An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety
Abstract
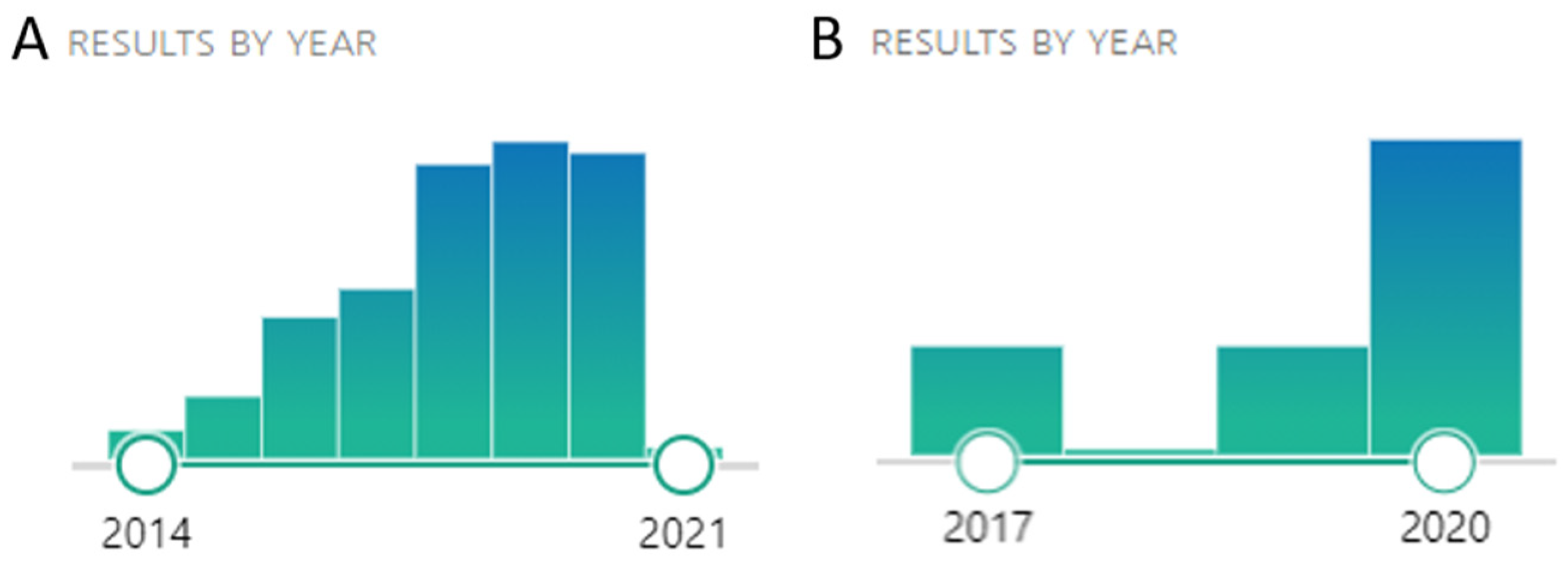
1. Introduction
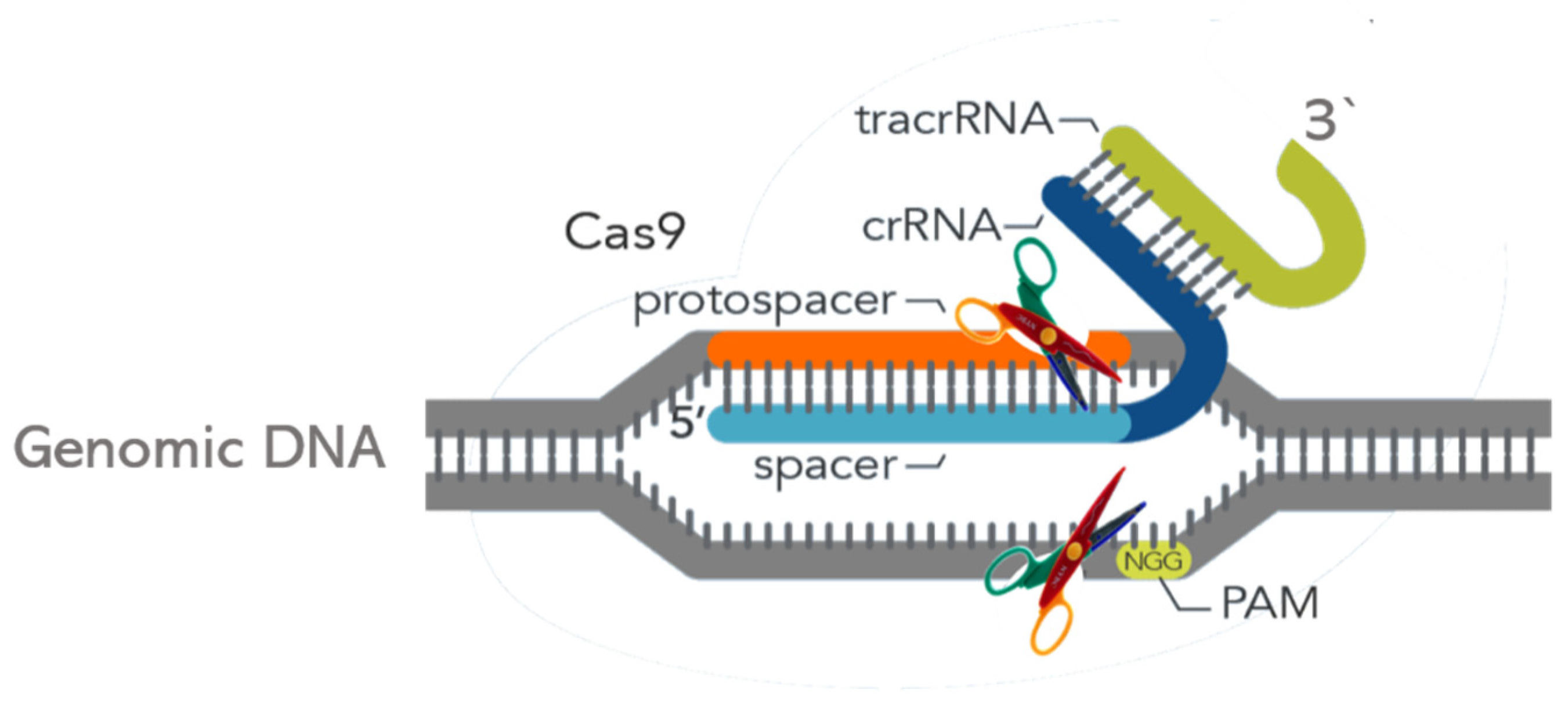
2. What Is CRISPR and How Does It Work?
3. CRISPR in the Vineyard
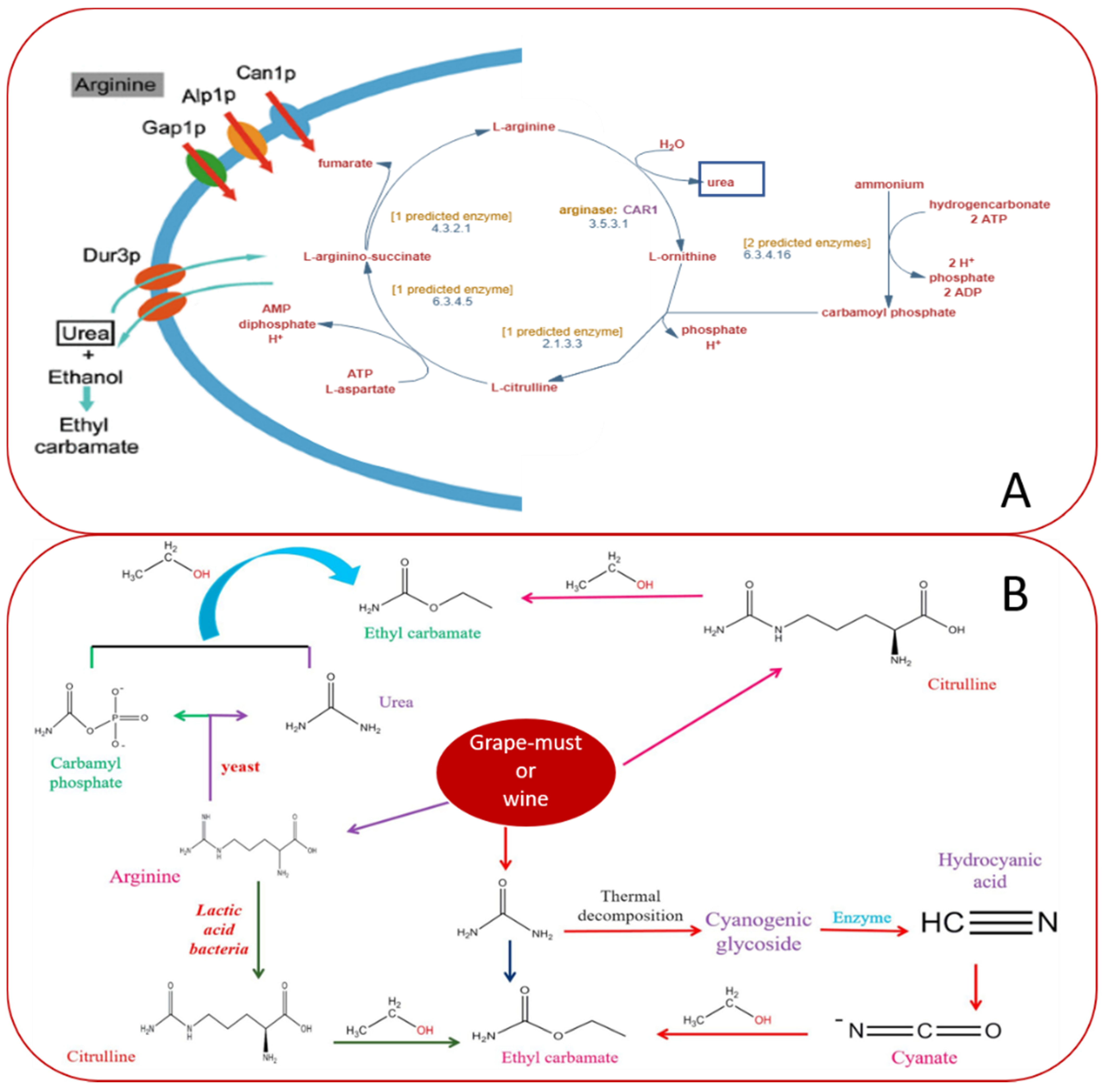
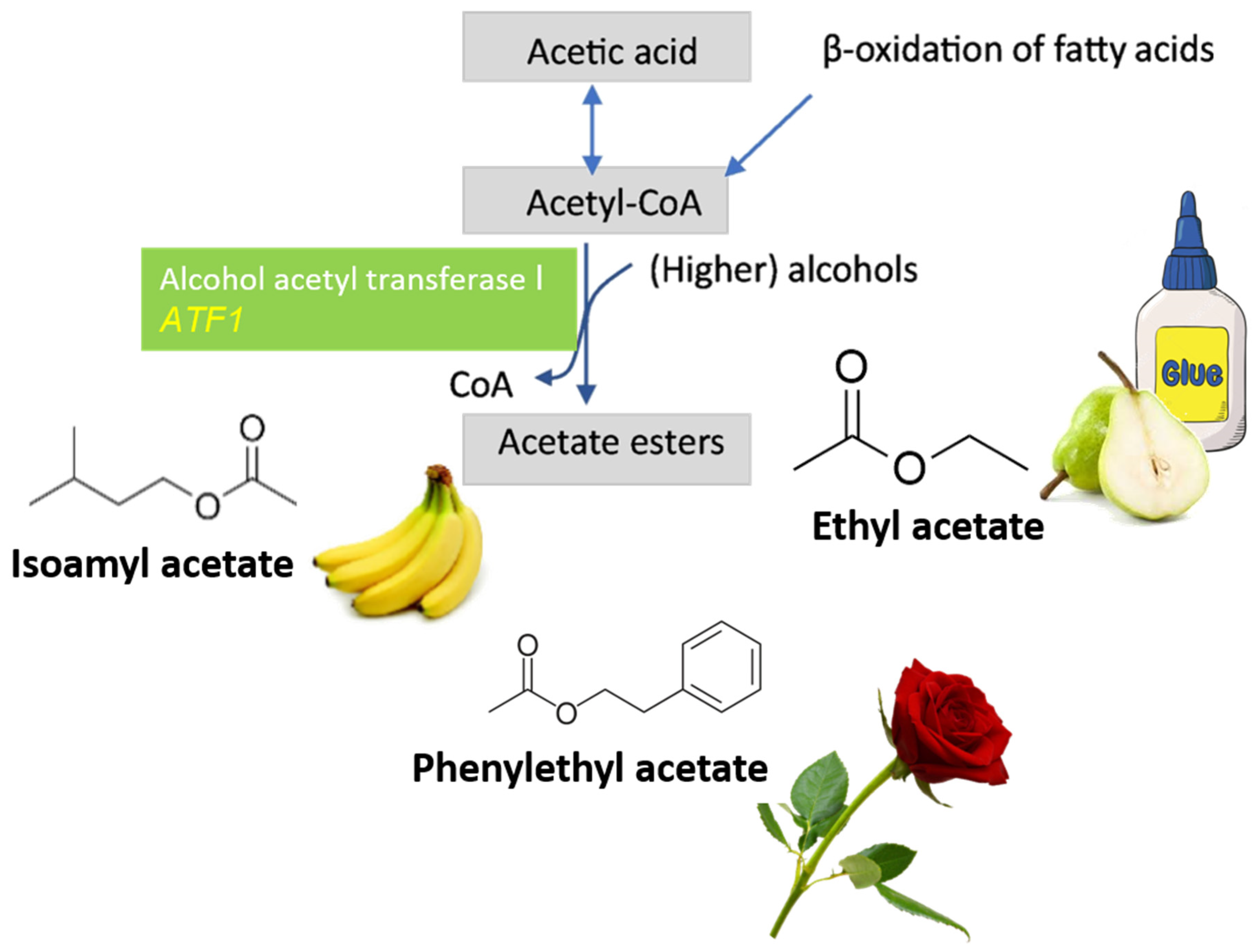
4. CRISPR System Applied to Wine Yeasts
5. Final Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, S.-J.; Noble, A.C. Characterization of odor-active compounds in Californian Chardonnay wines using GC-olfactometry and GC-mass spectrometry. J. Agric. Food Chem. 2003, 51, 8036–8044. [Google Scholar] [CrossRef] [PubMed]
- Sell, C.S. The Mechanism of Olfaction. In Chemistry and the Sense of Smell; Sell, C.S., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Vilela, A.; Inês, A.; Cosme, F. Is wine savory? Umami taste in wine. SDRP J. Food Sci. Technol. 2016, 1, 100–105. [Google Scholar] [CrossRef][Green Version]
- Pickering, G.J.; Robert, G. Perception Of Mouthfeel Sensations Elicited By Red Wine Are Associated With Sensitivity To 6-N-Propylthiouracil. J. Sens. Stud. 2006, 21, 249–265. [Google Scholar] [CrossRef]
- Vilela, A. Modulating Wine Pleasantness throughout Wine-Yeast Co-Inoculation or Sequential Inoculation. Fermentation 2020, 6, 22. [Google Scholar] [CrossRef]
- Capece, A.; Romano, P. Yeasts and Their Metabolic Impact on Wine Flavour. In Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G., Eds.; Springer: New York, NY, USA, 2019; pp. 43–80. [Google Scholar] [CrossRef]
- Swiegers, J.; Bartowsky, E.; Henschke, P.; Pretorius, I. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Benito, S. The Management of Compounds that Influence Human Health in Modern Winemaking from an HACCP Point of View. Fermentation 2019, 5, 33. [Google Scholar] [CrossRef]
- Vilela, A. Non-Saccharomyces Yeasts and Organic Wines Fermentation: Implications on Human Health. Fermentation 2020, 6, 54. [Google Scholar] [CrossRef]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving Industrial Yeast Strains: Exploiting Natural and Artificial Diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Marsit, S.; Dequin, S. Diversity and Adaptive Evolution of Saccharomyces Wine Yeast: A Review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The Emergence of Adaptive Laboratory Evolution as an Efficient Tool for Biological Discovery and Industrial Biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Kutyna, D.R.; Varela, C.; Stanley, G.A.; Borneman, A.R.; Henschke, P.A.; Chambers, P.J. Adaptive Evolution of Saccharomyces cerevisiae to Generate Strains with Enhanced Glycerol Production. Appl. Microbiol. Biotechnol. 2012, 93, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Vilela, A. Biological Demalication and Deacetification of Musts and Wines: Can Wine Yeasts Make the Wine Taste Better? Fermentation 2017, 3, 51. [Google Scholar] [CrossRef]
- Betlej, G.; Bator, E.; Oklejewicz, B.; Potocki, L.; Górka, A.; Slowik-Borowiec, M.; Czarny, W.; Domka, W.; Kwiatkowska, A. Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast. Genes 2020, 11, 576. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Mardanov, A.V. Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae. Genes 2020, 11, 964. [Google Scholar] [CrossRef]
- Remize, F.; Roustan, J.L.; Sablayrolles, J.M.; Barre, P.; Dequin, S. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 1999, 65, 143–149. [Google Scholar] [CrossRef]
- Dequin, S. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl. Microbiol. Biotechnol. 2001, 56, 577–588. [Google Scholar] [CrossRef]
- van Wyk, N.; Grossmann, M.; Wendland, J.; von Wallbrunn, C.; Pretorius, I.S. The whiff of wine yeast innovation: Strategies for enhancing aroma production by yeast during wine fermentation. J. Agric. Food Chem. 2019, 67, 13496–13505. [Google Scholar] [CrossRef]
- Goold, H.D.; Kroukamp, H.; Williams, T.C.; Paulsen, I.T.; Varela, C.; Pretorius, I.S. Yeast’s balancing act between ethanol and glycerol production in low-alcohol wines. Microb. Biotechnol. 2017, 10, 264–278. [Google Scholar] [CrossRef]
- Lilly, M.; Bauer, F.; Lambrechts, M.; Swiegers, J.; Coxxolino, D.; Pretorius, I. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 2006, 23, 641–659. [Google Scholar] [CrossRef]
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; van Vuuren, H.J.J. Metabolic Engineering of Malolactic Wine Yeast. Metab. Eng. 2006, 8, 315–323. [Google Scholar] [CrossRef]
- Coulon, J.; Husnik, J.I.; Inglis, D.L.; van der Merwe, G.K.; Lonvaud, A.; Erasmus, D.J.; van Vuuren, H.J.J. Metabolic Engineering of Saccharomyces cerevisiae to Minimize the Production of Ethyl Carbamate in Wine. Am. J. Enol. Vitic. 2006, 57, 113–124. [Google Scholar]
- Alperstein, L.; Gardner, J.M.; Sundstrom, J.F.; Sumby, K.M.; Jiranek, V. Yeast bioprospecting versus synthetic biology—which is better for innovative beverage fermentation? Appl. Microbiol. Biotechnol. 2020, 104, 1939–19536. [Google Scholar] [CrossRef] [PubMed]
- Raschmanová, H.; Weninger, A.; Glieder, A.; Kovar, K.; Vogl, T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: Current state and future prospects. Biotechnol. Adv. 2018, 36, 641–665. [Google Scholar] [CrossRef] [PubMed]
- van Wyk, N.; Kroukamp, H.; Espinosa, M.I.; von Wallbrunn, C.; Wendland, J.; Pretorius, I.S. Blending wine yeast phenotypes with the aid of CRISPR DNA editing technologies. Int. J. Food Microbiol. 2020, 324, 108615. [Google Scholar] [CrossRef]
- Terns, R.M.; Terns, M.P. CRISPR-based technologies: Prokaryotic defense weapons repurposed. Trends Genet. 2014, 30, 111–118. [Google Scholar] [CrossRef]
- Barrangou, R. The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr. Opin. Immunol. 2015, 32, 36–41. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. 2016, 101, 213–215. [Google Scholar] [CrossRef]
- Semenova, E.; Severinov, K. Interdependencies between the Adaptation and Interference Modules Guide Efficient CRISPR-Cas Immunity. In Evolutionary Biology: Self/Nonself, Evolution, Species and Complex Traits Evolution, Methods and Concepts; Pontarotti, P., Ed.; Springer: Cham, Switzerland, 2017; pp. 51–62. [Google Scholar] [CrossRef]
- Mohanraju, P.; Makarova, K.S.; Zetsche, B.; Zhang, F.; Koonin, E.V.; van der Oost, J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016, 353, aad5147. [Google Scholar] [CrossRef]
- Zhang, F.; Wen, Y.; Guo, X. CRISPR/Cas9 for genome editing: Progress, implications and challenges. Hum. Mol. Genet. 2014, 23, 40–46. [Google Scholar] [CrossRef]
- The Nobel Prize in Chemistry 2020. NobelPrize.org. Nobel Media AB 2020. Sat. 24 October 2020. Available online: https://www.nobelprize.org/prizes/chemistry/2020/summary/ (accessed on 15 November 2020).
- Kirchner, M.; Schneider, S. CRISPR-Cas: From the bacterial adaptive immune system to a versatile tool for genome engineering. Angew. Chem. Int. Ed. Engl. 2015, 54, 13508–13514. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Graham, A.E.; Studena, L.; Ledesma-Amaro, R. Multiplexed crispr technologies for gene editing and transcriptional regulation. Nat. Commun. 2020, 11, 1281. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Bouby, L.; Figueiral, I.; Bouchette, A.; Rovira, N.; Ivorra, S.; Lacombe, T.; Pastor, T.; Picq, S.; Marinval, P.; Terral, J.F. Bioarchaeological insights into the process of domestication of grapevine (Vitis vinifera L.) during Roman times in Southern France. PLoS ONE 2013, 8, e63195. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, C.; Destrac-Irvine, A. Modified grape composition under climate change conditions requires adaptations in the vineyard. OENO One 2017, 51, 147–154. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef]
- Li, M.Y.; Jiao, Y.T.; Wang, Y.T.; Zhang, N.; Wang, B.B.; Liu, R.Q.; Yin, X.; Xu, Y.; Liu, G.T. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Ueda, M. CRISPR Nickase-Mediated Base Editing in Yeast. In Methods in Molecular Biology Yeast Protocols; Xiao, W., Ed.; Humana: New York, NY, USA, 2021; Volume 2196. [Google Scholar] [CrossRef]
- Vigentini, I.; Gebbia, M.; Belotti, A.; Foschino, R.; Roth, F.P. CRISPR/Cas9 System as a Valuable Genome Editing Tool for Wine Yeasts with Application to Decrease Urea Production. Front. Microbiol. 2017, 8, 2194. [Google Scholar] [CrossRef] [PubMed]
- Fleiss, A.; O’Donnell, S.; Fournier, T.; Lu, W.; Agier, N.; Delmas, S.; Schacherer, J.; Fischer, G. Reshuffling yeast chromosomes with CRISPR/Cas9. PLoS Genet. 2019, 15, e1008332. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Xie, W.; Li, X.; Cai, G.; Lu, J.; Xie, G. Metabolic engineering of Saccharomyces cerevisiae using the CRISPR/Cas9 system to minimize ethyl carbamate accumulation during Chinese rice wine fermentation. Appl. Microbiol. Biotechnol. 2020, 104, 4435–4444. [Google Scholar] [CrossRef]
- Varela, C.; Bartel, C.; Onetto, C.; Borneman, A. Targeted gene deletion in Brettanomyces bruxellensis with an expression-free CRISPR-Cas9 system. Appl. Microbiol. Biotechnol. 2020, 104, 7105–7115. [Google Scholar] [CrossRef]
- Lachenmeier, D.W.; Lima, M.C.; Nóbrega, I.C.; Pereira, J.A.; Kerr-Corrêa, F.; Kanteres, F.; Rehm, J. Cancer risk assessment of ethyl carbamate in alcoholic beverages from Brazil with special consideration to the spirits cachaça and tiquira. BMC Cancer 2010, 10, 266. [Google Scholar] [CrossRef]
- Mira De Orduña, R.; Patchett, M.L.; Liu, S.Q.; Pilone, G.J. Growth and arginine metabolism of the wine lactic acid bacteria Lactobacillus buchneri and Oenococcus oeni at different pH values and arginine concentrations. Appl. Environ. Microbiol. 2001, 67, 1657–1662. [Google Scholar] [CrossRef]
- Opekarová, M.; Caspari, T.; Tanner, W. Unidirectional arginine transport in reconstituted plasma-membrane vesicles from yeast overexpressing CAN1. Eur. J. Biochem. 1993, 211, 683–688. [Google Scholar] [CrossRef]
- Opekarová, M.; Kubín, J. On the unidirectionality of arginine uptake in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1997, 152, 261–267. [Google Scholar] [CrossRef]
- Zhang, P.; Hu, X. Metabolic engineering of arginine permeases to reduce the formation of urea in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2018, 34, 47. [Google Scholar] [CrossRef]
- Benucci, I.; Fiorelli, V.; Lombardelli, C.; Liburdi, K.; Esti, M. Kinetic characterization of arginase from Saccharomyces cerevisiae during alcoholic fermentation at different temperatures. LWT-Food Sci. Technol. 2017, 82, 268–273. [Google Scholar] [CrossRef]
- Zhao, X.R.; Du, G.C.; Zou, H.J.; Fu, J.W.; Zhou, J.W.; Chen, J. Progress in preventing the accumulation of ethyl carbamate in alcoholic beverages. Trends Food Sci. Tech. 2013, 32, 97–107. [Google Scholar] [CrossRef]
- Zhang, P.; Du, G.; Zou, H.; Chen, J.; Xie, G.; Shi, Z.; Zhou, J. Effects of three permeases on arginine utilization in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, e20910. [Google Scholar] [CrossRef] [PubMed]
- Gowd, V.; Su, H.; Karlovsky, P.; Chen, W. Ethyl carbamate: An emerging food and environmental toxicant. Food Chem. 2018, 248, 312–321. [Google Scholar] [CrossRef]
- Jiao, Z.; Dong, Y.; Chen, Q. Ethyl Carbamate in Fermented Beverages: Presence, Analytical Chemistry, Formation Mechanism, and Mitigation Proposals. Compr. Rev. Food Sci. Food Saf. 2014, 13, 611–626. [Google Scholar] [CrossRef]
- Butzke, C.A.; Bisson, F.L. F.D.A. Ethyl Carbamate Preventative Action Manual. 1997. Available online: https://www.fda.gov/food/chemicals/ethyl-carbamate-preventative-action-manual#part3 (accessed on 22 November 2020).
- Wu, D.H.; Li, X.M.; Lu, J.; Chen, J.; Xie, G.F.; Zhang, L. The overexpression of DUR1,2 and deletion of CAR1 in an industrial Saccharomyces cerevisiae strain and effects on nitrogen catabolite repression in Chinese rice wine production. J. Inst. Brew. 2016, 122, 480–485. [Google Scholar] [CrossRef]
- Ansell, R.; Granath, K.; Hohmann, S.; Thevelein, J.M.; Adler, L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997, 16, 2179–2187. [Google Scholar] [CrossRef]
- Laguna, L.; Álvarez, M.D.; Simone, E.; Moreno-Arribas, M.V.; Bartolomé, B. Oral Wine Texture Perception and Its Correlation with Instrumental Texture Features of Wine-Saliva Mixtures. Foods 2019, 8, 190. [Google Scholar] [CrossRef]
- Albers, E.; Larsson, C.; Liden, G.; Niklasson, C.; Gustafsson, L. Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth and product formation. Appl. Environ. Microbiol. 1996, 62, 3187–3195. [Google Scholar] [CrossRef]
- Eriksson, P.; Andre, L.; Ansell, R.; Blomberg, A.; Adler, L. Cloning and characterization of GPD2, a second gene encoding in-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995, 17, 95–107. [Google Scholar] [CrossRef]
- Larsson, K.; Ansell, R.; Eriksson, P.; Adler, L. A gene encoding sn-glycerol 3-phosphate deydrogenase complements an osmosensitive mutant of Saccharomyces cerevisiae. Mol. Microbiol. 1993, 67, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Pahlman, A.K.; Granath, K.; Ansell, R.; Hohmann, S.; Adler, L. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic and oxidative stress. J. Biol. Chem. 2001, 276, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Remize, F.; Cambon, B.; Barnavon, L.; Dequin, S. Glycerol formation during wine fermentation is mainly linked to Gpd1p and is only partially controlled by the HOG pathway. Yeast 2003, 20, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Nagasawa, N.; Iwamatsu, A.; Bogaki, T.; Tamai, Y.; Hamachi, M. Molecular cloning, sequence analysis and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 1994, 60, 2786–2792. [Google Scholar] [CrossRef]
- Fujii, T.; Yoshimoto, H.; Tamai, T. Acetate ester production by Saccharomyces cerevisiae lacking the ATF1 gene encoding the alcohol acetyltransferase. J. Ferment. Bioeng. 1996, 81, 538–542. [Google Scholar] [CrossRef]
- Lilly, M.; Lambrechts, M.G.; Pretorius, I.S. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl. Environ. Microbiol. 2000, 66, 744–753. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.P.; Winderickx, J.; Pretorius, I.S.; Thevelein, J.M.; Delvaux, F.R. The Saccharomyces cerevisiae alcohol acetyl transferase gene ATF1 is a target of the cAMP/PKA and FGM nutrient-signalling pathways. FEMS Yeast Res. 2003, 4, 285–296. [Google Scholar] [CrossRef]
- Muysson, J.; Miller, L.; Allie, R.; Inglis, D.L. The Use of CRISPR-Cas9 Genome Editing to Determine the Importance of Glycerol Uptake in Wine Yeast during Icewine Fermentation. Fermentation 2019, 5, 93. [Google Scholar] [CrossRef]
- Ferreira, C.; von Voorst, F.; Martins, A.; Neves, L.; Oliveira, R.; Kielland-Brandt, M.C.; Lucas, C.; Brandt, A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005, 16, 2068–2076. [Google Scholar] [CrossRef]
- Carvalho, B.T.; Holt, S.; Souffriau, B.; Brandão, R.L.; Foulquié-Moreno, M.R.; Thevelein, J.M. Identification of Novel Alleles Conferring Superior Production of Rose Flavor Phenylethyl Acetate Using Polygenic Analysis in Yeast. mBio 2017, 8, e01173-17. [Google Scholar] [CrossRef]
- Zhang, L.H.; Zhang, H.B.; Liu, Y.F.; Zhou, J.Y.; Shen, W.; Liu, L.M.; Li, Q.; Chen, X.Z. A CRISPR-Cas9 system for multiple genome editing and pathway assembly in Candida tropicalis. Biotechnol. Bioeng. 2019, 117, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.Y.; Sun, L.; Lu, X.Y.; Zong, H.; Bin, Z.G. Establishment of a transient CRISPR-Cas9 genome editing system in Candida glycerinogenes for co-production of ethanol and xylonic acid. J. Biosci. Bioeng. 2019, 128, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Shi, X.N.; Song, L.L.; Liu, H.F.; Zhou, X.S.; Wang, Q.Y.; Zhang, Y.X.; Cai, M.H. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Factories 2019, 18, 144. [Google Scholar] [CrossRef] [PubMed]
- Numamoto, M.; Maekawa, H.; Kaneko, Y. Efficient genome editing by CRISPR/Cas9 with a tRNA-sgRNA fusion in the methylotrophic yeast Ogataea polymorpha. J. Biosci. Bioeng. 2017, 124, 487–492. [Google Scholar] [CrossRef]
- Cao, M.F.; Gao, M.R.; Ploessl, D.; Song, C.J.; Shao, Z.Y. CRISPR-mediated genome editing and gene repression in Scheffersomyces stipitis. Biotechnol. J. 2018, 13, e1700598. [Google Scholar] [CrossRef]
- Tran, V.G.; Cao, M.F.; Fatma, Z.; Song, X.F.; Zhao, H.M. Development of a CRISPR/Cas9-based tool for gene deletion in Issatchenkia orientalis. mSphere 2019, 4, e00345-19. [Google Scholar] [CrossRef]
- Horwitz, A.A.; Walter, J.M.; Schubert, M.G.; Kung, S.H.; Hawkins, K.; Platt, D.M.; Hernday, A.D.; Mahatdejkul-Meadows, T.; Szeto, W.; Chandran, S.S.; et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 2015, 1, 88–96. [Google Scholar] [CrossRef]
- Juergens, H.; Varela, J.A.; de Vries, A.R.G.; Perli, T.; Gast, V.J.M.; Gyurchev, N.Y.; Rajkumar, A.S.; Mans, R.; Pronk, J.T.; Morrissey, J.P.; et al. Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res. 2018, 18, foy012. [Google Scholar] [CrossRef]
- Jacobs, J.Z.; Ciccaglione, K.M.; Tournier, V.; Zaratiegui, M. Implementation of the CRISPR-Cas9 system in fission yeast. Nat. Commun. 2014, 5, 5344. [Google Scholar] [CrossRef]
- Ganesan, V.; Spagnuolo, M.; Agrawal, A.; Smith, S.; Gao, D.F.; Blenner, M. Advances and opportunities in gene editing and gene regulation technology for Yarrowia lipolytica. Microb. Cell Factories 2019, 18, 208. [Google Scholar] [CrossRef]
- Milheiro, J.; Filipe-Ribeiro, L.; Vilela, A.; Cosme, F.; Nunes, F.M. 4-Ethylphenol, 4-ethylguaiacol and 4-ethylcatechol in red wines: Microbial formation, prevention, remediation and overview of analytical approaches. Crit. Rev. Food Sci. Nutr. 2019, 59, 1367–1391. [Google Scholar] [CrossRef] [PubMed]
- Neto, A.; Pestana-Calsa, M.C.; de Morais, M.A., Jr.; Calsa, T., Jr. Proteome responses to nitrate in bioethanol production contaminant Dekkera bruxellensis. J. Proteom. 2014, 104, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Curtin, C.; Varela, C.; Borneman, A. Harnessing improved understanding of Brettanomyces bruxellensis biology to mitigate the risk of wine spoilage. Aust. J. Grape Wine 2015, 21, 680–692. [Google Scholar] [CrossRef]
- Reis, A.L.S.; de Souza, R.D.F.R.; Torres, R.R.N.B.; Leite, F.C.B.; Paiva, P.M.G.; Vidal, E.E.; de Morais, M.A.J. Oxygen-limited cellobiose fermentation and the characterization of the cellobiase of an industrial Dekkera/Brettanomyces bruxellensis strain. SpringerPlus 2014, 3, 38–39. [Google Scholar] [CrossRef][Green Version]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
- Urnov, F.D.; Ronald, P.C.; Carroll, D. A call for science-based review of the European court’s decision on gene-edited crops. Nat. Biotechnol. 2018, 36, 800–802. [Google Scholar] [CrossRef]
- Hanlon, P.; Sewalt, V. GEMs: Genetically engineered microorganisms and the regulatory oversight of their uses in modern food production. Crit. Rev. Food Sci. Nutr. 2020, 1–12, ahead-of-print. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilela, A. An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety. Fermentation 2021, 7, 5. https://doi.org/10.3390/fermentation7010005
Vilela A. An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety. Fermentation. 2021; 7(1):5. https://doi.org/10.3390/fermentation7010005
Chicago/Turabian StyleVilela, Alice. 2021. "An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety" Fermentation 7, no. 1: 5. https://doi.org/10.3390/fermentation7010005
APA StyleVilela, A. (2021). An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety. Fermentation, 7(1), 5. https://doi.org/10.3390/fermentation7010005




