Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Media
2.2. DNA Manipulation
2.3. SA Production under Conditions of Oxygen Deprivation
2.4. Analytical Techniques
2.5. Detection of Biotinylated Protein
3. Results
3.1. Construction of a Plasmid-Free Strain for SA Production
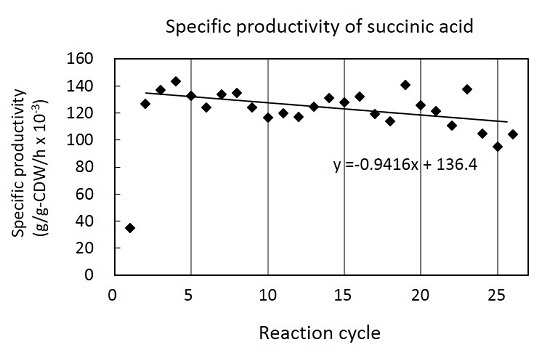
3.2. Cell Recycle Fed-Batch Reactions
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McKinlay, J.B.; Vieille, C.; Zeikus, J.G. Prospects for a bio-based succinate industry. Appl. Microbiol. Biotechnol. 2007, 76, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Thakker, C.; Martinez, I.; San, K.Y.; Bennett, G.N. Succinate production in Escherichia coli. Biotechnol. J. 2012, 7, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Succinity. Succinity Produces First Commercial Quantities of Biobased Succinic Acid. Available online: http://www.succinity.com/9-news/9-first-commercial-succinic-acid (accessed on 1 December 2015).
- Ikeda, M.; Takeno, S. Amino acid production by Corynebacterium glutamicum. In Corynebacterium Glutamicum; Yukawa, H., Inui, M., Eds.; Springer: Heidelberg, Germany, 2013; Volume 23, pp. 107–147. [Google Scholar]
- Inui, M.; Kawaguchi, H.; Murakami, S.; Vertès, A.A.; Yukawa, H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004, 8, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Jojima, T.; Noburyu, R.; Sasaki, M.; Tajima, T.; Suda, M.; Yukawa, H.; Inui, M. Metabolic engineering for improved production of ethanol by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2015, 99, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Suda, M.; Niimi, S.; Inui, M.; Yukawa, H. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol. Bioeng. 2013, 110, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Cho, K.M.; Liao, J.C. Engineering Corynebacterium glutamicum for isobutanol production. Appl. Microbiol. Biotechnol. 2010, 87, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Blombach, B.; Riester, T.; Wieschalka, S.; Ziert, C.; Youn, J.W.; Wendisch, V.F.; Eikmanns, B.J. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl. Environ. Microbiol. 2011, 77, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, Y.; Yamamoto, S.; Suda, M.; Inui, M.; Yukawa, H. Reactions upstream of glycerate-1,3-bisphosphate drive Corynebacterium glutamicum d-lactate productivity under oxygen deprivation. Appl. Microbiol. Biotechnol. 2013, 97, 6693–6703. [Google Scholar] [CrossRef] [PubMed]
- Mimitsuka, T.; Sawai, H.; Hatsu, M.; Yamada, K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci. Biotechnol. Biochem. 2007, 71, 2130–2135. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Wendisch, V.F. Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2010, 88, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Jojima, T.; Inui, M.; Yukawa, H. Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 2010, 86, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kim, B.; Kim, H.; Kweon, Y.; Lee, S.; Lee, J. Industrial Production of 2,3-Butanediol from the Engineered Corynebacterium glutamicum. Appl. Biochem. Biotechnol. 2015, 176, 2303–2313. [Google Scholar] [CrossRef] [PubMed]
- Radoš, D.; Turner, D.L.; Fonseca, L.L.; Carvalho, A.L.; Blombach, B.; Eikmanns, B.J.; Neves, A.R.; Santos, H. Carbon flux analysis by 13C nuclear magnetic resonance to determine the effect of CO2 on anaerobic succinate production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 2014, 80, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhu, N.; Xia, H. Aerobic production of succinate from arabinose by metabolically engineered Corynebacterium glutamicum. Bioresour. Technol. 2014, 151, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Litsanov, B.; Brocker, M.; Bott, M. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2013, 6, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Litsanov, B.; Kabus, A.; Brocker, M.; Bott, M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2012, 5, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Xia, H.; Wang, Z.; Zhao, X.; Chen, T. Engineering of acetate recycling and citrate synthase to improve aerobic succinate production in Corynebacterium glutamicum. PLoS ONE 2013, 8, e60659. [Google Scholar] [CrossRef] [PubMed]
- Litsanov, B.; Brocker, M.; Bott, M. Toward homosuccinate fermentation: Metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl. Environ. Microbiol. 2012, 78, 3325–3337. [Google Scholar] [CrossRef] [PubMed]
- Okino, S.; Noburyu, R.; Suda, M.; Jojima, T.; Inui, M.; Yukawa, H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 2008, 81, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, H.; Cai, H.; Zhou, Z.; Chen, Y.; Ouyang, P. Succinic acid production from corn cob hydrolysates by genetically engineered Corynebacterium glutamicum. Appl. Biochem. Biotechnol. 2014, 172, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Hirasawa, T.; Nishii, M.; Furusawa, C.; Shimizu, H. Enhanced acetic acid and succinic acid production under microaerobic conditions by Corynebacterium glutamicum harboring Escherichia coli transhydrogenase gene pntAB. J. Gen. Appl. Microbiol. 2014, 60, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Xia, H.; Yang, J.; Zhao, X.; Chen, T. Improved succinate production in Corynebacterium glutamicum by engineering glyoxylate pathway and succinate export system. Biotechnol. Lett. 2014, 36, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Murakami, S.; Okino, S.; Kawaguchi, H.; Vertès, A.A.; Yukawa, H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004, 7, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Okino, S.; Inui, M.; Yukawa, H. Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 2005, 68, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, G.N.; Eiteman, M.A.; Altman, E. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl. Environ. Microbiol. 2002, 68, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Sarks, C.; Jin, M.; Sato, T.K.; Balan, V.; Dale, B.E. Studying the rapid bioconversion of lignocellulosic sugars into ethanol using high cell density fermentations with cell recycle. Biotechnol. Biofuels 2014, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, H.; Omumasaba, C.A.; Nonaka, H.; Kos, P.; Okai, N.; Suzuki, N.; Suda, M.; Tsuge, Y.; Watanabe, J.; Ikeda, Y.; et al. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 2007, 153, 1042–1058. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Okayama, S.; Nonaka, H.; Tsuge, Y.; Inui, M.; Yukawa, H. Large-scale engineering of the Corynebacterium glutamicum genome. Appl. Environ. Microbiol. 2005, 71, 3369–3372. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Nonaka, H.; Tsuge, Y.; Inui, M.; Yukawa, H. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl. Environ. Microbiol. 2005, 71, 8472–8480. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Inui, M.; Yukawa, H. Site-directed integration system using a combination of mutant lox sites for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2007, 77, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Pátek, M.; Nešvera, J. Promoters and Plasmid Vectors of Corynebacterium glutamicum. In Corynebacterium glutamicum; Yukawa, H., Inui, M., Eds.; Springer: Heidelberg, Germany, 2013; Volume 23, pp. 51–88. [Google Scholar]
- Gande, R.; Dover, L.G.; Krumbach, K.; Besra, G.S.; Sahm, H.; Oikawa, T.; Eggeling, L. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J. Bacteriol. 2007, 189, 5257–5264. [Google Scholar] [CrossRef] [PubMed]
- Peters-Wendisch, P.G.; Wendisch, V.F.; Paul, S.; Eikmanns, B.J.; Sahm, H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology 1997, 143, 1095–1103. [Google Scholar] [CrossRef]
- Kim, M.I.; Kim, N.J.; Shang, L.; Chang, Y.K.; Lee, S.Y.; Chang, H.N. Continuous production of succinic acid using an external membrane cell recycle system. J. Microbiol. Biotechnol. 2009, 19, 1369–1373. [Google Scholar] [CrossRef] [PubMed]


| Strain and Plasmid | Relevant Characteristics | Source or Reference |
|---|---|---|
| Strain | ||
| R | Wild type | Yukawa et al. (2007) |
| CRZ1 | ldhA:markerless | Inui et al. (2004) |
| CRZ19 | Kmr; two sets of Pnative-pyc gene recombinantly integrated into SSI 2 of C. glutamicum CRZ1 strain | This work |
| CRZ20 | Markerless Pnative-pyc gene recombinantly integrated into SSI 1, 2, 3 and 4 of C. glutamicum CRZ1 strain | This work |
| CRZ21 | Markerless Pnative-pyc gene recombinantly integrated into SSI 1, 2, 3, 4, 5, 6, 8, 9 and 11 of C. glutamicum CRZ1 strain | This work |
| Plasmids | ||
| pCRA717 | Cmr; pCRB1 with a 3.8-kb PCR fragment containing Pnative-pyc gene | Inui et al. (2004) |
| Strain | No. of pyc b | SA Volumetric Productivity (g/L/h) c |
|---|---|---|
| CRZ19 | 2 | 2.9 |
| CRZ20 | 4 | 3.6 ± 0.2 |
| CRZ21 | 9 | 3.7 ± 0.4 |
| CRZ1-pCRA717 | 10-30 (plasmid) | 4.0 ± 0.5 |
| A: Produced succinic acid | Consumed glucose (g) | Reaction yield | Process yield | |
|---|---|---|---|---|
| (g) | B: for cell preparation a | C: for SA production b | A/C | A/(B + C) |
| 634 | 48 | 891 | 0.71 | 0.68 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jojima, T.; Noburyu, R.; Suda, M.; Okino, S.; Yukawa, H.; Inui, M. Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum. Fermentation 2016, 2, 5. https://doi.org/10.3390/fermentation2010005
Jojima T, Noburyu R, Suda M, Okino S, Yukawa H, Inui M. Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum. Fermentation. 2016; 2(1):5. https://doi.org/10.3390/fermentation2010005
Chicago/Turabian StyleJojima, Toru, Ryoji Noburyu, Masako Suda, Shohei Okino, Hideaki Yukawa, and Maysayuki Inui. 2016. "Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum" Fermentation 2, no. 1: 5. https://doi.org/10.3390/fermentation2010005
APA StyleJojima, T., Noburyu, R., Suda, M., Okino, S., Yukawa, H., & Inui, M. (2016). Improving Process Yield in Succinic Acid Production by Cell Recycling of Recombinant Corynebacterium glutamicum. Fermentation, 2(1), 5. https://doi.org/10.3390/fermentation2010005