Biological Stoichiometric Analysis during Substrate Utilization and Secondary Metabolite Production by Non-Saccharomyces Yeasts Using Grape Pomace Extract as Fermentation Medium
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms Selection, Culture Condition, and Inoculum Preparation
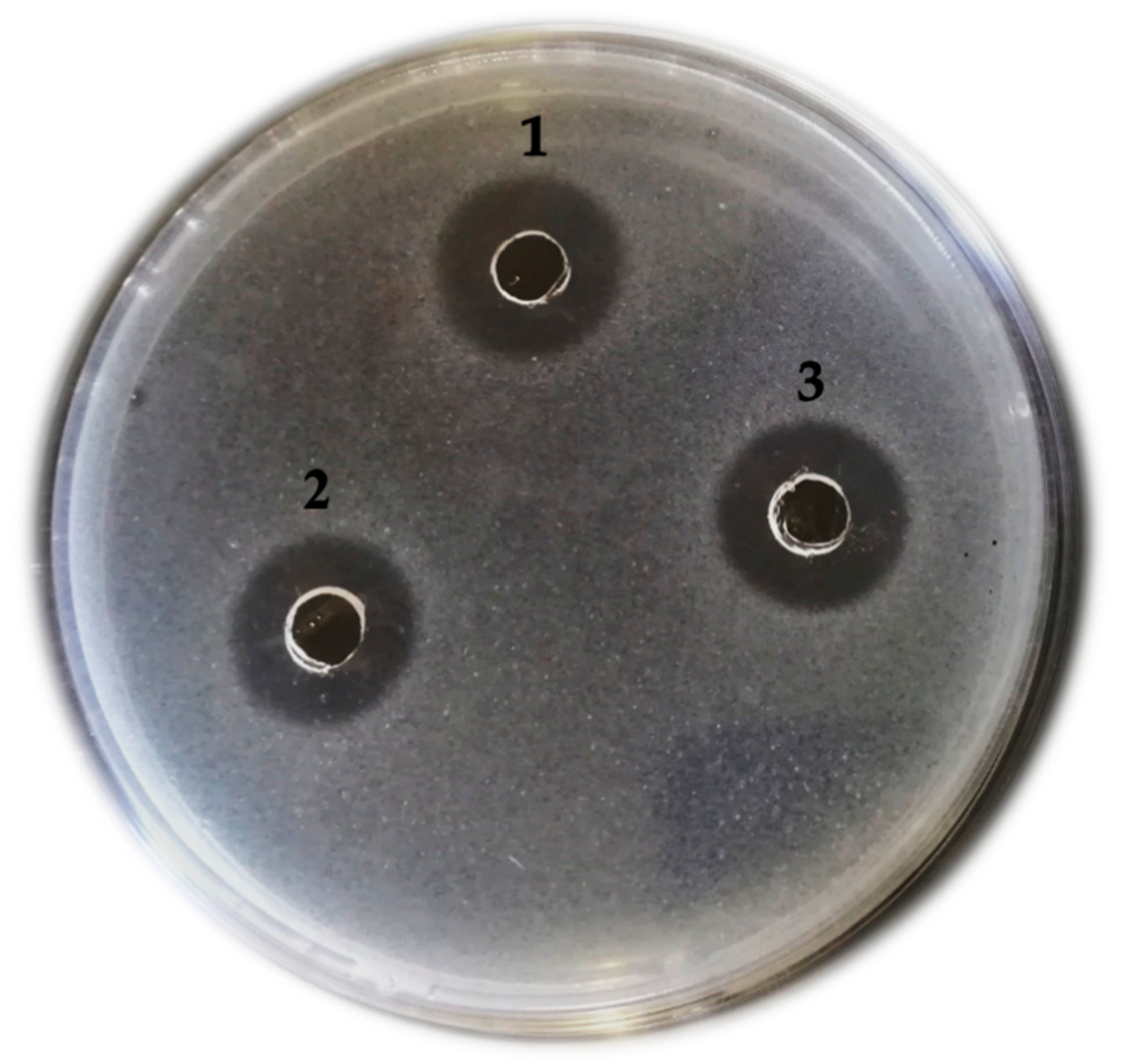
2.2. Medium Preparation, Fermentation Condition, and Confirmation of Growth Inhibition Activity
2.3. Biomass Preparation and Elemental Analysis
2.4. Heat of Combustion Determination
2.5. Determination of Stoichiometric Molar Coefficients
3. Results
3.1. Confirmation of Growth Inhibition Activity
3.2. Biomass Elemental Analysis
3.3. Determination of Stoichiometric Molar Coefficients
3.4. Bioenergetic Parameters Determination
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ciani, M.; Fatichenti, F. Killer Toxin of Kluyveromyces Phaffii DBVPG 6076 as a Biopreservative Agent To Control Apiculate Wine Yeasts. Appl. Environ. Microbiol. 2001, 67, 3058–3063. [Google Scholar] [CrossRef] [PubMed]
- Gürakan, G.C. Biopreservation by Lactic Acid Bacteria. In Metabolism and Applications of Lactic Acid Bacteria; Özer, B., Ed.; Research Signpost: Trivandrum, India, 2007; pp. 15–31. [Google Scholar]
- Nally, M.C.; Pesce, V.M.; Maturano, Y.P.; Assaf, L.R.; Toro, M.E.; de Figueroa, L.C.; Vazquez, F. Antifungal Modes of Action of Saccharomyces and Other Biocontrol Yeasts against Fungi Isolated from Sour and Grey Rots. Int. J. Food Microbiol. 2015, 204, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Nadai, C.; Junior, W.J.F.L.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol Activity of Starmerella Bacillaris yeast against Blue Mold Disease on Apple Fruit and Its Effect on Cider Fermentation. PLoS ONE 2018, 13, e0204350. [Google Scholar] [CrossRef] [PubMed]
- Abbey, J.A.; Percival, D.; Abbey, L.; Asiedu, S.K.; Prithiviraj, B.; Schilder, A. Biofungicides as Alternative to Synthetic Fungicide Control of Grey Mould (Botrytis Cinerea)–Prospects and Challenges. Biocontrol Sci. Technol. 2019, 29, 207–228. [Google Scholar] [CrossRef]
- Mewa-Ngongang, M.; du Plessis, H.W.; Ntwampe, S.K.; Chidi, B.S.; Hutchinson, U.F.; Mekuto, L.; Jolly, N.P. Grape Pomace Extracts as Fermentation Medium for the Production of Potential Biopreservation Compounds. Foods 2019, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- von Stockar, U.; Vojinović, V.; Maskow, T.; Liu, J. Can Microbial Growth Yield Be Estimated Using Simple Thermodynamic Analogies to Technical Processes? Chem. Eng. Process. Process Intensif. 2008, 47, 980–990. [Google Scholar] [CrossRef]
- Duboc, P.; Marison, I.; Von Stockar, U. Quantitative Calorimetry and Biochemical Engineering. Handb. Therm. Anal. Calorim. 1999, 4, 267–365. [Google Scholar]
- Liu, J.-S.; Vojinović, V.; Patiño, R.; Maskow, T.; von Stockar, U. A Comparison of Various Gibbs Energy Dissipation Correlations for Predicting Microbial Growth Yields. Thermochim. Acta 2007, 458, 38–46. [Google Scholar] [CrossRef]
- Heijnen, J.J. Impact of Thermodynamic Principles in Systems Biology. In Biosystems Engineering II; Springer: Berlin/Heidelberg, Germany, 2010; pp. 139–162. [Google Scholar]
- Akinpelu, E.A.; Ntwampe, S.K.O.; Chen, B.-H. Biological Stoichiometry and Bioenergetics of Fusarium Oxysporum EKT01/02 Proliferation Using Different Substrates in Cyanidation Wastewater. Can. J. Chem. Eng. 2018, 96, 537–544. [Google Scholar] [CrossRef]
- Battley, E.H. A Comparison of Energy Changes Accompanying Growth Processes by Saccharomyces Cerevisiae. J. Therm. Anal. Calorim. 2011, 104, 193–200. [Google Scholar] [CrossRef]
- Battley, E.H. A Theoretical Study of the Thermodynamics of Microbial Growth Using Saccharomyces Cerevisiae and a Different Free Energy Equation. Q. Rev. Biol. 2013, 88, 69–96. [Google Scholar] [CrossRef] [PubMed]
- Mewa-Ngongang, M.; du Plessis, H.W.; Hutchinson, U.F.; Mekuto, L.; Ntwampe, S.K. Kinetic Modelling and Optimisation of Antimicrobial Compound Production by Candida Pyralidae KU736785 for Control of Candida guilliermondii. Food Sci. Technol. Int. 2017, 23, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Thornton, W.M. XV. The Relation of Oxygen to the Heat of Combustion of Organic Compounds. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1917, 33, 196–203. [Google Scholar] [CrossRef]
| Parameter | Equation |
|---|---|
| Biological Stoichiometry | |
| Catabolic Reaction | |
| Anabolic Reaction |
| Substance | Formula | |
|---|---|---|
| Glucose | −1263.07 | |
| Ammonia | −80.29 | |
| Oxygen | −12.09 | |
| Water | −285.83 |
| Yeast | [C] | [H] | [N] | [O] | [S] | |
|---|---|---|---|---|---|---|
| C. pyralidae Y1117 | % | 41.435 | 6.582 | 7.200 | 44.501 | 0.282 |
| Average MW | 3.453 | 6.582 | 0.514 | 2.781 | 0.009 | |
| Unit-carbon | 1.000 | 1.906 | 0.149 | 0.805 | 0.003 | |
| P. kluyveri Y1125 | % | 41.157 | 6.484 | 6.333 | 45.713 | 0.313 |
| Average MW | 3.430 | 6.484 | 0.452 | 2.857 | 0.010 | |
| Unit-carbon | 1.000 | 1.891 | 0132 | 0.833 | 0.003 | |
| P. kluyveri Y1164 | % | 44.149 | 6.871 | 7.530 | 41.104 | 0.346 |
| Average MW | 3.679 | 6.871 | 0.538 | 2.569 | 0.011 | |
| Unit-carbon | 1.000 | 1.868 | 0.146 | 0.698 | 0.003 |
| C. pyralidae Y1117 | P. kluyveri Y1125 | P. kluyveri Y1164 | |
|---|---|---|---|
| Empirical Formula | |||
| Elemental Formula | |||
| MW of Unit Carbon (g/C-mol) | 28.968 | 29.163 | 27.176 |
| C. pyralidae Y1117 |
| Catabolism: |
Anabolism: Metabolism: |
| P. kluyveri Y1125 |
| Catabolism: |
Anabolism: Metabolism: |
| P. kluyveri Y1164 |
| Catabolism: |
Anabolism: Metabolism: |
| C. pyralidae Y1117 |
| P. kluyveri Y1125 |
| P. kluyveri Y1164 |
| Yeast | Combustion Equations |
|---|---|
| C. pyralidae Y1117 | |
| P. kluyveri Y1125 | |
| P. kluyveri Y1164 |
| Yeast | ||||
|---|---|---|---|---|
| C pyralidae Y1117 | −193.22 | −5006.75 | −5141.07 | −0.45 |
| P. kluyveri Y1125 | −138.463 | −5160.48 | −5223.87 | −0.21 |
| P. kluyveri Y1164 | −195.23 | −8283.54 | −8441.26 | −0.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mewa-Ngongang, M.; du Plessis, H.W.; Ntwampe, S.K.O.; Akinpelu, E.A.; Hutchinson, U.F.; Chidi, B.S.; Okudoh, V.I.; Jolly, N.P. Biological Stoichiometric Analysis during Substrate Utilization and Secondary Metabolite Production by Non-Saccharomyces Yeasts Using Grape Pomace Extract as Fermentation Medium. Fermentation 2021, 7, 89. https://doi.org/10.3390/fermentation7020089
Mewa-Ngongang M, du Plessis HW, Ntwampe SKO, Akinpelu EA, Hutchinson UF, Chidi BS, Okudoh VI, Jolly NP. Biological Stoichiometric Analysis during Substrate Utilization and Secondary Metabolite Production by Non-Saccharomyces Yeasts Using Grape Pomace Extract as Fermentation Medium. Fermentation. 2021; 7(2):89. https://doi.org/10.3390/fermentation7020089
Chicago/Turabian StyleMewa-Ngongang, Maxwell, Heinrich W. du Plessis, Seteno K. O. Ntwampe, Enoch A. Akinpelu, Ucrecia F. Hutchinson, Boredi S. Chidi, Vincent I. Okudoh, and Neil P. Jolly. 2021. "Biological Stoichiometric Analysis during Substrate Utilization and Secondary Metabolite Production by Non-Saccharomyces Yeasts Using Grape Pomace Extract as Fermentation Medium" Fermentation 7, no. 2: 89. https://doi.org/10.3390/fermentation7020089
APA StyleMewa-Ngongang, M., du Plessis, H. W., Ntwampe, S. K. O., Akinpelu, E. A., Hutchinson, U. F., Chidi, B. S., Okudoh, V. I., & Jolly, N. P. (2021). Biological Stoichiometric Analysis during Substrate Utilization and Secondary Metabolite Production by Non-Saccharomyces Yeasts Using Grape Pomace Extract as Fermentation Medium. Fermentation, 7(2), 89. https://doi.org/10.3390/fermentation7020089










