New Halophilic Community Degrades Plastics: A Metagenomic Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample
2.2. Media and Plastic Degradation Conditions
2.3. Estimation of the Gravimetric Weight of the Plastics
2.4. Esterase Activity Determination
2.5. Polycaprolactone Biodegradation
2.6. Biofilm Formation Estimation
2.7. Bacterial Hydrophobicity Assay
2.8. Scanning Electron Microscopy (SEM)
2.9. DNA Isolation, Metagenome Sequencing, and Bioinformatics Analysis
3. Results
3.1. Isolation of the Plastic-Degrading Halophilic Community
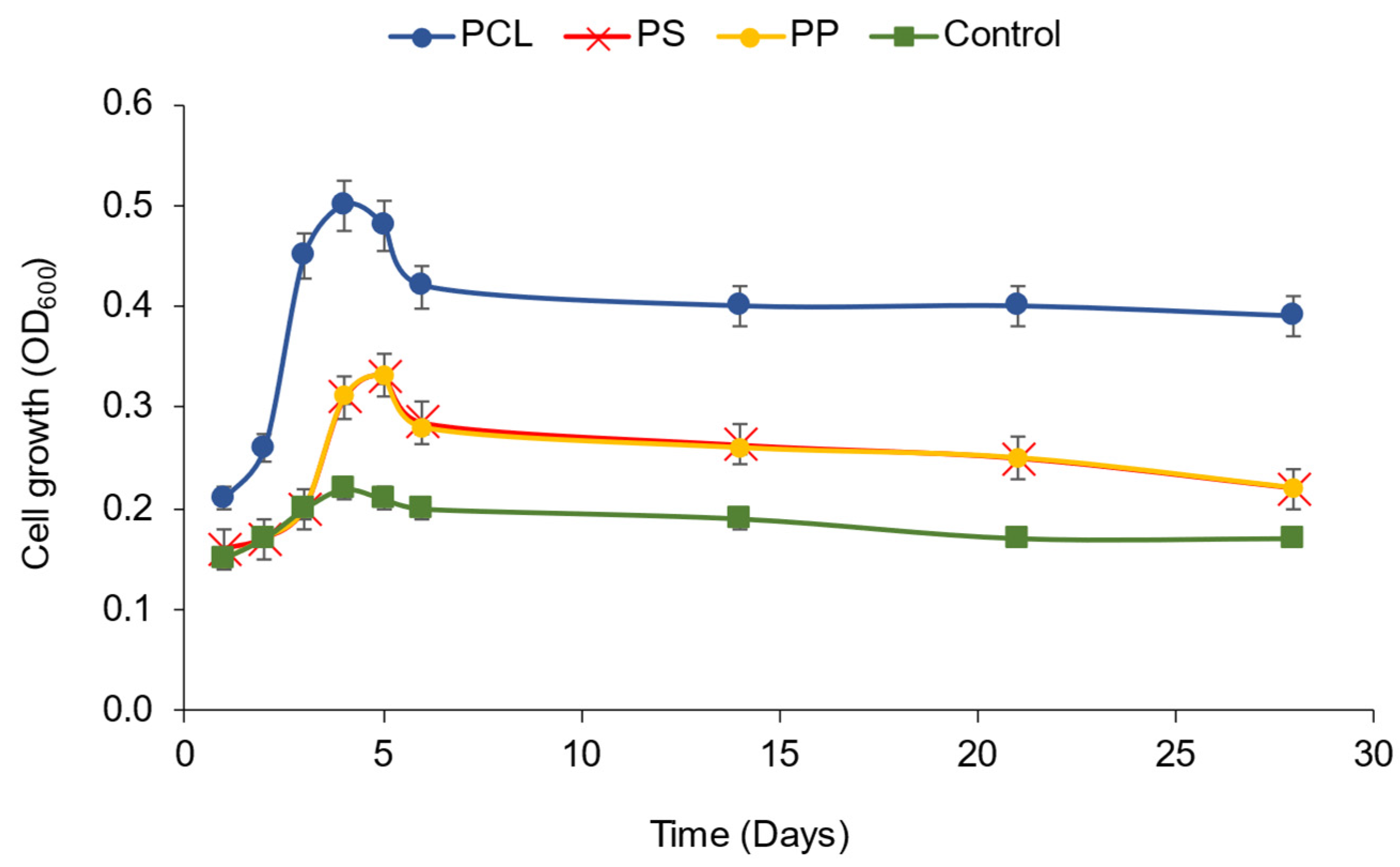
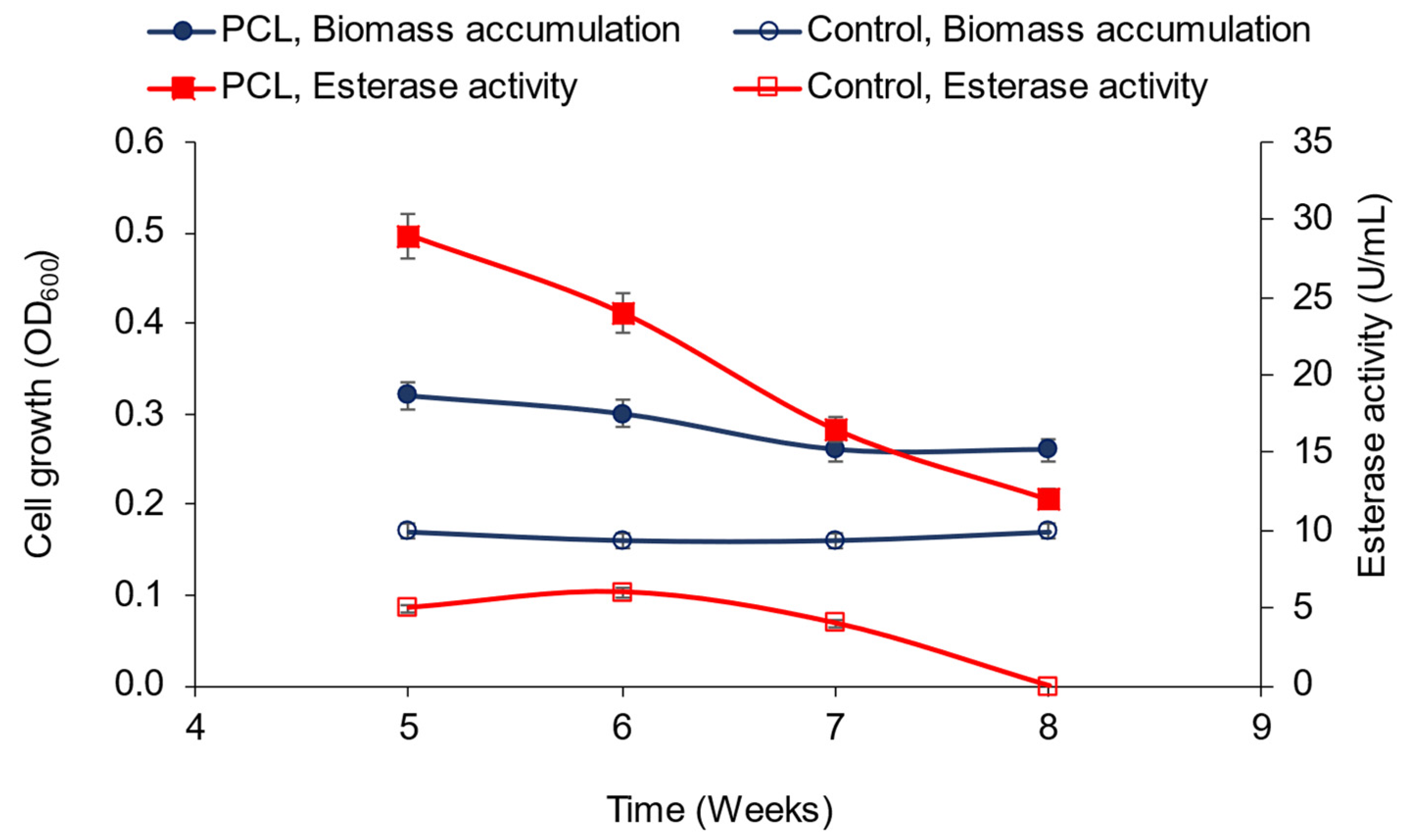
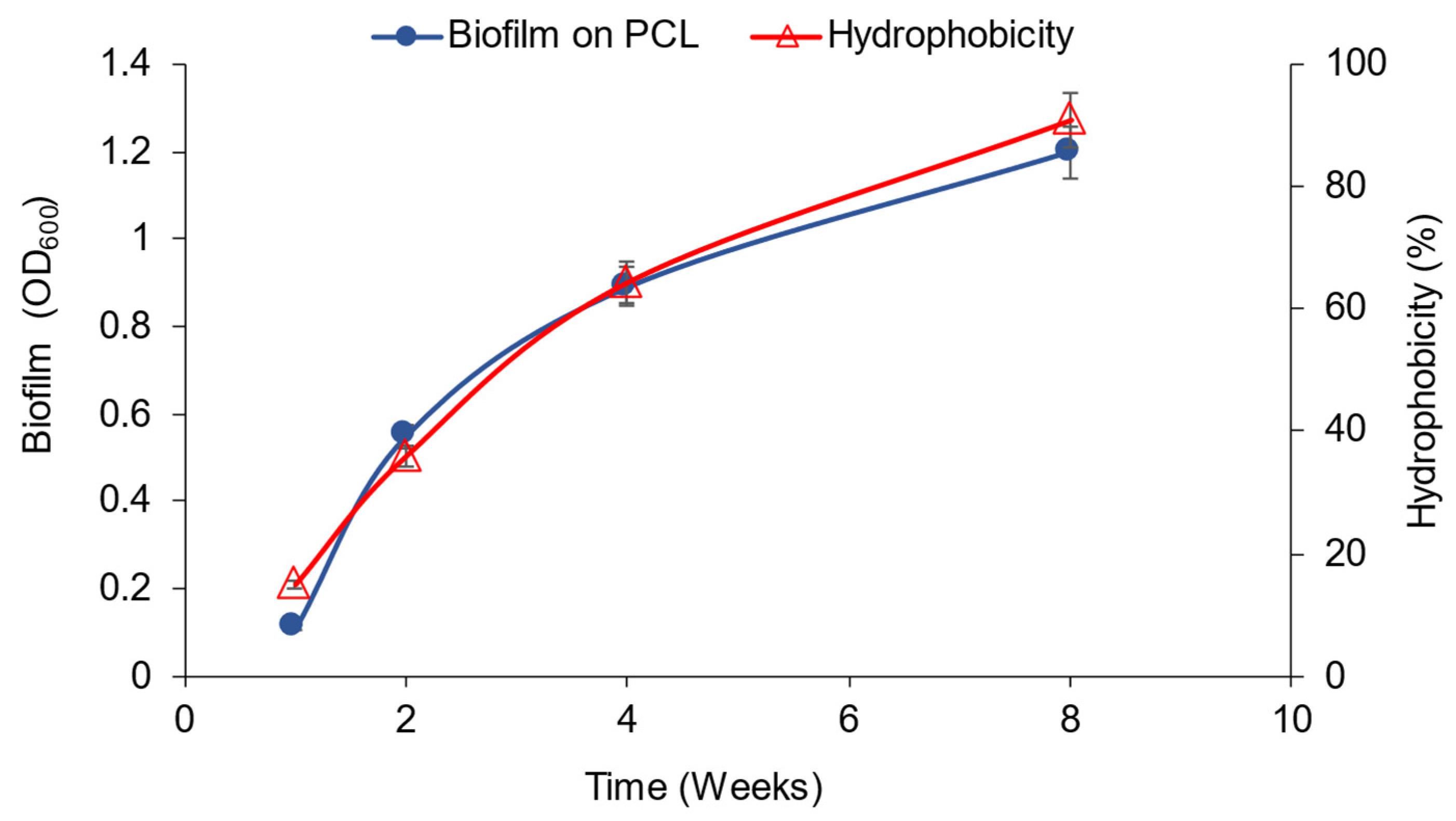
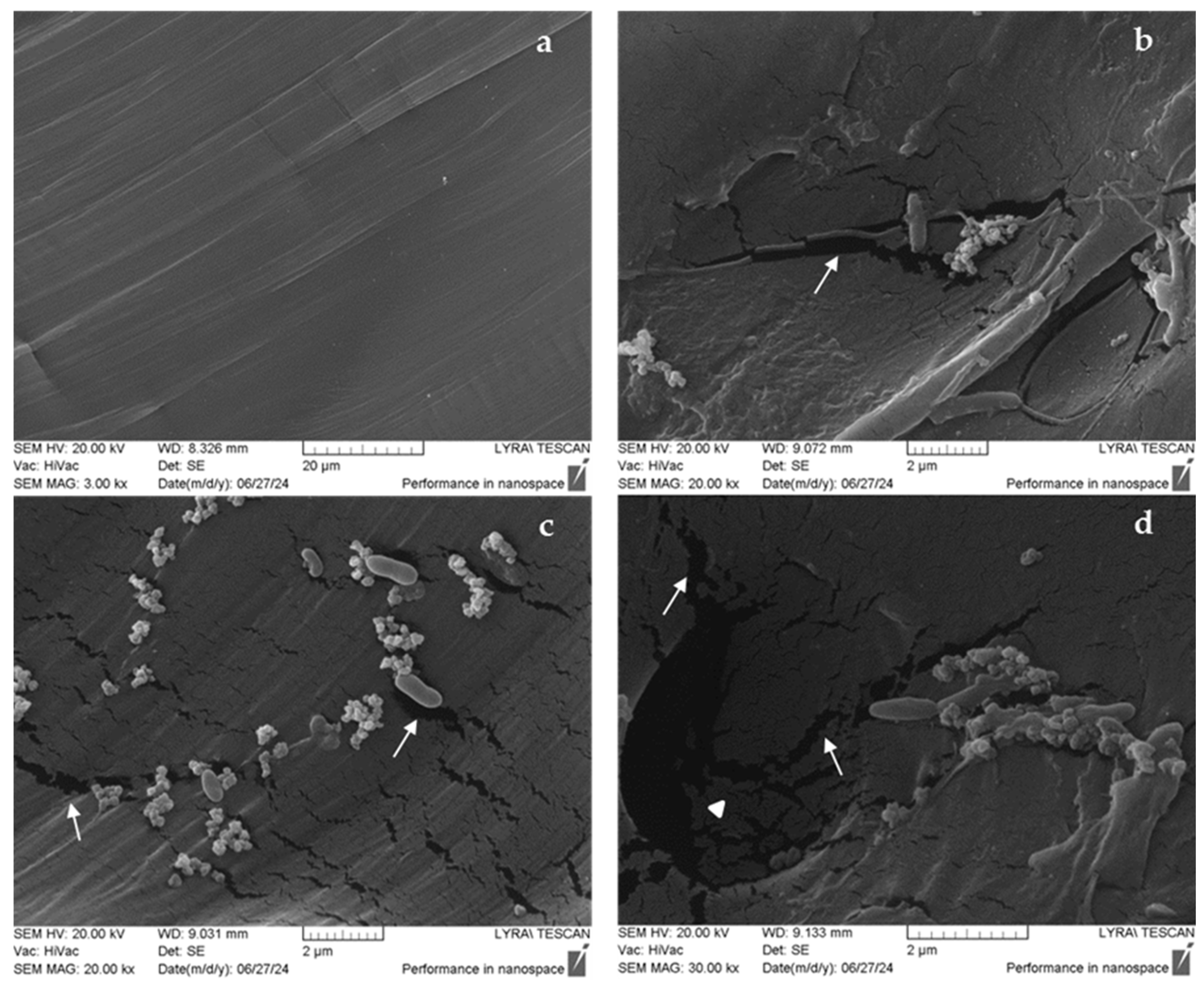
3.2. Polycaprolactone (PCL) Degradation by the Novel Halophilic Community
3.3. Bacterial Biodiversity of the Halophilic Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pilapitiya, P.G.C.N.T.; Ratnayake, A.S. The world of plastic waste: A review. Clean. Mater. 2024, 11, 100220. [Google Scholar] [CrossRef]
- The Market Value of Plastics Worldwide Will Be in 2023, with a Forecast for 2033. Available online: https://www.statista.com/statistics/1060583/global-market-value-of-plastic/ (accessed on 28 January 2025).
- Carpenter, E.J.; Smith, K.L. Plastics on the Sargasso sea surface. Science 1972, 175, 1240–1241. [Google Scholar] [CrossRef]
- Williams, A.T.; Rangel-Buitrago, N. The past, present, and future of plastic pollution. Mar. Pollut. Bull. 2022, 176, 113429. [Google Scholar] [CrossRef]
- Terrazas-López, R.; Guadarrama-Guzman, P.; Sujitha, S.B.; Arreola-Mendoza, L.; Ponniah, J.M. The Occurrence of Microplastics in the Marine Food Web in Latin America: Insights on the Current State of Knowledge and Future Perspectives. Sustainability 2024, 16, 5905. [Google Scholar] [CrossRef]
- Thushari, G.G.N.; Senevirathna, J.D.M. Plastic pollution in the marine environment. Heliyon 2020, 6, e04709. [Google Scholar] [CrossRef]
- Viel, T.; Manfra, L.; Zupo, V.; Libralato, G.; Cocca, M.; Costantini, M. Biodegradation of Plastics Induced by Marine Organisms: Future Perspectives for Bioremediation Approaches. Polymers 2023, 15, 2673. [Google Scholar] [CrossRef]
- Boldrini, A.; Gaggelli, N.; Falcai, F.; Polvani, A.; Talarico, L.; Galgani, L.; Cirrone, R.; Liu, X.; Loiselle, S. Emerging Contaminants from Bioplastic Pollution in Marine Waters. Water 2024, 16, 3676. [Google Scholar] [CrossRef]
- Hurley, R.; Horton, A.; Lusher, A.; Nizzetto, L. Chapter 7—Plastic waste in the terrestrial environment. In Plastic Waste and Recycling; Letcher, T.M., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 163–193. [Google Scholar] [CrossRef]
- Rai, M.; Pant, G.; Pant, K.; Aloo, B.N.; Kumar, G.; Singh, H.B.; Tripathi, V. Microplastic Pollution in Terrestrial Ecosystems and Its Interaction with Other Soil Pollutants: A Potential Threat to Soil Ecosystem Sustainability. Resources 2023, 12, 67. [Google Scholar] [CrossRef]
- Heris, Y.S. Bacterial biodegradation of synthetic plastics: A review. Bull. Natl. Res. Cent. 2024, 48, 87. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Takehana, T.; Yoshida, S.; Hiraga, K.; Oda, K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate). Int. J. Syst. Evol. Microbiol. 2016, 66, 2813–2818. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Tibpromma, S.; Dai, D.; Xu, R.; Suwannarach, N.; Stephenson, S.L.; Dao, C.; Karunarathna, S.C. A review of the fungi that degrade plastic. J. Fungi 2022, 8, 772. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2017, 38, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Longo, C.; Savaris, M.; Zeni, M.; Brandalise, R.N.; Grisa, A.M.C. Degradation study of polypropylene (PP) and bioriented polypropylene (BOPP) in the environment. Mater. Res. 2011, 14, 442–448. [Google Scholar] [CrossRef]
- Lefèvre, C.; Tidjani, A.; Wauven, C.V.; David, C. The interaction mechanism between microorganisms and substrate in the biodegradation of polycaprolactone. J. Appl. Pol. Sci. 2001, 83, 1334–1340. [Google Scholar] [CrossRef]
- Ghatge, S.; Yang, Y.; Ahn, J.-H.; Hur, H.-G. Biodegradation of polyethylene: A brief review. Appl. Biol. Chem. 2020, 63, 27. [Google Scholar] [CrossRef]
- Pometto, A.L.; Lee, B.T.; Johnson, K.E. Production of an extracellular polyethylene-degrading enzyme(s) by Streptomyces species. Appl. Environ. Microbiol. 1992, 58, 731–733. [Google Scholar] [CrossRef]
- Roman, E.K.B.; Ramos, M.A.; Tomazetto, G.; Foltran, B.B.; Galvão, M.H.; Ciancaglini, I.; Tramontina, R.; de Almeida Rodrigues, F.; da Silva, L.S.; Sandano, A.L.H.; et al. Plastic-degrading microbial communities reveal novel microorganisms, pathways, and biocatalysts for polymer degradation and bioplastic production. Sci. Total Environ. 2024, 949, 174876. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Labrenz, M. Marine microbial assemblages on microplastics: Diversity, adaptation, and role in degradation. Ann. Rev.Mar. Sci. 2020, 12, 209–232. [Google Scholar] [CrossRef]
- Skariyachan, S.; Patil, A.A.; Shankar, A.; Manjunath, M.; Bachappanavar, N.; Kiran, S. Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Deg. Stab. 2018, 149, 52–68. [Google Scholar] [CrossRef]
- Atanasova, N.; Stoitsova, S.; Paunova-Krasteva, T.; Kambourova, M. Plastic Degradation by Extremophilic Bacteria. Int. J. Mol. Sci. 2021, 22, 5610. [Google Scholar] [CrossRef]
- Atanasova, N.; Paunova-Krasteva, T.; Stoitsova, S.; Radchenkova, N.; Boyadzhieva, I.; Petrov, K.; Kambourova, M. Degradation of Poly(ε-caprolactone) by a Thermophilic Community and Brevibacillus thermoruber Strain 7 Isolated from Bulgarian Hot Spring. Biomolecules 2021, 11, 1488. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.; Paunova-Krasteva, T.; Kambourova, M.; Boyadzhieva, I. A Thermostable Lipase Isolated from Brevibacillus thermoruber Strain 7 Degrades Ɛ-Polycaprolactone. BioTech 2023, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Rathi, P.; Gupta, R. Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal. Biochem. 2002, 311, 98–99. [Google Scholar] [CrossRef]
- Almeida, E.L.; Carrillo Rincón, A.F.; Jackson, S.A.; Dobson, A.D. In silico screening and heterologous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone (PCL)-degrading activity from the marine sponge-derived strain Streptomyces sp. SM14. Front. Microbiol. 2019, 10, 2187. [Google Scholar] [CrossRef]
- Rosenberg, M. Bacterial adherence to hydrocarbons: A useful technique for studying cell surface hydrophobicity. FEMS Microbiol. Lett. 1984, 22, 289–295. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Carney Almroth, B.; Eggert, H. Marine Plastic Pollution: Sources, Impacts, and Policy Issues. Rev. Environ. Econ. Policy 2019, 13, 317–326. [Google Scholar] [CrossRef]
- Zandieh, M.; Griffiths, E.; Waldie, A.; Li, S.; Honek, J.; Rezanezhad, F.; Van Cappellen, P.; Liu, J. Catalytic and biocatalytic degradation of microplastics. Exploration 2023, 4, 20230018. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K. Biodegradation of PET: Current status and application aspects. ACS Catal. 2019, 9, 4089–4105. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef]
- Atanasova, N.; Paunova-Krasteva, T.; Stoitsova, S.; Kambourova, M.; Shapagin, A.; Matveev, V.; Provotorova, E.; Elcheninov, A.; Sokolov, T.; Bonch-Osmolovskaya, E. Plastic Degradation by Extremophilic Microbial Communities Isolated from Bulgaria and Russia. Ecol. Balk. 2021, 13, 211–222. [Google Scholar]
- Suzuki, M.; Tachibana, Y.; Kasuya, K. Biodegradability of poly(3-hydroxyalkanoate) and poly(ε-caprolactone) via biological carbon cycles in marine environments. Polym. J. 2021, 53, 47–66. [Google Scholar] [CrossRef]
- Yoon, Y.; Park, H.; An, S.; Ahn, J.H.; Kim, B.; Shin, J.; Kim, Y.E.; Yeon, J.; Chung, J.H.; Kim, D.; et al. Bacterial degradation kinetics of poly(Ɛ-caprolactone) (PCL) film by Aquabacterium sp. CY2-9 isolated from plastic-contaminated landfill. J. Environ. Manag. 2023, 335, 117493. [Google Scholar] [CrossRef]
- Kotova, I.B.; Taktarova, Y.V.; Tsavkelova, E.A.; Egorova, M.A.; Bubnov, I.A.; Malakhova, D.V.; Shirinkina, L.I.; Sokolova, T.G.; Bonch-Osmolovskaya, E.A. Microbial Degradation of Plastics and Approaches to Make it More Efficient. Microbiology 2021, 90, 671–701. [Google Scholar] [CrossRef]
- Tiwari, N.; Bansal, M.; Santhiya, D.; Sharma, J.G. Insights into microbial diversity on plastisphere by multi-omics. Arch. Microbiol. 2022, 204, 216. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef]
- Ghosh, S.; Qureshi, A.; Purohit, H.J. Microbial degradation of plastics: Biofilms and degradation pathways. In Contaminants in Agriculture and Environment: Health Risks and Remediation; Kumar, V., Kumar, R., Singh, J., Kumar, P., Eds.; Agro Environ Media: Haridwar, India, 2019; Volume 1, pp. 184–199. [Google Scholar] [CrossRef]
- Bardají, D.K.R.; Moretto, J.A.S.; Furlan, J.P.R.; Stehling, E.G. A mini-review: Current advances in polyethylene biodegradation. World J. Microbiol. Biotechnol. 2020, 36, 32. [Google Scholar] [CrossRef]
- Baumgarten, T.; Sperling, S.; Seifert, J.; Von Bergen, M.; Steiniger, F.; Wick, L.Y.; Heipieper, H.J. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 2012, 78, 6217–6224. [Google Scholar] [CrossRef] [PubMed]
- Delacuvellerie, A.; Cyriaque, V.; Gobert, S.; Benali, S.; Wattiez, R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the degradation. J. Hazard. Mater. 2019, 380, 120899. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.F.; Xu, K.W.; Wu, J. Synergism Between Multi-Pseudomonas and Cutinase for Biodegradation of Crude Oil-Based Derivatives. Curr. Microbiol. 2022, 80, 30. [Google Scholar] [CrossRef]
- Okshevsky, M.; Gautier, E.; Farner, J.M.; Schreiber, L.; Tufenkji, N. Biofilm formation by marine bacteria is impacted by concentration and surface functionalization of polystyrene nanoparticles in a species-specific manner. Environ. Microbiol. Rep. 2020, 12, 203–213. [Google Scholar] [CrossRef]
- Howard, S.A.; McCarthy, R.R. Modulating biofilm can potentiate activity of novel plastic-degrading enzymes. npj Biofilms Microbiomes 2023, 9, 72. [Google Scholar] [CrossRef]
- Zampolli, J.; Vezzini, D.; Brocca, S.; Di Gennaro, P. Insights into the biodegradation of polycaprolactone through genomic analysis of two plastic-degrading Rhodococcus bacteria. Front. Microbiol. 2024, 14, 1284956. [Google Scholar] [CrossRef]
- Inglis, G.D.; Yanke, L.J.; Selinger, L.B. Cutinolytic esterase activity of bacteria isolated from mixed-plant compost and characterization of a cutinase gene from Pseudomonas pseudoalcaligenes. Can. J. Microbiol. 2011, 57, 902–913. [Google Scholar] [CrossRef]
- Shin, N.; Kim, S.H.; Cho, J.Y.; Hwang, J.H.; Kim, H.J.; Oh, S.J.; Park, S.-H.; Park, K.; Bhatia, S.K.; Yanget, Y.-H. Fast Degradation of Polycaprolactone/Poly(butylene adipate-co-terephthalate) Blends by Novel Bacillus Strain NR4 with Broad Degrading Activity. J. Polym. Environ. 2024, 32, 898–912. [Google Scholar] [CrossRef]
- Wang, Z.; Hou, X.; Shang, G.; Deng, G.; Luo, K.; Peng, M. Exploring Fatty Acid β-Oxidation Pathways in Bacteria: From General Mechanisms to DSF Signaling and Pathogenicity in Xanthomonas. Curr. Microbiol. 2024, 81, 336. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Sharma, R.; Thakur, V.; Sharma, M.; Birkeland, N.K. Biocatalysis through thermostable lipases: Adding flavor to chemistry. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Springer: Dordrecht, The Netherlands, 2013; pp. 905–927. [Google Scholar] [CrossRef]
- Oda, Y.; Oida, N.; Urakami, T.; Tonomura, K. Polycaprolactone depolymerase produced by the bacterium Alcaligenes faecalis. FEMS Microbiol. Lett. 1997, 152, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Khandare, S.D.; Chaudhary, D.R.; Jha, B. Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic. Biodegradation 2021, 32, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yue, Y.; Chen, J.; Zhou, J.; Li, Y.; Zhang, Q. Biodegradation mechanism of polycaprolactone by a novel esterase MGS0156: A QM/MM approach. Environ. Sci. Proc. Impacts 2020, 22, 2332–2344. [Google Scholar] [CrossRef] [PubMed]
- Simon-Colin, C.; Raguénès, G.; Cozien, J.; Guezennec, J.G. Halomonas profundus sp. nov., a new PHA-producing bacterium isolated from a deep-sea hydrothermal vent shrimp. J. Appl. Microbiol. 2008, 104, 1425–1432. [Google Scholar] [CrossRef]
- Okeyode, A.H.; Al-Thukair, A.; Chanbasha, B.; Nazal, M.K.; Afuecheta, E.; Musa, M.M.; Algarni, S.; Nzila, A. Degradation of the highly complex polycyclic aromatic hydrocarbon coronene by the halophilic bacterial strain Halomonas caseinilytica, 10SCRN4D. Arch. Environ. Prot. 2023, 49, 78–86. [Google Scholar] [CrossRef]
- Kumar, S.; Tao, Y. Coronenes, Benzocoronenes and Beyond: Modern aspects of their syntheses, properties, and applications. Chem. Asian J. 2021, 16, 621–647. [Google Scholar] [CrossRef]
- Rivadeneyra, M.A.; Martín-Algarra, A.; Sánchez-Navas, A.; Martín-Ramos, D. Carbonate and Phosphate Precipitation by Chromohalobacter marismortui. Geomicrobiol. J. 2005, 23, 1–13. [Google Scholar] [CrossRef]
- Rivadeneyra, M.A.; Martín-Algarra, A.; Sánchez-Román, M.; Sánchez-Navas, A.; Martín-Ramos, J.D. Amorphous Ca-phosphate precursors for Ca-carbonate biominerals mediated by Chromohalobacter marismortui. ISME J. 2010, 4, 922–932. [Google Scholar] [CrossRef]
- Rogers, K.L.; Carreres Calabuig, J.A.; Nynke, K.; Shashoua, Y.; Posth, N. Potential Role of Metal Precipitation on Plastic Fate: Transport and Degradation. Abstract from Micro 2020 International Conference. Available online: https://pure.kb.dk/en/publications/potential-role-of-metal-precipitation-on-plastic-fate-transport-a (accessed on 12 February 2025).
- Dodhia, M.S.; Rogers, K.L.; Fernández-Juárez, V.; Carreres-Calabuig, J.A.; Löscher, C.R.; Tisserand, A.A.; Keulen, N.; Riemann, L.; Shashoua, Y.; Posth, N.R. Microbe-mineral interactions in the Plastisphere: Coastal biogeochemistry and consequences for degradation of plastics. Front. Mar. Sci. 2023, 10, 1134815. [Google Scholar] [CrossRef]
- Beutling, D.M.; Peçonek, J.; Stan-Lotter, H. Chromohalobacter beijerinckii: A psychrophilic, extremely halotolerant and enzymatically active microbe from salted food with the capacity for biogenic amine production. Europ. Food Res. Technol. 2009, 229, 725–730. [Google Scholar] [CrossRef]
- Derippe, G.; Philip, L.; Lemechko, P.; Eyheraguibel, B.; Meistertzheim, A.L.; Pujo-Pay, M.; Conan, P.; Barbe, V.; Bruzaud, S.; Ghiglione, J.F. Marine biodegradation of tailor-made polyhydroxyalkanoates (PHA) influenced by the chemical structure and associated bacterial communities. J. Hazard. Mater. 2024, 462, 132782. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Verdú, À.; Latorre-Pérez, A.; Molina-Menor, E.; Baixeras, J.; Peretó, J.; Porcar, M. Living in a bottle: Bacteria from sediment-associated Mediterranean waste and potential growth on polyethylene terephthalate. Microbiol. Open 2022, 11, e1259. [Google Scholar] [CrossRef] [PubMed]
- Dhaka, V.; Singh, S.; Anil, A.G.; Naik, T.S.S.K.; Garg, S.; Samuel, J.; Kumar, M.; Ramamurthy, P.C.; Singh, J. Occurrence, toxicity and remediation of polyethylene terephthalate plastics. A review. Environ. Chem. Lett. 2022, 20, 1777–1800. [Google Scholar] [CrossRef] [PubMed]
- Moosvi, S.A.; Pacheco, C.C.; McDonald, I.R.; De Marco, P.; Pearce, D.A.; Kelly, D.P.; Wood, A.P. Isolation and properties of methanesulfonate-degrading Afipia felis from Antarctica and comparison with other strains of A. felis. Environ. Microbiol. 2004, 7, 22–33. [Google Scholar] [CrossRef]
- Zhang, P.; Hozalski, R.M.; Leach, L.H.; Camper, A.K.; Goslan, E.H.; Parsons, S.A.; Xie, Y.F.; LaPara, T.M. Isolation and characterization of haloacetic acid-degrading Afipia spp. from drinking water. FEMS Microbiol. Lett. 2009, 297, 203–208. [Google Scholar] [CrossRef]
- Bao, Z.; Wang, C.; Cao, J.; Zhang, T.; Guo, Y.; Sato, Y.; Nishizawa, T.; Ohta, H. Tardiphaga alba sp. nov., a heavy-metal-tolerant bacterium isolated from garden soil. Int. J. Syst. Evol. Microbiol. 2024, 74, 006238. [Google Scholar] [CrossRef]
- Attaelmanan, A.G.; Aslam, H.; Ali, T.; Dronjak, L. Mapping of heavy metal contamination associated with microplastics marine debris—A case study: Dubai, UAE. Sci. Total Environ. 2023, 891, 164370. [Google Scholar] [CrossRef]
- Lemboub, C.; Chekireb, D. Rock Phosphate Solubilization and Heavy Metals Resistance of Rhizobacteria Isolated from Nodules of Lathyrus occurs. J. Pure Appl. Microbiol. 2018, 12, 473–482. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krumov, N.; Atanasova, N.; Boyadzhieva, I.; Paunova-Krasteva, T.; Berberov, K.; Petrov, K.; Petrova, P. New Halophilic Community Degrades Plastics: A Metagenomic Study. Fermentation 2025, 11, 227. https://doi.org/10.3390/fermentation11040227
Krumov N, Atanasova N, Boyadzhieva I, Paunova-Krasteva T, Berberov K, Petrov K, Petrova P. New Halophilic Community Degrades Plastics: A Metagenomic Study. Fermentation. 2025; 11(4):227. https://doi.org/10.3390/fermentation11040227
Chicago/Turabian StyleKrumov, Nikolay, Nikolina Atanasova, Ivanka Boyadzhieva, Tsvetelina Paunova-Krasteva, Kaloyan Berberov, Kaloyan Petrov, and Penka Petrova. 2025. "New Halophilic Community Degrades Plastics: A Metagenomic Study" Fermentation 11, no. 4: 227. https://doi.org/10.3390/fermentation11040227
APA StyleKrumov, N., Atanasova, N., Boyadzhieva, I., Paunova-Krasteva, T., Berberov, K., Petrov, K., & Petrova, P. (2025). New Halophilic Community Degrades Plastics: A Metagenomic Study. Fermentation, 11(4), 227. https://doi.org/10.3390/fermentation11040227







