 7 | −1 | 0 | −1 | 0.2200 | 0.2437 | −0.0237 | 0.2300 | 0.2250 | 0.0050 |
| 8 | −1 | 1 | 0 | 0.6000 | 0.5800 | 0.0200 | 0.4200 | 0.5062 | −0.0862 |
| 9 | 0 | 0 | 0 | 0.2500 | 0.2500 | 0.0000 | 0.3200 | 0.3200 | 0.0000 |
| 10 | 0 | −1 | 1 | 0.4200 | 0.4237 | −0.0037 | 0.3200 | 0.4012 | −0.0812 |
| 11 | 0 | 0 | 0 | 0.2500 | 0.2500 | 0.0000 | 0.3200 | 0.3200 | 0.0000 |
| 12 | 1 | 1 | 0 | 0.4400 | 0.4075 | 0.0325 | 0.4900 | 0.5187 | −0.0287 |
| 13 | 0 | −1 | −1 | 0.4600 | 0.4037 | 0.0562 | 0.4500 | 0.4837 | −0.0337 |
| 14 | 0 | 0 | 0 | 0.2500 | 0.2500 | 0.0000 | 0.3200 | 0.3200 | 0.0000 |
| 15 | 1 | 0 | −1 | 0.2500 | 0.2862 | −0.0362 | 0.2800 | 0.3325 | −0.0525 |
| Run No. | Variables (unit) | Carbon Source CMC | Carbon Source FC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| pH | T (°C) | [CS] (%) | FPase Activity (IU/mL) | FPase Activity (IU/mL) | |||||
| Observed | Predicted | Residual Value | Observed | Predicted | Residual Value | ||||
| 1 | 0 | 1 | 1 | 0.1900 | 0.19 | −0.0025 | 0.1500 | 0.1587 | −0.0087 |
| 2 | −1 | 0 | 1 | 0.0900 | 0.0775 | 0.0125 | 0.1300 | 0.1012 | 0.0287 |
| 3 | 1 | −1 | 0 | 0.1200 | 0.1100 | 0.0100 | 0.1700 | 0.1500 | 0.0200 |
| 4 | −1 | −1 | 0 | 0.1600 | 0.1700 | −0.0100 | 0.1500 | 0.1575 | −0.0075 |
| 5 | 0 | 1 | −1 | 0.2000 | 0.1975 | 0.0025 | 0.2100 | 0.1887 | 0.0212 |
| 6 | 1 | 0 | 1 | 0.0900 | 0.0975 | −0.0075 | 0.0900 | 0.0887 | 0.0012 |
| 7 | −1 | 0 | −1 | 0.0900 | 0.0825 | 0.0075 | 0.0800 | 0.0812 | −0.0012 |
| 8 | −1 | 1 | 0 | 0.1200 | 0.1300 | −0.0100 | 0.1400 | 0.1600 | −0.0200 |
| 9 | 0 | 0 | 0 | 0.1100 | 0.1100 | 0.0000 | 0.0700 | 0.0700 | 0.0000 |
| 10 | 0 | −1 | 1 | 0.1600 | 0.1625 | −0.0025 | 0.1300 | 0.1512 | −0.0212 |
| 11 | 0 | 0 | 0 | 0.1100 | 0.1100 | 0.0000 | 0.0700 | 0.0700 | 0.0000 |
| 12 | 1 | 1 | 0 | 0.2300 | 0.2200 | 0.0100 | 0.2100 | 0.2025 | 0.0075 |
| 13 | 0 | −1 | −1 | 0.1600 | 0.1575 | 0.0025 | 0.1500 | 0.1412 | 0.0087 |
| 14 | 0 | 0 | 0 | 0.1100 | 0.1100 | 0.0000 | 0.0700 | 0.0700 | 0.0000 |
| 15 | 1 | 0 | −1 | 0.0800 | 0.0925 | −0.0125 | 0.1000 | 0.1287 | −0.0287 |
| SS | df | MS | F Value | p Value | Effect | |
|---|---|---|---|---|---|---|
| Model | 0.181529 | 9 | 0.020170 | 7.984293 | 0.017037 | Significant |
| Interception | 0.088287 | 1 | 0.88287 | 34.94865 | 0.001973 | Significant |
| pH | 0.000656 | 1 | 0.000656 | 0.25966 | 0.632041 | Not significant |
| pH2 | 0.004206 | 1 | 0.004206 | 1.66486 | 0.253394 | Not significant |
| T °C | 0.114625 | 1 | 0.114625 | 45.37443 | 0.001094 | Significant |
| T °C2 | 0.138606 | 1 | 0.138606 | 54.86738 | 0.000706 | Significant |
| [CS] | 0.005124 | 1 | 0.005124 | 2.02839 | 0.213687 | Not significant |
| [CS]CMC2 | 0.000751 | 1 | 0.000751 | 0.29742 | 0.608940 | Not significant |
| pH * T °C | 0.009025 | 1 | 0.009025 | 3.57256 | 0.117342 | Not significant |
| pH * [CS] | 0.014645 | 1 | 0.014645 | 5.79720 | 0.061034 | Not significant |
| T °C * [CS] | 0.000249 | 1 | 0.000249 | 0.09863 | 0.766162 | Not significant |
| Error | 0.012631 | 5 | 0.002526 |
| SS | df | MS | F Value | p Value | Effect | |
|---|---|---|---|---|---|---|
| Model | 0.181847 | 9 | 0.020205 | 3.165695 | 0.108702 | Not significant |
| Interception | 0.075109 | 1 | 0.075109 | 11.76784 | 0.018627 | Significant |
| pH | 0.000341 | 1 | 0.000341 | 0.05346 | 0.826305 | Not significant |
| pH2 | 0.001477 | 1 | 0.001477 | 0.23140 | 0.65080 | Not significant |
| T °C | 0.092586 | 1 | 0.092586 | 14.50613 | 0.012517 | Significant |
| T °C2 | 0.091592 | 1 | 0.091592 | 14.35040 | 0.012782 | Significant |
| [CS] | 0.010401 | 1 | 0.010401 | 1.62955 | 0.257840 | Not significant |
| [CS]CMC2 | 0.000873 | 1 | 0.000873 | 0.13672 | 0.726713 | Not significant |
| pH * T °C | 0.000625 | 1 | 0.000625 | 0.09792 | 0.766968 | Not significant |
| pH * [CS] | 0.015876 | 1 | 0.015876 | 2.48743 | 0.175586 | Not significant |
| T °C * [CS] | 0.031411 | 1 | 0.031411 | 4.92139 | 0.077282 | Not significant |
| Error | 0.031913 | 5 | 0.006383 |
| Dependent Variable | R | R2 | R2 Adjusted |
|---|---|---|---|
| CMC | 0.966926 | 0.934946 | 0.817847 |
| FC | 0.922338 | 0.850707 | 0.581980 |
| SS | df | MS | F Value | p Value | Effect | |
|---|---|---|---|---|---|---|
| Model | 0.028706 | 9 | 0.003190 | 18.38383 | 0.002549 | Significant |
| Interception | 0.018335 | 1 | 0.018335 | 105.6788 | 0.000150 | Significant |
| pH | 0.000545 | 1 | 0.000545 | 3.1400 | 0.136593 | Non-significant |
| pH2 | 0.001667 | 1 | 0.001667 | 9.6100 | Significant | |
| T °C | 0.020584 | 1 | 0.020584 | 118.6394 | 0.000113 | Significant |
| T °C2 | 0.017452 | 1 | 0.017452 | 100.5892 | 0.000169 | Significant |
| [CS] | 0.000014 | 1 | 0.000014 | 0.0802 | 0.788331 | Non-significant |
| [CS]2 | 0.000006 | 1 | 0.000006 | 0.0331 | 0.862854 | Non-significant |
| pH * T °C | 0.005625 | 1 | 0.005625 | 32.4213 | 0.002330 | Significant |
| pH * [CS] | 0.000008 | 1 | 0.000008 | 0.0442 | 0.841779 | Non-significant |
| T °C * [CS] | 0.000025 | 1 | 0.000025 | 0.1432 | 0.720645 | Non-significant |
| Error | 0.000867 | 5 | 0.000173 |
| SS | df | MS | F Value | p Value | Effect | |
|---|---|---|---|---|---|---|
| Model | 0.027945 | 9 | 0.003105 | 4.442367 | 0.057607 | Non-significant |
| Interception | 0.021001 | 1 | 0.021001 | 30.04645 | 0.002756 | Significant |
| pH | 0.001536 | 1 | 0.001536 | 2.19739 | 0.198345 | Non-significant |
| pH2 | 0.001298 | 1 | 0.001298 | 1.85716 | 0.231119 | Non-significant |
| T °C | 0.022677 | 1 | 0.022677 | 32.44350 | 0.002327 | Significant |
| T °C2 | 0.022898 | 1 | 0.022898 | 32.76035 | 0.002277 | Significant |
| [CS] | 0.000594 | 1 | 0.000594 | 0.84956 | 0.398968 | Non-significant |
| [CS]2 | 0.000512 | 1 | 0.000512 | 0.73207 | 0.431294 | Non-significant |
| pH * T °C | 0.000625 | 1 | 0.000625 | 0.89419 | 0.387765 | Non-significant |
| pH * [CS] | 0.001086 | 1 | 0.001086 | 1.55370 | 0.267803 | Non-significant |
| T °C * [CS] | 0.000344 | 1 | 0.000344 | 0.49252 | 0.514120 | Non-significant |
| Error | 0.003495 | 5 | 0.000699 |
| Dependent Variable | R | R2 | R2 Adjusted |
|---|---|---|---|
| CMC | 0.985224 | 0.970667 | 0.917867 |
| FC | 0.942785 | 0.888843 | 0.688760 |
| Enzymes | Activity (UI/mL) | Specific Activity (U/mg) |
|---|---|---|
| CMCase | 0.525 | 1.876 |
| Xylanase | 0.942 | 3.365 |
| ß-glucosidase | 0.121 | 0.430 |
| ß-Xylosidase | 0.004 | 0.013 |
| Proteins (mg/mL) | Enzymatic Activities | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CMCase | Xylanase | Β-Glucosidase | Β-Xylosidase | ||||||
| UI/mL | U/mg | UI/mL | U/mg | UI/mL | U/mg | UI/mL | U/mg | ||
| Pre-purified extract | 0.28 | 0.438 | 1.563 | 0.785 | 2.804 | 0.106 | 0.380 | 0.0035 | 0.0125 |
| Collection 1 | 0.00790 | nd* | nd* | nd* | nd* | 0.000261 | 0.033019 | 0.000261 | 0.0330 |
| Collection 2 | 0.09217 | 0.0671 | 0.729 | 0.124 | 1.348 | 0.00417 | 0.045 | 0.000470 | 0.00510 |
| Collection 3 | 0.21566 | 0.0152 | 0.141 | 0.107 | 0.498 | 0.00867 | 0.0402 | nd* | nd* |
| Collection 4 | 0.19851 | 0.00136 | 0.0137 | 0.126 | 0.636 | 0.0302 | 0.1520 | 0.00933 | 0.04702 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touijer, H.; Benchemsi, N.; Irfan, M.; Tramice, A.; Slighoua, M.; Mothana, R.A.; Alanzi, A.R.; Dalila, B.; Bekkari, H. Statistical Optimization and Purification of Cellulase Enzyme Production from Trichosporon insectorum. Fermentation 2024, 10, 453. https://doi.org/10.3390/fermentation10090453
Touijer H, Benchemsi N, Irfan M, Tramice A, Slighoua M, Mothana RA, Alanzi AR, Dalila B, Bekkari H. Statistical Optimization and Purification of Cellulase Enzyme Production from Trichosporon insectorum. Fermentation. 2024; 10(9):453. https://doi.org/10.3390/fermentation10090453
Chicago/Turabian StyleTouijer, Hanane, Najoua Benchemsi, Muhammad Irfan, Annabella Tramice, Meryem Slighoua, Ramzi A. Mothana, Abdullah R. Alanzi, Bousta Dalila, and Hicham Bekkari. 2024. "Statistical Optimization and Purification of Cellulase Enzyme Production from Trichosporon insectorum" Fermentation 10, no. 9: 453. https://doi.org/10.3390/fermentation10090453







