Abstract
The industrial potential of Clostridium beijerinckii for acetone–butanol–ethanol (ABE) fermentation is limited by oxygen sensitivity and suboptimal solvent productivity. Peroxide repressor (PerR), a key negative regulator protein, is reported to suppress the oxidative stress defense system in anaerobic clostridia, leading to poor survival of bacteria under aerobic conditions. However, the regulatory mechanism underlying this phenomenon remains unclear. This study demonstrates that targeted deletion of perR (Cbei_1336) in the solvent-deficient strain C. beijerinckii DS confers robust oxygen tolerance and enhances ABE fermentation performance. The engineered perR mutant exhibited unprecedented aerobic growth under atmospheric oxygen (21% O2), achieving a (3.79 ± 0.09)-fold increase in biomass accumulation, a (2.84 ± 0.12)-fold improvement in glucose utilization efficiency, a (57.23 ± 0.01)-fold elevation in butanol production, and a (32.78 ± 0.02)-fold amplification in acetone output compared to the parental strain. Transcriptomic analysis revealed that perR knockout simultaneously upregulated oxidative defense systems and activated ABE pathway-related genes. This genetic rewiring redirected carbon flux from acidogenesis to solventogenesis, yielding a (9.64 ± 0.90)-fold increase in total solvent titer (15.61 ± 0.89 vs. 1.62 ± 0.12 g/L) and a (2.71 ± 0.04)-fold rise in volumetric productivity (0.19 ± 0.01 vs. 0.07 ± 0.01 g/L/h). Our findings establish PerR as a master regulator of both oxygen resilience and metabolic reprogramming, providing a scalable engineering strategy for industrial oxygen-tolerant ABE bioprocessing toward low-cost biobutanol production.
1. Introduction
The increasing depletion of fossil fuels and the associated environmental issues, such as pollution and greenhouse gas emissions, have intensified the search for renewable and clean energy sources. Among these, bio-butanol has emerged as a promising alternative due to its superior fuel properties, positioning it as a more favorable renewable energy source compared to bio-ethanol [1]. The primary method for bio-butanol production is acetone–butanol–ethanol (ABE) fermentation using anaerobic Clostridium species, which degrade substrates such as sugars, starches, or lignocellulosic materials to produce solvents, including bio-butanol [2]. Compared to petrochemical-derived butanol, the relatively lower production efficiency and substrate conversion rate are key reasons for the higher production costs of bio-butanol and its lack of market competitiveness [2]. To address this, the genetic modification has been applied to optimize the solvent-producing Clostridium strains [2,3]. Significant improvements have been achieved in the anaerobic clostridia modification through genetic editing technologies (e.g., antisense RNA, TargeTron Technology, and CRISPR) and metabolic engineering, which resulted in the knock-in, knockout, and overexpression of genes such as cre, spo0A, cat1, adhE1/adhE2, scrA/scrB/scrK, and ctfB [4]. Although these modifications have contributed substantially to enhancing biobutanol production capabilities and elucidating relevant mechanisms, numerous functional genes in the solvent metabolic pathways remain to be elucidated.
In practice, two wild-type strains, Clostridium acetobutylicum ATCC 824 and Clostridium beijerinckii NCIMB 8052, are the two predominant industrial strains utilized for ABE fermentation. However, all genes responsible for butanol production in the ATCC 824 strain are pSOL1 plasmid-encoded, which may result in reduced butanol production due to plasmid loss. In contrast, the NCIMB 8052 carries chromosome-encoded genes essential for butanol production, showing a stable genetic trait [2]. Moreover, the presence of type II restriction-modification system Cac824I in ATCC 824 complicates the genetic manipulation, while NCIMB 8052 lacks these limitations and exhibits greater genetic stability and ease of manipulation [5]. Therefore, we select NCIMB 8052 for the subsequent genetic engineering.
As strictly anaerobic bacteria, the detrimental effects of oxygen (O2) and reactive oxygen species (ROS) on clostridia pose significant challenges for engineering high-performance strains and conducting ABE fermentation. Aerobic bacteria generate energy by breaking down carbohydrates via aerobic respiration, supporting their growth and metabolism. Importantly, ROSs such as superoxide anions (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH˙) are generated during these metabolic processes, causing cellular damage, mutations, and cell death [6,7]. In general, aerobic bacteria utilize defense systems (e.g., superoxide dismutase, catalase) to detoxify ROS and prevent oxidative damage. However, strictly anaerobic clostridia lack functional ROS defenses, rendering them highly susceptible to lethal O2/ROS exposure [8,9]. Therefore, the growth, metabolism, fermentation, and genetic manipulation of anaerobic clostridia must be conducted under strictly anaerobic conditions [10].
Although ABE fermentation in clostridia is anaerobic, the bacterial cells retain an oxidative stress defense system capable of eliminating O2 and ROS. Clostridia struggle to survive in aerobic environments due to the presence of peroxide repressor (PerR), which inhibits the activation of bacterial oxidative stress defense system [8,10]. PerR, a member of the ferric uptake regulator (Fur) protein family, binds to DNA and functions as a transcriptional repressor. It senses H2O2 by catalyzing the oxidation of histidine residues (His37 or His91) at the metal-binding domain with assistance of the Fe2+ cofactor. This oxidation triggers conformational changes in PerR, leading to the repression of genes involved in O2 and ROS tolerance [11,12]. Knocking out the PerR-encoding gene CA_C2634 in C. acetobutylicum ATCC 824 via type II intron insertion alleviates oxidative stress repression. This genetic modification enables aerobic growth with rapid oxygen consumption and permits successful electroporation under aerobic conditions [10,13]. However, few studies have investigated the impact of perR knockout on ABE fermentation performance and solvent (including butanol) production in the solvent-producing clostridial isolates. Therefore, it is a highly valuable research undertaking to determine whether the genetically stable and easily manipulable strain C. beijerinckii NCIMB 8052 possesses a gene encoding PerR—a negative regulator protein that suppresses the oxidative stress defense system—and if knockout of this gene can enhance the strain’s oxygen tolerance and solvent production capacity while elucidating the underlying mechanisms.
To address the aforementioned gap, we identified the gene Cbei_1336 encoding PerR in C. beijerinckii NCIMB 8052 through genomic BLASTp alignment. Cbei_1336 in a solvent-producing strain was knocked out using the type II intron insertion method. Results showed that the perR mutant was oxygen-tolerant with increased acetone and bio-butanol production. Transcriptomic profiling revealed that the perR mutant concurrently upregulated the oxidative defense system and activated genes associated with the ABE fermentation pathway. Collectively, these findings identified PerR as a central regulator coordinating oxygen resilience and metabolic reprogramming in this industrially relevant bacterium. By inactivating PerR, we provide an industrial biobutanol strategy enabling robust clostridia growth under ambient air (21% O2). This breakthrough achieves high-efficiency genetic editing and enhanced ABE fermentation performance while reducing production costs and advancing renewable biofuel technologies.
2. Materials and Methods
2.1. Strains, Plasmids, and Culturing Conditions
The strains and plasmids used in this study are shown in Table S1. Escherichia coli (E. coli trans1-T1, phage-resistant chemically competent cells, Takara 2011A, Dalian, Liaoning, China) was cultured in Luria-Bertani (LB) medium/agar (1.5% agar) with 15 μg/mL chloramphenicol at 34 °C. C. beijerinckii NCIMB 8052 (CGMCC 15077) was purchased from China General Microbiological Culture Collection Center (CGMCC). The C. beijerinckii solvent-producing degenerated strain (C. beijerinckii DS) was derived from the wild-type NCIMB 8052 by continuous subculturing every 12 h for 50 passages in MRCM liquid culture under strictly anaerobic conditions. The oxygen-tolerant mutant of C. beijerinckii DS, designated perR mutant, was engineered by targeted inactivation of the PerR-encoding gene Cbei_1336. The C. beijerinckii and its derived strains were stored as spores in anaerobic freeze-drying tubes with 15% skimmed milk.
The modified reinforced clostridial medium (MRCM) and P2 media [3,14] were used as the seed and fermentation culture media, respectively, for the strains of interest. Erythromycin (10 μg/mL) was used for single colony selection. All strains were cultured anaerobically at 34 °C with 150 rpm agitation (ELECTROTEK, 400SG, Shipley, West Yorkshire, UK).
2.2. Construction of Plasmid pMTL007-Cbei1336-Intron
The intron PCR template was utilized according to the protocol of the TargeTron® Gene Knockout System (Sigma TA0100, St. Louis, MO, USA), and PCR amplification was performed using primers 374|375s-IBS, 374|375s-EBS1d,374|375s-EBS2, and EBS universal (Table S2). A 353 bp TargeTron fragment containing perR (Cbei_1336) was cloned into shuttle plasmid pMTL007C-E2 [15] to obtain pMTL007-E2-Cbei1336-intron.
2.3. Electroporation and Identification of perR Mutant
C. beijerinckii DS competent cells were prepared by heat-shocking lyophilized spores (80 °C/10 min), cooling on ice (5 min), and inoculating into MRCM broth. Post-anaerobic incubation (34 °C, 150 rpm, OD600 = 0.8–1.0), bacterial cells were collected (4 °C, 4500× g for 10 min), sequentially washed with ice-cold H2O and ETB buffer (270 mM sucrose, 5 mM NaH2PO4, pH 7.4), and resuspended in ETB. Approximately 10 µL of pMTL007-E2-Cbei1336-intron plasmid (500 ng/mL) were mixed with 100 µL of C. beijerinckii DS competent cells and incubated in ice for 5 min. Electroporation was performed with a 0.4 × 1.0 cm cuvette in Gene Pulser Xcell™ (Bio-Rad Laboratories, Hercules, CA, USA) under parameters of 25 kV, 25 μF, and ∞Ω. Cells were recovered in MRCM (34 °C/4 h), then plated on 10 μg/mL erythromycin-containing MRCM agar and incubated anaerobically (34 °C/18–24 h) as previously described [16,17].
2.4. Aerobic Tolerance Assay of the perR Mutant
The perR mutant was inoculated into MRCM broth and anaerobically incubated at 34 °C for 18 h. Subsequently, 100 μL of aliquots were spread onto MRCM agar plates and incubated under both aerobic and anaerobic conditions at 34 °C for 24 h. The parental strain C. beijerinckii DS served as a control.
2.5. ABE Production Analysis of the Aerobic-Tolerant perR Mutant
Comparative analyses of ABE solvent production in the aerotolerant perR mutant, parental C. beijerinckii DS, and wild-type C. beijerinckii NCIMB 8052 were performed through shake-flask fermentation with P2 broth as previously described [14]. Briefly, bacteria were cultured in 5 mL of MRCM at 34 °C for 12 h (OD600 = 0.8–1.0), then the culture was inoculated into 100 mL of MRCM broth and cultivated at 34 °C for 8 h. Afterward, 6 mL of culture were transferred to 100 mL of P2 fermentation medium and cultivated at 34 °C with 150 rpm agitation in an Electrotek 400SG anaerobic workstation (Shipley, West Yorkshire, UK). Bacterial growth (OD600), pH value, and products, including acetate, butyrate, acetone, butanol, ethanol (ABE) solvent, and residual glucose concentration, were monitored during fermentation.
2.6. Analytical Methods
Biomass accumulation was quantified by measuring OD600 using an Eppendorf Biophotometer Plus spectrophotometer (Hamburg, Germany). Glucose concentrations were determined using the 3,5-dinitrosalicylic acid spectrometric method [14] with an SBA−40D biosensor (Institute of Biology, Shandong Academy of Sciences, Jinan, Shandong, China). Volatile acids (acetate, butyrate) and ABE solvents (acetone, butanol, ethanol) were analyzed using a PerkinElmer Clarus GC680 system (Waltham, MA, USA) equipped with a ZKAT-FFAP capillary column (50 m × 0.32 mm ×1 μm). The gas chromatograph operated under programmed temperature conditions as previously described [14]: (i) Temperature program: 55 °C (1 min)→3 °C/min→90 °C (1 min)→50 °C/min→180 °C (6 min); (ii) Injection/detection: 250 °C, N2 carrier (2 mL/min), H2 (40 mL/min), and air (400 mL/min).
2.7. RNA Sequencing
Bacterial pellets of C. beijerinckii DS and perR mutant were harvested at 12, 24, and 36 h post-inoculation. For each time point, 30 mL of broth (triplicate-pooled) were centrifuged (5000× g, 10 min, 4 °C), and pellets were washed with RNase-free water, frozen in liquid N2, and mechanically grinded in pre-chilled mortars. About 1 mL of TRIzol Reagent (Thermo Fisher, Waltham, MA, USA) was added, and the mixture was homogenized with vortexing (2500 rpm, 2 min). Total RNA isolation was performed using the modified Trizol (MT) protocol [18]. RNA quality was verified with NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Baden-Württemberg, Germany).
Ribosomal RNA was depleted using a Ribo-Zero rRNA Removal Kit for Prokaryotes (Illumina, San Diego, CA, USA). A strand-specific library was constructed using ALFA-SEQ RNA Library Prep Kit II (Bioyi Biotechnology, Wuhan, Hubei, China). RNA sequencing (RNA-seq) was carried out in the Illumina NovaSeq X Plus platform (2 × 150 bp paired-end) at Beijing Genomics Institution Co., Ltd. (Beijing, China). The low-quality reads were filtered, and those passing FastQC quality control were subsequently aligned using SOAPaligner/soap2. C. beijerinckii NCIMB 8052 (RefSeq: NC_009617.1) was used as the reference genome for alignment. Gene expression levels were quantified via RPKM (Reads Per Kilobase per Million mapped reads) normalization. Differential expression analysis between comparative groups was conducted using EdgeR 3.12.0. A differentially expressed gene (DEG) was defined as |log2 fold-change| (|log2FC|) >1 and p < 0.05. The |log2FC| threshold was used to minimize oversight of potentially functionally significant genes exhibiting modest expression changes following perR knockout. All filtered DEGs were used for subsequent analyses and visualization.
2.8. Real-Time Quantitative RT-PCR (qRT-PCR)
For qRT-PCR analysis, cDNA was synthesized from 1 μg of total RNA (same samples in RNA sequencing) using ReverTra Ace qPCR RT Master Mix with gDNA Remover (FSQ-301, TOYOBO, Osaka, Japan) following the protocol of manufacturer. Quantitative PCR was conducted with SYBR® Green Realtime PCR Master Mix Plus (QPK-212, TOYOBO, Osaka, Japan), following the procedure of 95 °C for 5 min, 40 cycles of 95 °C for 10 s, 55 °C for 10 s, and 72 °C for 20 s. Gene-specific primers used are shown in Table S2, and the expression level of a certain gene was normalized to that of the 16S rRNA gene.
2.9. Statistical Analysis
All experiments were repeated at least three times, and the data are expressed as the mean ± standard deviation (SD). Statistical comparisons were performed via the GraphPad Prism program version 10.5.0 (GraphPad Software, La Jolla, CA, USA). Student’s t test or one-way analysis of variance (ANOVA) was used to analyze differences among groups. A p value less than 0.05 was considered statistically significant.
3. Results and Discussion
3.1. perR Disruption Enables the Survival of C. beijerinckii DS Under Atmospheric Oxygen
Accumulated data have revealed that obligate anaerobic Clostridium species employed in ABE fermentation paradoxically retain oxidative stress defense systems capable of scavenging O2 and ROS. However, the constitutive expression of the transcriptional repressor PerR protein, which suppresses the activation of these detoxification pathways, inhibits organisms to sustain aerobic growth, to perform solventogenesis, or to be genetically manipulated under oxygen-exposed conditions [8,10].
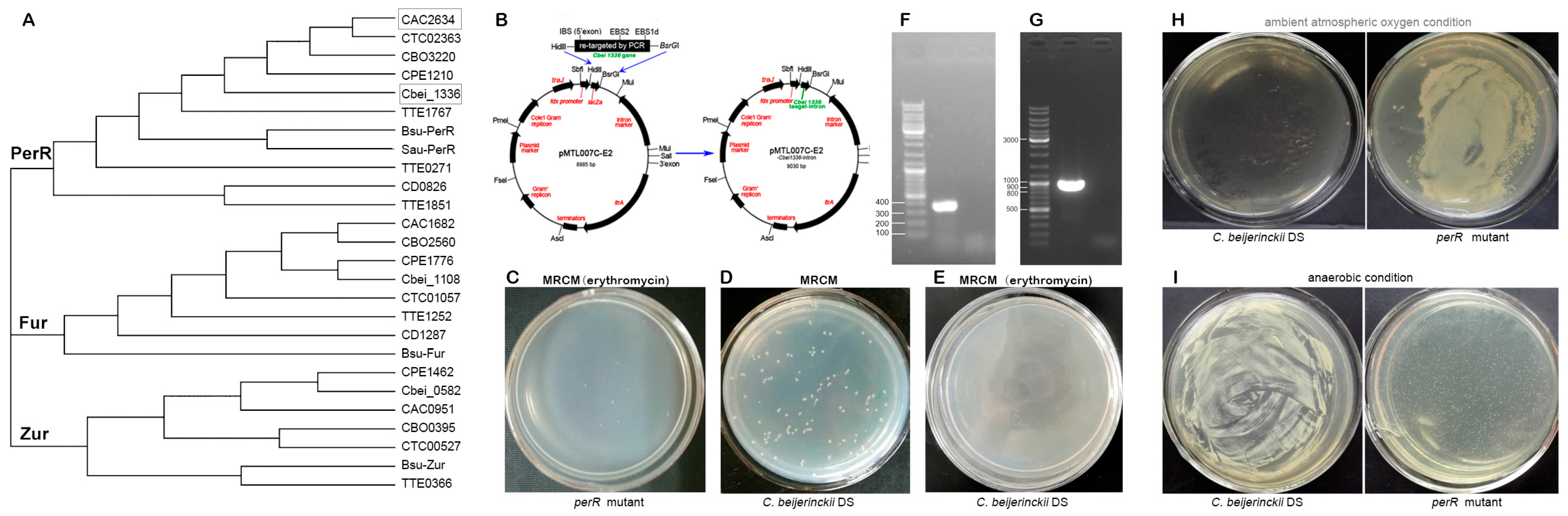
To identify oxidative stress PerR-like regulators in C. beijerinckii NCIMB 8052, BLASTp alignment of C. acetobutylicum ATCC 824 perR (CA_C2634) revealed three homolog genes: Cbei_1336, Cbei_0582, and Cbei_1108, which were subsequently screened for type II intron insertion sites through the Clostron platform (http://www.clostron.com; accessed on 5 January 2016). This analysis identified optimal insertion loci with the following highest prediction scores: 6.457 for Cbei_1336 at position 374|375s (sense strand), 4.256 for Cbei_0582 at position 170|171s, and 7.287 for Cbei_1108 at position 271|272s. Phylogenetic analysis placed Cbei_1336 of C. beijerinckii NCIMB 8052 with CA_C2634 (C. acetobutylicum ATCC 824) in the conserved PerR clade (Figure 1A) [10]. This evolutionary conservation and its high insertion score (6.457) justified targeting position 374|375s for gene inactivation.
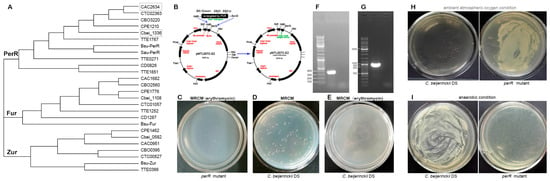
Figure 1.
Construction and characterization of the oxygen-resilient perR mutant. (A) Phylogenetic conservation analysis of PerR-, Fur-, and Zur-like proteins across Clostridium species. An unrooted cladogram of Fur-homologous proteins from representative clostridia and other Gram-positive bacteria was constructed using multiple sequence alignment. Protein identifiers combine open reading frame (ORF) numbers with species abbreviations as follows: Bsu: Bacillus subtilis; CAC: C. acetobutylicum ATCC 824; Cbei: C. beijerinckii NCIMB 8052; CBO: C. botulinum A str. ATCC 3502; CD: C. difficile 630; CPE: C. perfringens str. 13; CTC: C. tetani E88; TTE: Thermoanaerobacter tengcongensis [10]. (B) Construction of the perR (Cbei_1336) knockout vector pMTL007-Cbei1336-intron. (C) Viability of perR mutant on erythromycin-containing MRCM (10 μg/mL). (D) Colony morphology of C. beijerinckii DS on MRCM agar plates. (E) C. beijerinckii DS showed no colony formation on MRCM erythromycin plates (10 μg/mL). (F) Validation of transformants by colony PCR. Specific 381 bp fragments were amplified with specific primers (pMTL007-ltrA-up/down). (G) Molecular verification of intron-mediated perR disruption. A 981 bp amplicon was amplified from genomic DNA with intron-ermB-specific primers (pMTL007-ErmB-up/down). (H) Growth of C. beijerinckii DS and its perR mutant under (H) atmospheric oxygen (21% O2) and (I) anaerobic conditions. perR mutant exhibits facultative anaerobe-like growth in atmospheric oxygen, whereas parental strain C. beijerinckii DS remains obligately anaerobic.
Then, we constructed pMTL007C-E2-Cbei1336-intron shuttle plasmid (Figure 1B) and electroporated it into C. beijerinckii DS. Eight transformants grew on erythromycin-MRCM plates (10 μg/mL; 34 °C/24 h anaerobiosis) as white translucent colonies (1.0 ± 0.15 mm, Figure 1C), contrasting with opaque cream parental colonies (2.0 ± 0.18 mm; Figure 1D). The parental strain control (C. beijerinckii DS) showed no growth on antibiotic plates (Figure 1E). Colony PCR with pMTL007-ltrA-up/down primers amplified a 381 bp fragment (Figure 1F), confirming plasmid retention. Genomic PCR using pMTL007-ErmB-up/down junction primers generated a 981 bp amplicon (Figure 1G), validating integration of intron cassette in the Cbei_1336 gene.
To assess the oxygen tolerance conferred by perR (Cbei_1336) inactivation, randomly selected positive colonies of the perR mutant were inoculated in MRCM broth and plated onto solid medium. Parallel cultures were incubated under ambient O2 (21%) versus strict anaerobiosis (N2: H2: CO2 = 85: 5: 10; 34 °C/24 h), with the parent strain C. beijerinckii DS serving as the control. The mutant exhibited robust growth in both conditions, while the parent strain grew only anaerobically (Figure 1H,I). These results confirm that PerR (Cbei_1336-encoded) acts as a master redox-sensing repressor mediating obligate anaerobiosis.
3.2. perR Inactivation Enhances ABE Fermentation Performance of C. beijerinckii DS
To investigate the impact of perR knockout on ABE fermentation in C. beijerinckii DS, 96 h anaerobic shake-flask fermentation was conducted in P2 medium at 34 °C with 150 rpm agitation. Fermentation performance was quantitatively assessed using multiple parameters: biomass accumulation dynamics, pH fluctuations, glucose utilization rates, acetate and butyrate levels, and solventogenesis metrics.
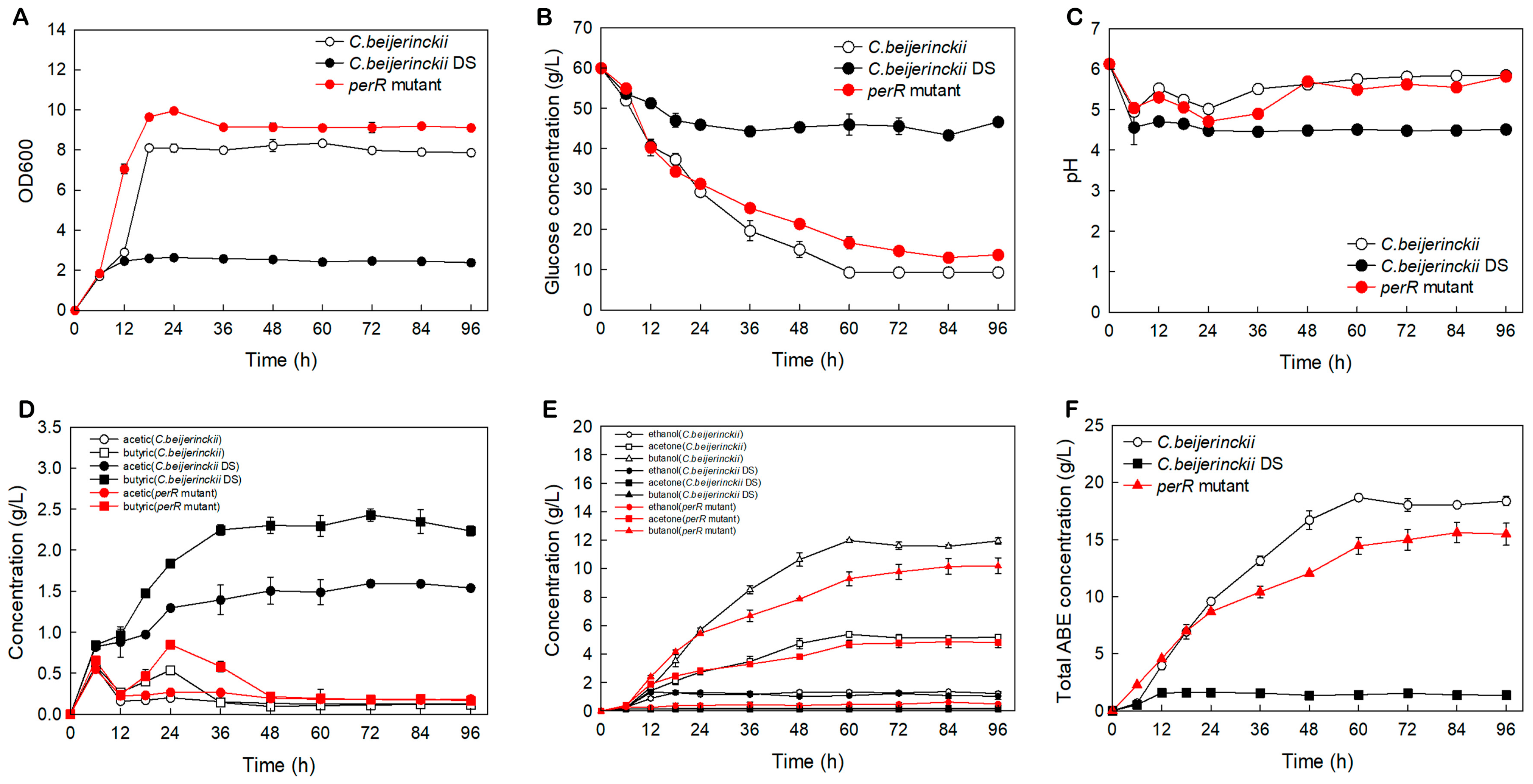
ABE fermentation performances of perR mutant, C. beijerinckii DS, and NCIMB 8052 were shown in Figure 2. The OD600 variation curves revealed that all three strains reached OD600 value of 1.8 at 6 h (Figure 2A), followed by divergence: the perR mutant underwent into logarithmic growth first during 6–18 h (OD600, 1.80 ± 0.02→9.64 ± 0.13), rose slightly at 24 h (9.96 ± 0.14), and stabilized at 9.14 ± 0.16 from 36 h until the endpoint; in contrast, the wild-type 8052 showed minimal OD600 increase during 6–12 h, reaching 2.89 ± 0.01 at 12 h, underwent logarithmic progression at 12–18 h (→8.11 ± 0.11), and plateaued thereafter; notably, the DS strain exhibited sluggish growth: from 1.85 ± 0.05 at 6 h to 2.63 ± 0.05 at 24 h with no further increase. Consequently, perR knockout in solvent-deficient DS strain significantly enhances biomass accumulation, achieving 9.14 ± 0.16 max OD600 and surpassing parental strain’s peak (8.11 ± 0.11) while advancing log-phase initiation by approximately 6 h.
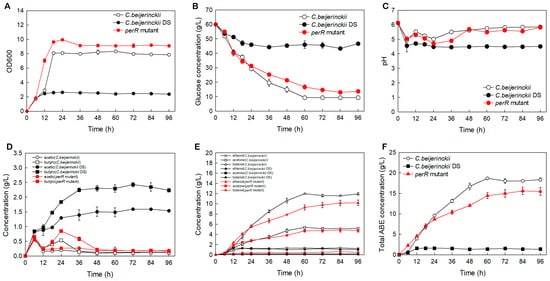
Figure 2.
ABE fermentation profiles of the perR mutant, C. beijerinckii DS, and C. beijerinckii NCIMB 8052 in P2 medium. (A) Cell density. (B) Glucose consumption. (C) pH variation. (D) Acetate and butyrate concentration. (E) Ethanol, acetone, and butanol production, and (F) Total ABE solvent production. The error bars represent standard deviations from three independent biological replicates.
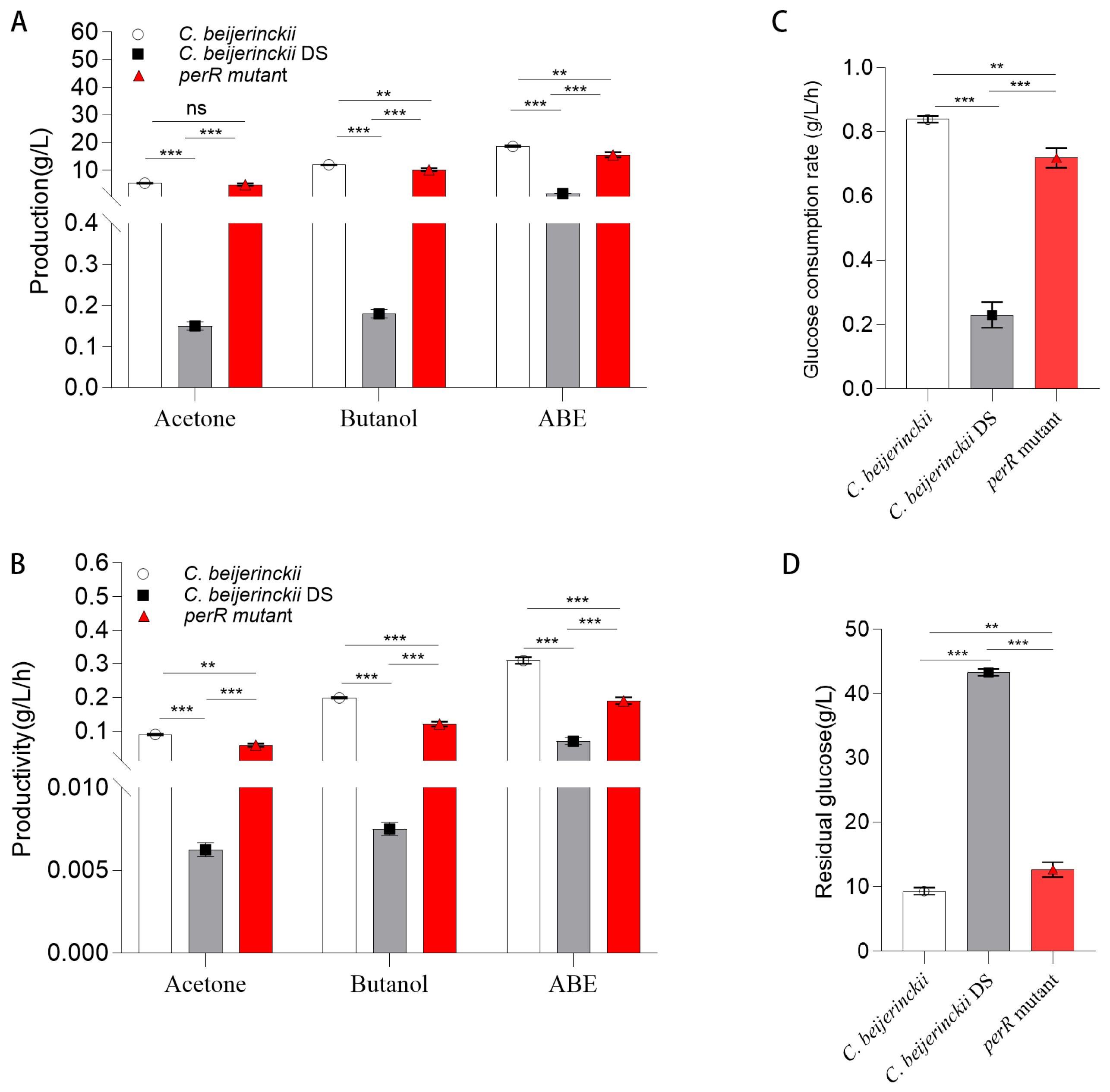
Glucose consumption (Figure 2B and Figure 3, Table 1) correlated with growth curves (Figure 2A), revealing hierarchical glucose metabolism patterns in these strains. Wild-type 8052 showed the highest total glucose consumption (50.67 ± 0.58 g/L), while the perR mutant reached 47.33 ± 1.15 g/L (representing a (2.84 ± 0.12)-fold increase over the degenerated DS strain with 16.67 ± 0.58 g/L). Glucose metabolic duration ranked as: engineered strain (84 h) > wild-type (60 h) > DS (36 h). Over the 0–60 h period, the wild-type strain exhibited the highest average consumption rate of 0.84 ± 0.01 g/L/h. The engineered strain (0.72 ± 0.03 g/L/h) operated at 85.7% of the wild-type efficiency but achieved a (3.13 ± 0.56)-fold higher rate than DS (0.23 ± 0.04 g/L/h). Collectively, the oxygen-tolerant perR mutant showed significantly enhanced total glucose consumption, extended duration, and increased rate compared with DS, yet remained marginally inferior to wild-type 8052 in consumption/rate. Notably, it surpassed wild type in utilization duration.
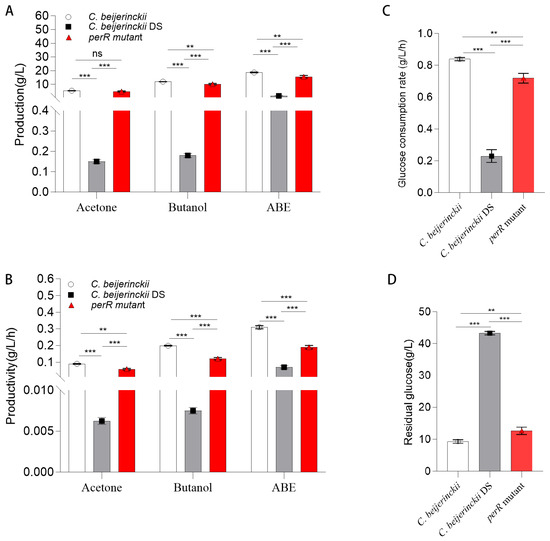
Figure 3.
Comparison of ABE production in shake-flask fermentation of C. beijerinckii, C. beijerinckii DS, and perR mutant. (A) Acetone, butanol, and total ABE solvent production (g/L). (B) Acetone, butanol, and total ABE solvent productivity (g/L/h). (C) Glucose consumption rate (g/L/h) of C. beijerinckii, C. beijerinckii DS, and perR mutant. (D) Residual glucose (g/L) after fermentation of C. beijerinckii, C. beijerinckii DS, and perR mutant. Error bars represent SD from three independent biological replicates, and asterisks denote statistical significance (ns = not significant; ** p < 0.01; *** p < 0.001).

Table 1.
Comparison of ABE production in shaking flask fermentation of C. beijerinckii, C. beijerinckii DS, and perR mutant.
pH and organic acid profiles (Figure 2C,D) showed all strains rapidly accumulated acetic/butyric acids (0–6 h), causing pH decline. During 6–12 h, acid re-assimilation substantially reduced concentrations in the perR mutant and wild-type 8052 (pH elevation), versus minimal changes in DS (relatively stable pH; impaired re-assimilation). At 12–24 h, butyric acid resurgence reduced pH in mutant/wild-type (acetic acid stable), while DS showed continuous acid accumulation (progressive pH decline). During 24–36 h, butyric acid re-assimilation resumed in mutant/wild-type (pH recovery) versus continued accumulation in DS (further acidification). From 36–96 h, acid concentrations stabilized in all strains (except mutant butyric acid, which declined until 48 h) with stable pH. Conclusively, C. beijerinckii DS’s impaired metabolic regulation caused persistent acidification, detrimental to growth/solvent production. The perR mutant restores dynamic acid regulation, maintaining optimal pH for proliferation and biosynthesis.
Significant metabolic divergence was present in the tested strains (Figure 2E and Figure 3A,B; Table 1). The oxygen-tolerant perR mutant achieved peak acetone (4.85 ± 0.39 g/L) and butanol (10.19 ± 0.56 g/L) concentrations, representing increases of 4.70 ± 0.40 g/L and 10.01 ± 0.57 g/L, respectively, over the solvent-deficient degenerated DS strain (acetone: 0.15 ± 0.01 g/L; butanol: 0.18 ± 0.01 g/L). Comparatively, these values were only 0.85 ± 0.52 g/L (acetone) and 1.79 ± 0.65 g/L (butanol) below those of the wild-type 8052 levels (5.38 ± 0.13 g/L acetone; 11.98 ± 0.09 g/L butanol). As shown in Figure 2F, the DS strain exhibited premature termination of solventogenesis at 12 h, reaching maximal total solvent yield (sum of acetone, ethanol, and butanol) of 1.62 ± 0.12 g/L by 24 h, with a production efficiency of 0.07 ± 0.01 g/L/h. In contrast, the wild-type 8052 achieved peak solvent accumulation (18.68 ± 0.26 g/L) at 60 h, corresponding to a superior production efficiency of 0.31 ± 0.01 g/L/h. The perR mutant displayed prolonged fermentation kinetics, attaining maximum solvent titer (15.61 ± 0.89 g/L) at 84 h with intermediate efficiency (0.19 ± 0.01 g/L/h). Therefore, perR knockout in degenerated C. beijerinckii DS reversed its solvent-deficient phenotype, elevating acetone/butanol production. This genetic modification confers oxygen tolerance and significantly enhances anaerobic ABE fermentation performance.
Moreover, the perR mutant exhibited a 453% increase in biomass over the solventogenesis-impaired progenitor C. beijerinckii DS (OD600: 9.96 ± 0.14 vs. 1.8 ± 0.02) while surpassing the wild-type C. beijerinckii by 22.8% (8.11 ± 0.11), confirming successful metabolic rewiring. This hyperproliferative phenotype enhanced ABE solvent titers in the mutant. However, prolonged fermentation (84 vs. 60 h) and a 61% reduction in productivity (0.19 vs. 0.31 g/L/h) relative to the wild-type strain (Table 1; Figure 3B) versus the wild-type represented significant improvement opportunities for industrial scale-up. This metabolic compensation suggests carbon flux may have been redistributed from solventogenesis to biomass formation, though the exact mechanism requires further investigation.
3.3. Transcriptomic Profiling Reveals Altered Expression of Detoxification and Redox Homeostasis Genes Following perR Deletion
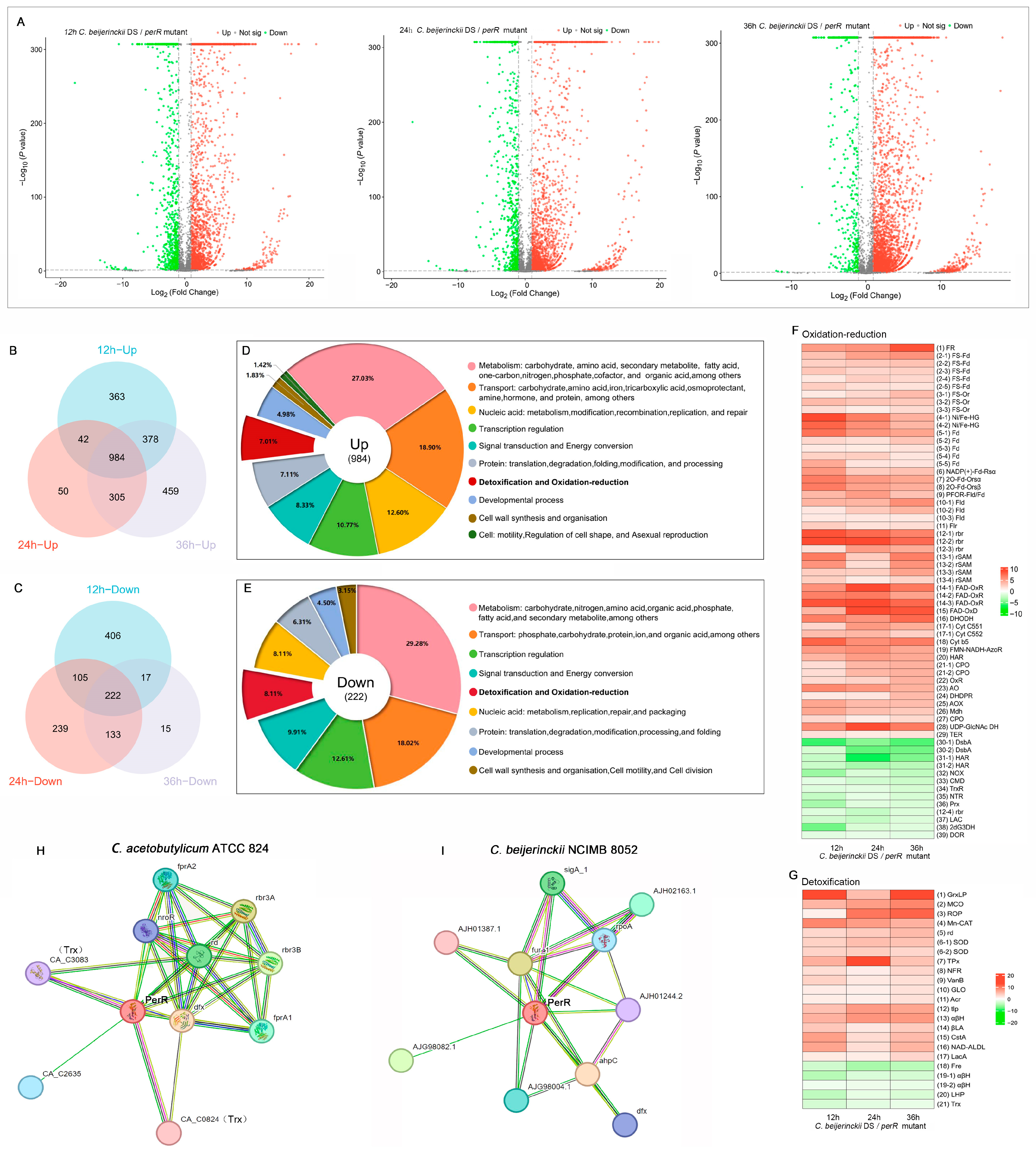
To delineate perR inactivation-induced transcriptional reprogramming, particularly its regulatory impact on oxidative stress defense systems and acidogenesis/solventogenesis pathways, we performed RNA-seq to compare the aerotolerant perR mutant with parental C. beijerinckii DS. RNA quality for sequencing met the requirements for HiSeq strand-specific transcriptome library construction. Specific values of the RNA integrity number (RIN) and concentration are provided in Table S3 and Figure S1. Using C. beijerinckii NCIMB 8052 (GenBank: CP000721.1) as the reference, RNA-seq analysis revealed dynamic transcriptional alterations during ABE fermentation phases. DEGs meeting the thresholds of |log2FC| >1 with p < 0.05 were subsequently subjected to downstream processing and visualization. In total, 3064 DEGs at 12 h (acidogenesis phase; 2162 upregulated and 902 downregulated), 2844 DEGs at 24 h (transitional phase; 1991 upregulated and 853 downregulated), and 3316 DEGs at 36 h (solventogenesis phase; 2845 upregulated and 471 downregulated) were identified (Figure 4A). Notably, perR deletion triggered global transcriptional reprogramming, with marked variations in DEGs abundance across diverse fermentation phases, reflecting phase-specific regulatory dynamics.
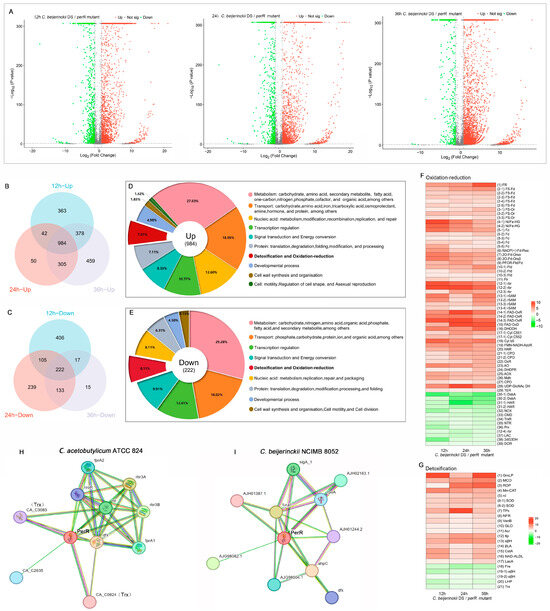
Figure 4.
PerR inactivation reprograms ABE fermentation in C. beijerinckii DS. (A) Volcano plots representing ABE fermentation transcriptome (12, 24, and 36 h) following perR deletion. Venn diagrams of core (B) co-upregulated and (C) co-downregulated DEGs across ABE fermentation stages after perR knockout. Gene Ontology (GO) functional classification of (D) co-upregulated and (E) co-downregulated DEGs post-perR knockout. Heatmap of differentially expressed (F) redox-associated and (G) detoxification-related DEGs. Color bar scale: log2FC (red: upregulated, green: downregulated). STRING database-predicted PerR-interaction proteins in (H) C. acetobutylicum ATCC 824 and (I) C. beijerinckii NCIMB 8052.
3.3.1. Temporal Profiling of Co-Regulated Detoxification and Redox Genes Across Fermentation Phases in the perR Mutant
To systematically investigate conserved oxidative stress reprogramming under PerR inactivation, we conducted comparative transcriptomics across fermentation phases (12/24/36 h) using intersecting Venn diagrams. After filtering hypothetical protein-encoding genes, 984 co-upregulated and 222 co-downregulated genes were identified across all phases (Figure 4B,C). Functional enrichment revealed significant association with discrete biological processes (Figure 4D,E), with 7.01% of co-upregulated genes associated with detoxification pathways and 8.11% of co-downregulated genes linked to redox homeostasis (Tables S1 and S2). The heatmaps demonstrated coordinated expression dynamics within these oxidative defense pathways (Figure 4F,G), showing hierarchical clustering of redox homeostasis and detoxification genes.
3.3.2. Integrative Analysis of STRING-Predicted PerR Targets in Oxidative Stress Defense Across C. acetobutylicum and C. beijerinckii
Our integrated analysis combining STRING database interrogation, PerR structural-functional predictions, transcriptional profiling, and DNA-binding assays revealed distinct regulatory mechanisms in these clostridial strains. In C. acetobutylicum, PerR directly regulates key oxidative defense components: NADH-rubredoxin oxidoreductase (nroR), rubredoxin (rd), flavodiiron proteins (fprA1/fprA2), desulfoferrodoxin (dfx), and reverse rubrerythrins (rbr3A/rbr3B) (Figure 4H). These components constitute a coordinated oxidative defense system. NROR initiates electron transfer from NADH to Rd (cysteine-stabilized Fe-S protein), which distributes electrons to diverse substrates: FprA1/2, the NADH oxidase that processes O2→H2O via 4e− transfer (FprA2 also scavenges NO); Dfx that directs Rd-derived electrons to detoxify O2− → H2O2; Rbr, the NADH peroxidase that functions in H2O2→H2O. This integrated system enables hierarchical ROS scavenging through a unified electron transport architecture: NADH→NROR→Rd→terminal oxidoreductases [19,20,21,22].
In C. beijerinckii, PerR mediates oxidative stress adaptation by transcriptionally regulating other molecules (Figure 4I), such as the fur family transcriptional regulator (fur_1) that mediates metal sensing (Fe/Zn/Mn/Ni) and stress response homeostasis, enabling iron-oxidative stress crosstalk [12], alkyl hydroperoxide reductase C (ahpC) for detoxifying H2O2 [23], and desulfoferrodoxin (dfx) that can convert O2− → H2O2 in anaerobes [22].
3.3.3. Temporal Co-Regulation Analysis and STRING-Based Prediction Reveal Key Detoxification and Redox Homeostasis Gene Expression Alterations Following perR Knockout
Through integrative analyses of temporal expression profiling (12/24/36 h) of co-regulated DEGs in detoxification and redox pathways, STRING-predicted PerR-direct targets elucidated PerR-dependent regulatory architecture governing oxidative defenses in C. beijerinckii DS. Crucially, iron serves essential roles in anaerobic C. acetobutylicum metabolism, requiring multifaceted homeostasis mechanisms, including high-efficiency iron acquisition, iron storage proteins, oxidative stress resistance, hierarchical iron–protein regulation, and global iron-responsive networks [24,25]. Temporal transcriptional profiling across three phases revealed significant upregulation of iron-associated redox proteins within co-regulated DEGs (39.22% of upregulated pathway components; Table S4), such as ferric reductase (FR), 4Fe-4S ferredoxin (FS-Fd), Fe-S oxidoreductase (FS-Or), Ni/Fe hydrogenase (Ni/Fe-HG), ferredoxin (Fd), ferredoxin-NADP (+) reductase subunit alpha (NADP (+)-Fd-Rsα), ferredoxin oxidoreductase subunits alpha/beta (OFORα/β), and pyruvate flavodoxin/ferredoxin oxidoreductase domain-containing protein (PFOR-Fld/Fd) (Figure 4F).
Moreover, the enzymes that utilize NADPH to maintain reductive environments, quench catalytic radical intermediates, and mitigate oxidative damage risks were significantly upregulated (Figure 4F; Table S4), including flavodoxins (Flds) that can substitute for ferredoxins (Fds) under iron limitation to maintain metabolism and stress adaptability [26] and radical SAM proteins (rSAMs) that regulate radical generation via [4Fe-4S] clusters [27,28]. The genes encoding multicopper oxidases (MCOs) that catalyze O2→H2O reduction without ROS release [29] and superoxide dismutase (SOD) that dismutates O2−→H2O2 + O2 to alleviate oxidative damage [30] were also markedly upregulated after PerR inactivation (Figure 4G and Table S5).
Notably, the co-regulated DEGs encoding rubrerythrin (Rbr3A/Rbr3B) and rubredoxin (Rd) merited further investigation, as they are simultaneously predicted as PerR targets by STRING (Figure 4H). Rbr targets are key oxidative stress defense proteins in anaerobic bacteria, capable of neutralizing peroxides (e.g., H2O2) through iron-catalyzed mechanisms, collaborating with superoxide reductase to scavenge superoxide radicals, enhancing oxygen tolerance, and participating in electron transfer networks [31]. Rds are small iron–sulfur (Fe–S) cluster-containing proteins in anaerobes, serving as a central electron transfer hub in C. acetobutylicum and enabling efficient scavenging of O2, O2−, and H2O2 [22]. Therefore, transcriptomic analysis revealed that perR knockout induced significant upregulation of redox and detoxification-associated genes, with approximately 40% participating in iron/iron–sulfur cluster-dependent redox reactions. These coordinated transcriptional responses establish the core oxidative stress defense machinery in C. beijerinckii DS, orchestrating peroxide detoxification, radical scavenging, and iron-mediated electron transfer networks.
3.4. Transcriptomic Profiling Following perR Knockout Reveals Altered Expression of Genes in ABE Production-Related Metabolic Pathways
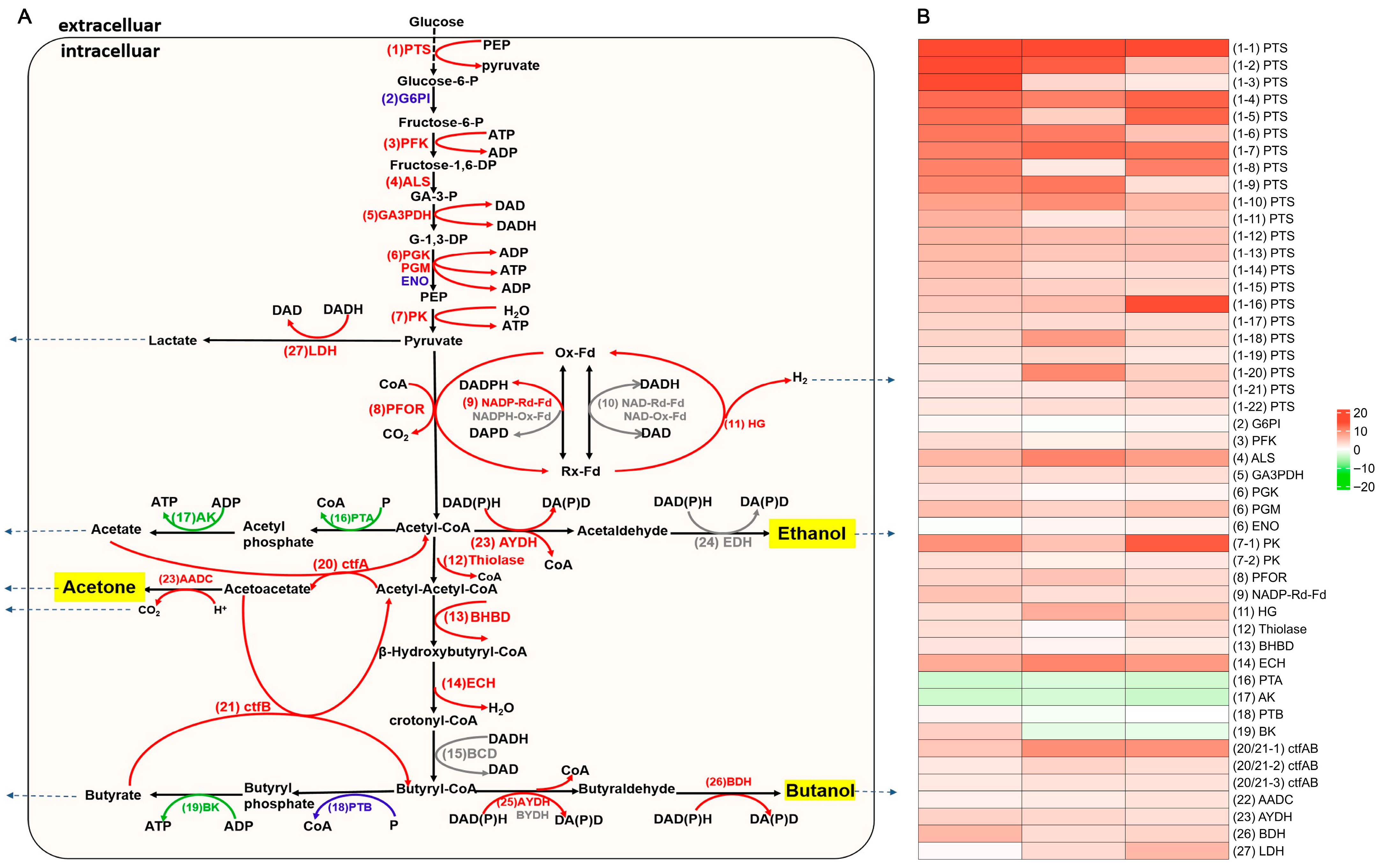
The genetic knockout of perR in the degenerated C. beijerinckii DS conferred two advantages: enabling proliferation under ambient oxygen conditions and restoring impaired solventogenesis capacity. These advantages enhanced ABE fermentation efficiency in the perR mutant. Consequently, we performed transcriptomic analysis of DEGs within the glucose-driven ABE biosynthesis pathway. As shown in Figure 5A, the glucose-to-ABE metabolic pathways highlighted 27 core enzymatic nodes, 23 enzyme systems (comprising 48 associated genes) with differential expression across 12/24/36 h phases after perR knockout (Table S6). These transcriptional alterations correlate with the perR knockout, suggesting its regulatory role in solventogenic metabolism.
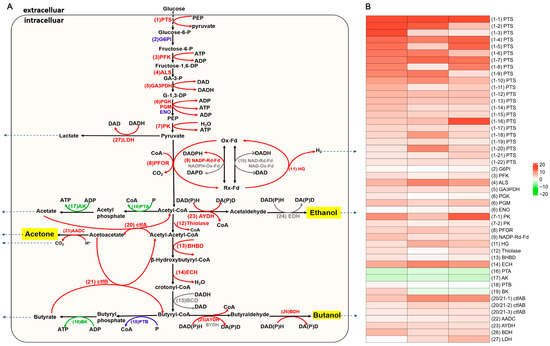
Figure 5.
Knockout of perR reprograms ABE fermentation network in C. beijerinckii DS. (A) Metabolic pathways of ABE fermentation from glucose to acetone, butanol, and ethanol in C. beijerinckii. Gene expression changes in perR mutant: red text/lines: upregulated core enzymes, green text/lines: downregulated molecules, blue text/lines: non-significant expression changed enzymes. and gray text/lines: undetected molecules in transcriptome. (B) Heatmap showing differentially expressed metabolic network genes. Color bar scale: log2FC, red: upregulated, green: downregulated.
3.4.1. perR Deletion Upregulates PTS Transporters and Core Glycolytic Enzymes for Metabolic Flux from Glucose to Pyruvate
In Clostridium, the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) primarily mediates carbohydrate uptake (glucose, sucrose, maltose, lactose, and mannitol) [32]. Transcriptomics analysis of the perR mutant revealed 22 significantly upregulated PTS genes (vs. parental strain) and 12 transporter subunits for carbohydrate utilization, including cellobiose (IIB, IIC, IIBC), maltose (IIBC), mannitol (IIB), galactitol (IIB), mannose (IIA, IID), fructose (IIA), lactose (IIC), β-glucoside (IIBC), sorbose (IIC, IIB), glucose (IIA), N-acetylgalactosamine (IIB), and sucrose (IIBC) (Figure 5B; Table S6). This transcriptional reprogramming correlates with the (2.84 ± 0.12)-fold increase in glucose consumption and (3.13 ± 0.56)-fold enhancement in uptake efficiency of the mutant (Figure 2B; Table S6). These findings indicate that PTS upregulation improves substrate acquisition in perR-deficient ABE fermentation.
Phosphofructokinase (PFK), a key Embden-Meyerhof-Parnas (EMP) pathway enzyme, catalyzes the phosphorylation of fructose-6-phosphate (F6P) to fructose-1,6-bisphosphate (F1,6-BP). In C. acetobutylicum, PFK and pyruvate kinase (PK) collectively regulate two irreversible EMP steps, controlling glycolytic flux [32]. Transcriptomics revealed that the PFK gene (CBEI_RS24735) and two PK genes (CBEI_RS07420, CBEI_RS24730) were significantly upregulated in the mutant compared with the parental strain (Figure 5B; Table S6). This coordinated overexpression correlates with increased glycolytic flux, strongly supporting enhanced glycolytic commitment.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the reversible phosphorylation of glyceraldehyde-3-phosphate (G3P) to glycerate-1,3-diphosphate (G-1,3-DP) in the EMP pathway with strict NAD specificity. Then, G-1,3-DP is sequentially converted by phosphoglycerate kinase (PGK), phosphoglycerate mutase (PGM), and enolase (ENO) to produce phosphoenolpyruvate (PEP) [32]. Transcriptomics revealed significant upregulation of key glycolytic genes in the mutant strain versus parental control, such as GAPDH-encoding CBEI_RS11815, PGM-encoding CBEI_RS16300, and PGK-encoding CBEI_RS03310 (Figure 5B; Table S6). This coordinated overexpression indicates enhanced flux through these critical glycolytic nodes.
3.4.2. perR Inactivation Upregulates Electron Carriers and Redox Enzymes to Amplify ABE Metabolic Flux
Pyruvate–ferredoxin oxidoreductase (PFOR) mediates the oxidative decarboxylation of pyruvate to acetyl-CoA and CO2 while concomitantly reducing ferredoxin, serving as the pivotal enzyme governing the acidogenesis–solventogenesis transition in Clostridium [32]. perR knockout significantly upregulated the PFOR-encoding gene CBEI_RS07645 versus parental control (Figure 5B; Table S6), enhancing pyruvate-to-acetyl-CoA conversion, a critical nodal point connecting interdependent fermentative pathways.
In the glucose-to-butanol pathway of obligate anaerobic Clostridium species (Figure 5A), ferredoxin (Fd) coordinates three core redox mechanisms critical for ABE metabolism: reduced Fd donates electrons to Fd-NAD (P) reductase for NAD (P)H generation, reduced Fd transfers electrons to hydrogenase for H2 production, and oxidized Fd cooperates with PFOR to cleave pyruvate into acetyl-CoA and CO2 [32]. Notably, NAD (P)H-Ox-Fd generates NAD (P)H by capturing reducing equivalents from ferredoxin, thereby suppressing hydrogen evolution and redirecting carbon flux toward butanol/butyrate biosynthesis [33]. Experimental evidence confirms that elevated activities of NADH-dependent butyraldehyde dehydrogenase and NAD (P)-Rx-Fd reductase promote reducing power channeling toward alcohol synthesis [34]. Comparative transcriptomics revealed significant upregulation of ferredoxin–NADP (+) reductase subunit α (CBEI_RS11300) in the perR mutant (Figure 5B; Table S6). These findings indicate that perR knockout upregulates ferredoxin-NADP (+) reductase to coordinate electron flux partitioning between acidogenesis/solventogenesis, enhancing butanol production as evidenced by ABE fermentation data.
In clostridia, hydrogenase regulates H2 production and electron partitioning, directly impacting acidogenesis/solventogenesis balance and intracellular redox state. It competes with ferredoxin-NAD (P) reductase for oxidizing electrons from reduced ferredoxin (Rd-Fd), while oxygen inactivates its H-cluster active site [32]. Transcriptomics revealed concurrent upregulation of ferredoxin-NADP (+) reductase (CBEI_RS11300) and hydrogenase (CBEI_RS03425) in the mutant. This coordinated overexpression likely stems from perR knockout enhancing oxidative stress defenses and O2 scavenging capacity.
3.4.3. perR Knockout Coordinates Upregulation of Thiolase/BHBD/Crotonase Nodes to Enhance Acetyl-CoA to Acetone/Butanol Conversion in the ABE Pathway
In C. acetobutylicum, thiolase (acetyl-CoA acetyltransferase) catalyzes the condensation of two acetyl-CoA molecules into acetoacetyl-CoA, the critical precursor for butanol/acetone biosynthesis. This enzyme competitively regulates metabolic flux by competing with both phosphotransacetylase (PTA) for acetyl-CoA during acidogenesis and aldehyde dehydrogenase (AYDH) during solventogenesis, thus modulating the butanol:acetone:ethanol ratio [32]. Transcriptomics revealed significant upregulation of acetyl-CoA acetyltransferase-encoding gene CBEI_RS02340 in the mutant versus parental strain (Figure 5B; Table S6).
β-Hydroxybutyryl-CoA dehydrogenase (BHBD) catalyzes the reduction of acetoacetyl-CoA to β-hydroxybutyryl-CoA, serving as the pivotal enzyme for butyrate/butanol biosynthesis by directly determining production efficiency [32]. Transcriptomics revealed significant upregulation of the 3-hydroxybutyryl-CoA dehydrogenase gene CBEI_RS01785 in the perR mutant (Figure 5B; Table S6).
In clostridial ABE fermentation (forward reaction), enoyl-CoA hydratase (crotonase) catalyzes the dehydration of β-hydroxybutyryl-CoA to crotonyl-CoA, a pivotal step in butanol biosynthesis [32]. Transcriptomics revealed significant upregulation of enoyl-CoA hydratase-encoding Cbei_RS01990 in the mutant versus parental strain (Figure 5B; Table S6).
Transcriptomic remodeling at three critical ABE metabolic nodes (thiolase, BHBD, and crotonase) in the oxygen-tolerant mutant strain significantly enhanced acetone and butanol production while substantially reducing acetate and butyrate accumulation. These metabolic shifts demonstrate that the perR mutant’s targeted regulation of these enzymatic nodes effectively circumvents solventogenesis limitations in degenerated strains through coordinated transcriptional reprogramming.
3.4.4. perR Knockout Modulates Acid Accumulation and Reassimilation
Phosphotransacetylase (PTA) catalyzes acetyl-CoA to acetyl-phosphate conversion during acidogenesis, which subsequently produces acetate and ATP by acetate kinase (AK) catalyzation. In C. acetobutylicum ABE fermentation, PTA and AK activities are significantly downregulated during the metabolic shift to solventogenesis, concurrent with reduced acetate production and decelerated cell growth [32]. Transcriptomics revealed marked downregulation of PTA- (CBEI_RS06155) and AK-encoding (CBEI_RS06160) genes in the perR mutant relative to the parental strain (Figure 5B; Table S6). Phenotypically, the perR mutant maintained consistently low acetate levels, contrasting with continuous acetate accumulation in the progenitor (Figure 2D). This transcriptional suppression likely facilitates acetate reassimilation, thereby redistributing metabolic flux toward solventogenesis.
Phosphotransbutyrylase (PTB) and butyrate kinase (BK) cooperatively catalyze butyryl-CoA to butyrate conversion during acidogenesis, with higher enzymic activity than in solventogenesis [32]. In contrast to the solventogenesis-impaired parental strain, the perR mutant showed no differential expression in the PTB-encoding gene (CBEI_RS01180), while the BK-encoding gene (CBEI_RS23520) exhibited significant upregulation at 12 h, followed by downregulation at 24 and 36 h (Figure 5B; Table S6). This expression pattern parallels the oxygen-tolerant mutant’s butyrate production, gradually increasing from 12 to 24 h, then progressively decreasing through 36 h (Figure 2D).
CoA transferase (ctfAB) catalyzes CoA transfer from acetoacetyl-CoA to acetate/butyrate, yielding acetoacetate with acetyl-/butyryl-CoA. This regulatory enzyme mitigates acid crash by reassimilating acids to drive solventogenesis, preferentially utilizing butyrate in C. acetobutylicum [32,35]. Transcriptomics revealed coordinated upregulation of three ctfAB genes (CBEI_RS16830, CBEI_RS17190, and CBEI_RS19555) in the mutant versus the solventogenesis-deficient parental strain during three fermentation phases (12/24/36 h) (Figure 5B; Table S6). Metabolic analysis demonstrated significantly reduced acetate/butyrate production in the mutant, along with active butyrate reassimilation during two critical intervals: 6–12 h and 24–48 h (Figure 2E). This phenomenon is likely associated with significant upregulation of three CoA transferase-encoding genes that enhance acid reutilization.
3.4.5. perR Knockout Modulates Transcriptional Regulation of Core Terminal Enzymes in Acetone, Ethanol, and Butanol Biosynthesis
In clostridia, the key enzyme for acetone generation from acetoacetyl-CoA is acetoacetate decarboxylase (AADC), which irreversibly decarboxylates acetoacetate to drive acetone synthesis and acetate/butyrate reabsorption. AADC activity increases 40-fold during solventogenesis [32]. Comparative transcriptomics revealed progressive upregulation of the AADC-encoding gene CBEI_RS19640 in the perR mutant (Table S6). This enhancement likely promoted acetone production and acid reassimilation during ABE fermentation (Figure 3A,B; Table 1).
Aldehyde dehydrogenase (AYDH) acts as the rate-limiting enzyme for ethanol/butanol synthesis in C. acetobutylicum and C. beijerinckii. It catalyzes the conversions of acetyl-CoA to acetaldehyde and butyryl-CoA to butyraldehyde with higher catalytic efficiency toward butyraldehyde synthesis than acetaldehyde. However, AYDH is oxygen-sensitive, which can be rapidly inactivated by ambient O2. Alternatively, butyraldehyde dehydrogenase (BYDH) specifically converts butyryl-CoA to butyraldehyde, followed by butanol dehydrogenase (BDH) catalyzation of the NAD (P)H-dependent butyraldehyde to butanol reduction [32,36]. Transcriptomics revealed significant upregulation of AYDH-encoding CBEI_RS01685 and BDH-encoding CBEI_RS11295 in the perR mutant versus parental strain (Figure 5B; Table S6), while BYDH and ethanol dehydrogenase (EDH)-encoding genes showed no differential expression. Phenotypic analysis showed the perR mutant maintained low ethanol production similar to the parental strain but exhibited substantially enhanced butanol synthesis (Figure 2E, Figure 3A,B; Table 1). These observations indicate that AYDH upregulation preferentially directs butyryl-CoA flux toward butyraldehyde, subsequently converted to butanol via amplified BDH activity, while unchanged EDH expression correlates with unaltered ethanol synthesis. The perR knockout-induced oxidative stress defense enhancement likely stabilizes oxygen-sensitive AYDH, further promoting butanol production.
3.5. perR Orchestrates Multilayered Redox Homeostasis and Fermentation Adaptation via Conserved Motif Networks
To explore the regulatory role of PerR in redox stress and ABE pathways in C. beijerinckii DS, we computationally analyzed putative PerR-binding candidates among annotated DEGs. The evolutionarily conserved PerR motif (TTATAATnATTATAA), which has been characterized in Bacillus subtilis, Staphylococcus aureus, and C. acetobutylicum [20], was employed in the scanning of 64 oxidation-reduction-related genes (Table S4), 23 detoxification-associated genes (Table S5), and 48 genes from ABE production-related metabolic pathways (Table S6) using the Find Individual Motif Occurrences (FIMO) tool (v5.5.7). Results showed that 34 conserved genes with canonical PerR-binding elements (Table S7). Computational prediction revealed direct PerR regulation of 35.9% (23/64) redox genes, 17.4% (4/23) detoxification genes, and 14.6% (7/48) ABE pathway genes. These findings not only confirm the central role of PerR in orchestrating oxidative stress defense systems but also suggest potential multilayered regulatory mechanisms extending beyond conventional transcription factor-target gene interactions.
3.6. qRT-PCR Verification of PerR Regulation in C. beijerinckii DS
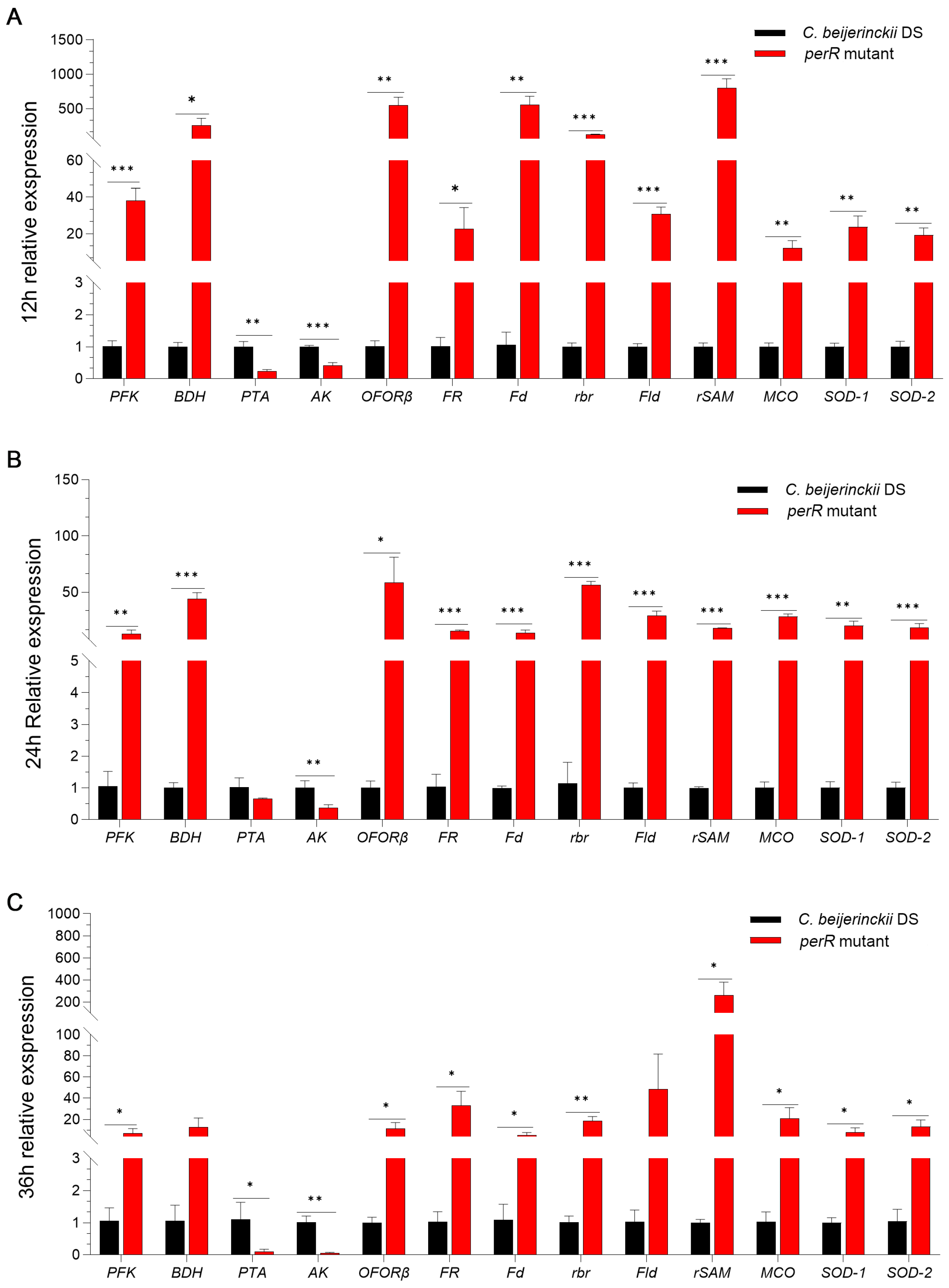
To validate the transcriptional effect of PerR on oxidative stress response and ABE metabolism genes in C. beijerinckii DS, we performed qRT-PCR using RNAs derived from the perR mutant and parental strain collected at 12, 24, and 36 h of fermentation. Key redox, detoxification, and ABE biosynthetic pathways’ DEGs from transcriptomics were analyzed. As shown in Figure 6, the results demonstrated consistent expression patterns for 13 genes across three time points (76% concordance with transcriptome data). Key ABE pathway enzymes (PFK, BDH, PTA, and AK) exhibited significant expression shifts. PFK/BDH upregulation enhances glycolysis/butanol production, while PTA/AK downregulation redirects flux from acetate to solventogenesis [32]. Five redox-associated genes encode enzymes (OFORβ, FR, Fd, rbr, Fld, and rSAM) controlling electron transfer [26,27,28], and three detoxification enzymes (MCO, SOD-1, SOD-2) reduce O2 and ROS and scavenge radicals [29,30], collectively boosting oxidative stress defense.
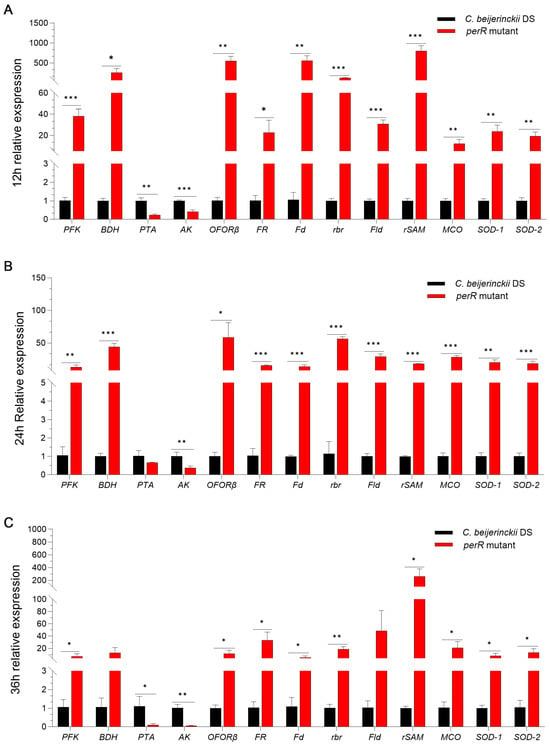
Figure 6.
Multi-temporal qRT-PCR validation of transcriptomic data showing coordinated expression shifts in ABE pathway enzymes, redox regulators, and oxidative defense machinery in the perR mutant and its parental strain collected at (A) 12 h, (B) 24 h, and (C) 36 h of fermentation. Asterisks denote statistical significance (* p < 0.05; ** p < 0.01; *** p < 0.001) determined by unpaired Student’s t-test.
In summary, the Cbei_1336 was first characterized as perR in C. beijerinckii NCIMB 8052. Deletion of perR in a solvent-deficient variant C. beijerinckii DS significantly promoted the oxygen tolerance of the mutant, as evidenced by robust growth under ambient aerobic conditions (21% O2). Moreover, perR deletion reverted the solvent-deficient phenotype of C. beijerinckii DS. While enhanced aerotolerance production was present in the perR knockout C. acetobutylicum ATCC 824 [13], the molecular mechanism underlying PerR regulation remains to be elucidated. Our transcriptomic profiling demonstrated at least two roles of PerR in C. beijerinckii NCIMB 8052: negative regulation of bacterial oxygen resilience and metabolic reprogramming that redirects carbon flux toward butanol/acetone biosynthesis. Overall, this study provides a clostridial chassis for both robust proliferation under aerobic conditions and butanol/acetone production based on ABE fermentation.
4. Conclusions
Targeted perR knockout in C. beijerinckii DS dramatically enhanced oxygen tolerance and ABE fermentation, enabling unprecedented proliferation under atmospheric oxygen (21% O2) where parental/wild-type 8052 strains failed. This genetic intervention revitalized the potential of a degenerated strain, with a (3.79 ± 0.09)-fold increase in bacterial biomass, a (9.64 ± 0.90)-fold enhancement in total solvent yield, and a (2.71 ± 0.04)-fold improvement in volumetric productivity. The perR knockout strain demonstrated reduced volumetric productivity and yield compared to the wild-type industrial strain 8052, achieving 0.61 ± 0.04-fold (0.19 ± 0.01 vs. 0.31 ± 0.01 g/L/h) and 0.89 ± 0.06-fold (0.33 ± 0.02 vs. 0.37 ± 0.01 g/g) of the respective metrics. Transcriptomic analysis revealed that the perR knockout orchestrates dual transcriptional reprogramming: unified oxidative defense activation through peroxide detoxification, radical scavenging, and iron-mediated electron transfer and metabolic rewiring toward solventogenesis by amplifying glycolytic flux, enhancing electron transfer efficiency, redirecting carbon toward butanol/acetone biosynthesis, suppressing acidogenic side pathways, and activating terminal solvent-forming enzymes. By genetically uncoupling PerR-mediated repression, we provide an industrial biobutanol production strategy.
Our findings enable robust clostridial proliferation under ambient aerobic conditions (21% O2), simultaneously achieving high-efficiency genetic editing and enhanced ABE fermentation performance. Importantly, the acquired oxygen tolerance reduces the substantial costs during industrial ABE fermentation. Concurrently, redirected carbon flux toward acetone/butanol synthesis improves substrate-to-product conversion yield. Such a transformative advancement is promising in cost-effective biobutanol manufacturing and sustainable biofuel development. However, this study only provided initial insights rather than comprehensive validation: (i) While we identified PerR’s crucial role in regulating oxidative stress defense and ABE metabolic reprogramming, the functional validation of downstream genes requires further experimental characterization; (ii) The genetic stability of the engineered strain remains to be systematically evaluated through serial passaging studies; and (iii) Our validation of enhanced ABE fermentation performance was confined to small-scale flask experiments, while the scaled-up pilot-scale validation is essential prior to industrial implementation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation11090526/s1, Figure S1: RNA quality assessment by Agilent 2100 Bioanalyzer; Table S1: Bacterial strains and plasmids used in this study; Table S2: Primers used in this study; Table S3: RNA sample and characterization; Table S4: Oxidation-reduction-related DEGs in ABE fermentation of C. beijerinckii DS following perR knockout; Table S5: Detoxification-related DEGs in ABE fermentation of C. beijerinckii DS following perR knockout; Table S6: perR deletion resulted in dysregulation of genes involved in glucose→ABE metabolism; Table S7: Predicted PerR-targeted genes and motifs in C. beijerinckii DS.
Author Contributions
Conceptualization, L.L., X.R., and L.Y.; Data curation, J.D., N.Z., S.D., and L.Y.; Formal analysis, C.X., J.D., N.Z., L.L., and S.D.; Funding acquisition, C.X. and L.Y.; Investigation, C.X., J.D., N.Z., and S.D.; Methodology, C.X. and L.L.; Project administration, L.Y.; Supervision, X.R.; Validation, C.X. and X.R.; Writing-original draft, C.X.; Writing-review and editing, J.D., N.Z., X.R., and L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation Project of Chongqing, Chongqing Science and Technology Commission (cstc2021jcyj-msxmX0483).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw RNA sequencing data generated in this study are available in the NCBI SRA database under BioProject accession number PRJNA1284305. All other original contributions are included in the article and Supplementary Material. Further inquiries should be directed to the corresponding authors.
Acknowledgments
We thank Nigel P. Minton, University of Nottingham, UK, for kindly providing plasmid pMTL007C-E2.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Guo, Y.; Liu, Y.; Guan, M.; Tang, H.; Wang, Z.; Lin, L.; Pang, H. Production of butanol from lignocellulosic biomass: Recent advances, challenges, and prospects. RSC Adv. 2022, 12, 18848–18863. [Google Scholar] [CrossRef]
- García, V.; Päkkilä, J.; Ojamo, H.; Muurinen, E.; Keiski, R.L. Challenges in biobutanol production: How to improve the efficiency? Renew. Sust. Energ. Rev. 2011, 15, 964–980. [Google Scholar] [CrossRef]
- Xiao, H.; Li, Z.; Jiang, Y.; Yang, Y.; Jiang, W.; Gu, Y.; Yang, S. Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid. Metab. Eng. 2012, 14, 569–578. [Google Scholar] [CrossRef]
- Riaz, S.; Mazhar, S.; Abidi, S.H.; Syed, Q.; Abbas, N.; Saleem, Y.; Nadeem, A.A.; Maryam, M.; Essa, R.; Ashfaq, S. Biobutanol production from sustainable biomass process of anaerobic ABE fermentation for industrial applications. Arch. Microbiol. 2022, 204, 672. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhang, Y.; Dai, Z.; Li, Y. Engineering clostridium strain to accept unmethylated DNA. PLoS ONE 2010, 5, e9038. [Google Scholar] [CrossRef]
- Czerska, M.; Mikołajewska, K.; Zieliński, M.; Gromadzińska, J.; Wąsowicz, W. Today’s oxidative stress markers. Med. Pr. 2015, 66, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Malych, R.; Fussy, Z.; Zeniskova, K.; Arbon, D.; Hampl, V.; Hrdy, I.; Sutak, R. The response of Naegleria gruberi to oxidative stress. Metallomics 2022, 14, mfac009. [Google Scholar] [CrossRef] [PubMed]
- Morvan, C.; Folgosa, F.; Kint, N.; Teixeira, M.; Martin-Verstraete, I. Responses of Clostridia to oxygen: From detoxification to adaptive strategies. Environ. Microbiol. 2021, 23, 4112–4125. [Google Scholar] [CrossRef]
- Kint, N.; Morvan, C.; Martin-Verstraete, I. Oxygen response and tolerance mechanisms in Clostridioides difficile. Curr. Opin. Microbiol. 2022, 65, 175–182. [Google Scholar] [CrossRef]
- Hillmann, F.; Fischer, R.J.; Saint-Prix, F.; Girbal, L.; Bahl, H. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol. Microbiol. 2008, 68, 848–860. [Google Scholar] [CrossRef]
- Ji, C.J.; Yang, Y.M.; Kim, J.H.; Ryu, S.H.; Youn, H.; Lee, J.W. The roles of two O-donor ligands in the Fe2+-binding and H2O2-sensing by the Fe2+-dependent H2O2 sensor PerR. Biochem. Biophys. Res. Commun. 2018, 501, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Pinochet-Barros, A.; Helmann, J.D. Bacillus subtilis Fur Is a Transcriptional Activator for the PerR-Repressed pfeT Gene, Encoding an Iron Efflux Pump. J. Bacteriol. 2020, 202, e00697-19. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Tao, W.; Zhu, L.; Zhang, Y.; Li, Y. CAC2634-disrupted mutant of Clostridium acetobutylicum can be electrotransformed in air. Lett. Appl. Microbiol. 2011, 53, 379–382. [Google Scholar] [CrossRef]
- Xiao, C.; Fan, W.; Du, S.; Liu, L.; Wang, C.; Guo, M.; Zhang, L.; Zhang, M.; Yu, L. A novel glycosylated solution from Dioscorea zingiberensis C.H. Wright significantly improves the solvent productivity of Clostridium beijerinckii. Bioresour. Technol. 2017, 241, 317–324. [Google Scholar] [CrossRef]
- Heap, J.T.; Kuehne, S.A.; Ehsaan, M.; Cartman, S.T.; Cooksley, C.M.; Scott, J.C.; Minton, N.P. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 2010, 80, 49–55. [Google Scholar] [CrossRef]
- Mermelstein, L.D.; Welker, N.E.; Bennett, G.N.; Papoutsakis, E.T. Expression of cloned homologous fermentative genes in Clostridium acetobutylicum ATCC 824. Biotechnology 1992, 10, 190–195. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Milne, C.B.; Janssen, H.; Lin, W.; Phan, G.; Hu, H.; Jin, Y.S.; Price, N.D.; Blaschek, H.P. Development of a gene knockout system using mobile group II introns (Targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii. Appl. Environ. Microbiol. 2013, 79, 5853–5863. [Google Scholar] [CrossRef]
- Li, L.Q.; Fu, C.H.; Zhao, C.F.; Xia, J.; Wu, W.J.; Yu, L.J. Efficient extraction of RNA and analysis of gene expression in a long-term Taxus cell culture using real-time RT-PCR. Z. Naturforsch C J. Biosci. 2009, 64, 125–130. [Google Scholar] [CrossRef]
- Guedon, E.; Petitdemange, H. Identification of the Gene Encoding NADH-Rubredoxin Oxidoreductase in Clostridium acetobutylicum. Biochem. Biophys. Res. Commun. 2001, 285, 496–502. [Google Scholar] [CrossRef]
- Hillmann, F.; Riebe, O.; Fischer, R.J.; Mot, A.; Caranto, J.D.; Kurtz, D.M., Jr.; Bahl, H. Reductive dioxygen scavenging by flavo-diiron proteins of Clostridium acetobutylicum. FEBS Lett. 2009, 583, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Riebe, O.; Fischer, R.J.; Wampler, D.A.; Kurtz, D.M.; Bahl, H. Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology (Read.) 2009, 155 Pt 1, 16–24. [Google Scholar] [CrossRef]
- Kawasaki, S.; Sakai, Y.; Takahashi, T.; Suzuki, I.; Niimura, Y. O2 and Reactive Oxygen Species Detoxification Complex, Composed of O2-Responsive NADH: Rubredoxin Oxidoreductase-Flavoprotein A2-Desulfoferrodoxin Operon Enzymes, Rubperoxin, and Rubredoxin, in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2009, 75, 1021–1029. [Google Scholar] [CrossRef]
- Li, J.; Yan, Y.; Yang, L.; Ding, S.; Zheng, Y.; Xiao, Z.; Yang, A.; Liang, W. Duality of H2O2 detoxification and immune activation of Ralstonia solanacearum alkyl hydroperoxide reductase C (AhpC) in tobacco. Int. J. Biol. Macromol. 2024, 279 Pt 1, 135138. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Vasileva, D.; Janssen, H.; Hönicke, D.; Ehrenreich, A.; Bahl, H. Effect of iron limitation and fur gene inactivation on the transcriptional profile of the strict anaerobe Clostridium acetobutylicum. Microbiology (Read.) 2012, 158 Pt 7, 1918–1929. [Google Scholar] [CrossRef]
- Troitzsch, D.; Knop, R.; Dittmann, S.; Bartel, J.; Zühlke, D.; Möller, T.A.; Trän, L.; Echelmeyer, T.; Sievers, S. Characterizing the flavodoxin landscape in Clostridioides difficile. Microbiol. Spectr. 2024, 12, e0189523. [Google Scholar] [CrossRef]
- Sofia, H.J.; Chen, G.; Hetzler, B.G.; Reyes-Spindola, J.F.; Miller, N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001, 29, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Mathew, L.G.; Brimberry, M.; Lanzilotta, W.N. Class C Radical SAM. Methyltransferases Involved in Anaerobic Heme Degradation. ACS Bio Med. Chem. Au 2022, 2, 655. [Google Scholar] [CrossRef]
- Komori, H.; Higuchi, Y. Structural insights into the O2 reduction mechanism of multicopper oxidase. J. Biochem. 2015, 158, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y.; Zhang, Y.; Zhang, Y.; Wang, W.; Han, H.; Yang, C.; Dong, X. Superoxide dismutase ameliorates oxidative stress and regulates liver transcriptomics to provide therapeutic benefits in hepatic inflammation. PeerJ 2023, 11, e15829. [Google Scholar] [CrossRef]
- Knop, R.; Keweloh, S.; Pukall, J.; Dittmann, S.; Zühlke, D.; Sievers, S. A rubrerythrin locus of Clostridioides difficile encodes enzymes that efficiently detoxify reactive oxygen species. Anaerobe 2025, 92, 102941. [Google Scholar] [CrossRef] [PubMed]
- Gheshlaghi, R.; Scharer, J.M.; Moo-Young, M.; Chou, C.P. Metabolic pathways of clostridia for producing butanol. Biotechnol. Adv. 2009, 27, 764–781. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Thakker, C.; Zhu, F.; Pena, M.; San, K.Y.; Bennett, G.N. Improvement of butanol production in Clostridium acetobutylicum through enhancement of NAD (P)H availability. J. Ind. Microbiol. Biotechnol. 2018, 45, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Girbal, L.; Soucaille, P. Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: Role of NADH/NAD ratio and ATP pool. J. Bacteriol. 1994, 176, 6433–6438. [Google Scholar] [CrossRef]
- Yu, L.; Zhao, J.; Xu, M.; Dong, J.; Varghese, S.; Yu, M.; Tang, I.C.; Yang, S.T. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production: Effects of CoA transferase. Appl. Microbiol. Biotechnol. 2015, 99, 4917–4930. [Google Scholar] [CrossRef]
- Yan, R.T.; Chen, J.S. Coenzyme A-acylating aldehyde dehydrogenase from Clostridium beijerinckii NRRL B592. Appl. Environ. Microbiol. 1990, 56, 2591–2599. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).