Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli
Abstract
:1. Introduction
2. Results
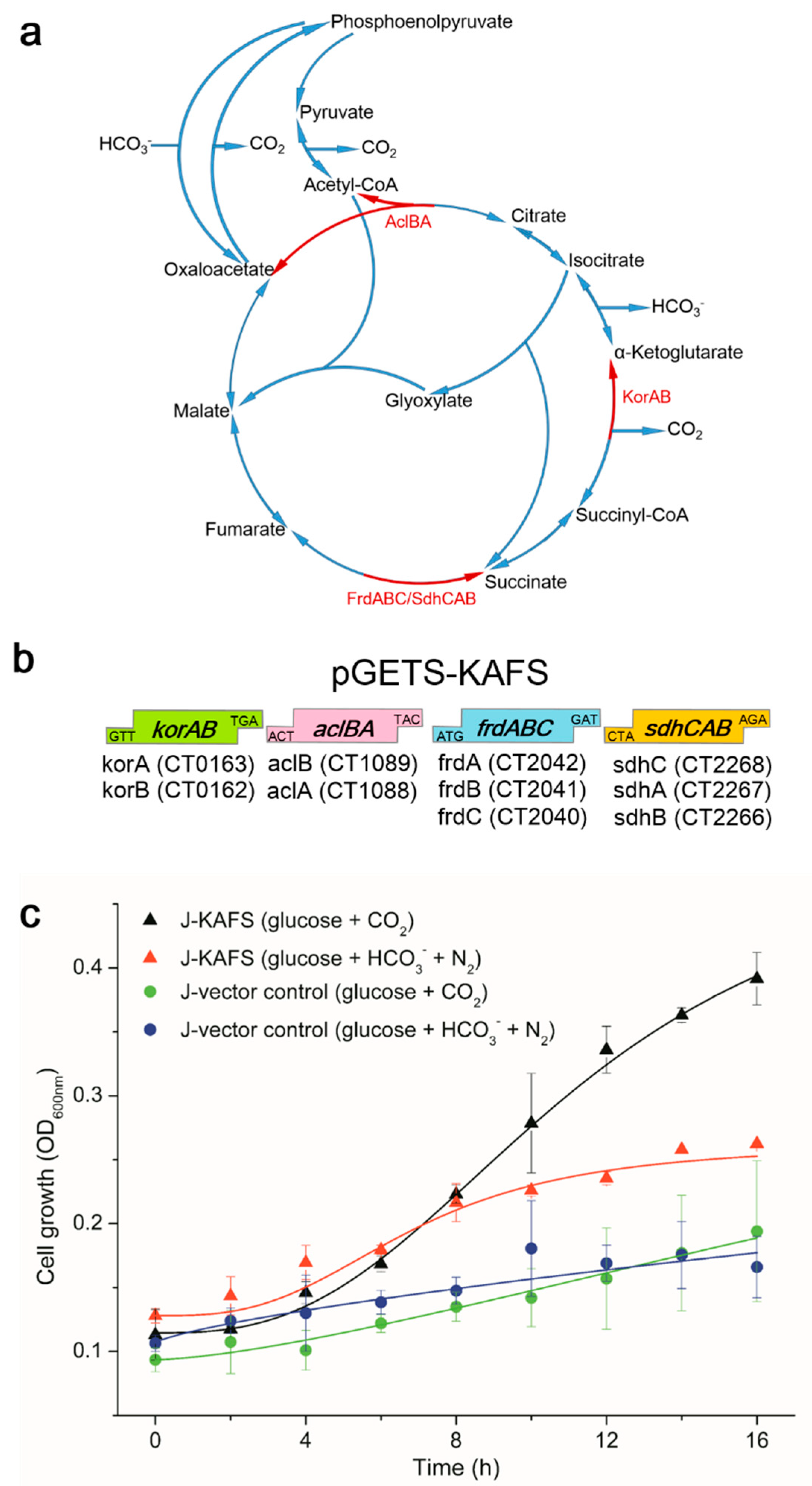
2.1. Genetic Cloning and Cell Growth
2.2. Transcriptome Analysis of Glucose Fermentation
2.3. Carbon Balance of Transgenic Strains under Glucose Fermentation Conditions
2.4. Growth of Transgenic Strains in Anaerobic Respiration Mode
2.5. Transcriptome Analysis of Transgenic Strains in Anaerobic Respiration Mode
2.6. Metabolomics Analysis and Adenylate Concentrations of Transgenic Strains in Anaerobic Respiration Mode
2.7. Relative Carbonic Anhydrase Activities of Transgenic Strains
3. Discussion
4. Conclusions
5. Methods
5.1. DNA Manipulation
5.2. Culture Conditions
5.3. Enzyme Assays
5.4. ATP-Dependent Citrate Lyase Activity Assays
5.5. α-Ketoglutarate Oxidoreductase and Fumarate Reductase Activity Assays
5.6. Carbonic Anhydrase Activity Assays
5.7. Metabolite Analysis
5.8. Transcriptome Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Oliver, N.J.; Rabinovitch-Deere, C.A.; Carroll, A.L.; Nozzi, N.E.; Case, A.E.; Atsumi, S. Cyanobacterial metabolic engineering for biofuel and chemical production. Curr. Opin. Chem. Biol. 2016, 35, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Heijstra, B.D.; Leang, C.; Juminaga, A. Gas fermentation: Cellular engineering possibilities and scale up. Microb. Cell Factories 2017, 16, 60. [Google Scholar] [CrossRef] [Green Version]
- Liew, F.; Martin, M.E.; Tappel, R.C.; Heijstra, B.D.; Mihalcea, C.; Köpke, M. Gas fermentation—A flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front. Microbiol. 2016, 7, 694. [Google Scholar] [CrossRef]
- Gleizer, S.; Ben-Nissan, R.; Bar-On, Y.M.; Antonovsky, N.; Noor, E.; Zohar, Y.; Jona, G.; Krieger, E.; Shamshoum, M.; Bar-Even, A.; et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 2019, 179, 1255–1263.e1212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.; Morowitz, H.J. Universality in intermediary metabolism. Proc. Natl. Acad. Sci. USA 2004, 101, 13168–13173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, M.C.; Buchanan, B.B.; Arnon, D.I. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 1966, 55, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Erb, T.J. Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 2011, 77, 8466–8477. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.-H.; Blankenship, R.E. Both forward and reverse TCA cycles operate in green sulfur bacteria. J. Biol. Chem. 2010, 285, 35848–35854. [Google Scholar] [CrossRef] [Green Version]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunoura, T.; Chikaraishi, Y.; Izaki, R.; Suwa, T.; Sato, T.; Harada, T.; Mori, K.; Kato, Y.; Miyazaki, M.; Shimamura, S.; et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuge, K.; Matsui, K.; Itaya, M. One step assembly of multiple DNA fragments with a designed order and orientation in Bacillus subtilis plasmid. Nucleic Acids Res. 2003, 31, e133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizaki, T.; Tsuge, K.; Itaya, M.; Doi, N.; Yanagawa, H. Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl. Env. Microbiol. 2007, 73, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, B.B.; Evans, M.C. The synthesis of alpha-ketoglutarate from succinate and carbon dioxide by a subcellular preparation of a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 1965, 54, 1212–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlin, C.; Masters, M.; McAteer, S.; Coulson, A. Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 2003, 185, 6415–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilous, P.T.; Weiner, J.H. Dimethyl sulfoxide reductase activity by anaerobically grown Escherichia coli HB101. J. Bacteriol. 1985, 162, 1151–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlier, D.; Nguyen Le Minh, P.; Roovers, M. Regulation of carbamoylphosphate synthesis in Escherichia coli: An amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 2018, 50, 1647–1661. [Google Scholar] [CrossRef] [Green Version]
- Charles, H.P.; Roberts, G.A. Carbon dioxide as a growth factor for mutants of Escherichia coli. Microbiology 1968, 51, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Dusenge, M.E.; Duarte, A.G.; Way, D.A. Plant carbon metabolism and climate change: Elevated CO2 and temperature impacts on photosynthesis, photorespiration and respiration. New Phytol. 2019, 221, 32–49. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Liu, E.J.; Chen, Y.L.; Ou-Yang, F.Y.; Li, S.Y. The comprehensive profile of fermentation products during in situ CO2 recycling by Rubisco-based engineered Escherichia coli. Microb. Cell Fact. 2016, 15, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.Y.-T.; Li, B.; Drennan, C.L.; Elliott, S.J. A reverse TCA cycle 2-oxoacid:ferredoxin oxidoreductase that makes C-C bonds from CO2. Joule 2019, 3, 595–611. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.T.; Jarrett, J.T. Electron acceptor specificity of ferredoxin (flavodoxin):NADP+ oxidoreductase from Escherichia coli. Arch. Biochem. Biophys. 2002, 406, 116–126. [Google Scholar] [CrossRef]
- Blaschkowski, H.P.; Knappe, J.; Ludwig-Festl, M.; Neuer, G. Routes of flavodoxin and ferredoxin reduction in Escherichia coli. Eur. J. Biochem. 1982, 123, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Mierendorf, R.C.; Morris, B.B.; Hammer, B.; Novy, R.E. Expression and purification of recombinant proteins using the pet system. Methods Mol. Med. 1998, 13, 257–292. [Google Scholar] [CrossRef]
- Eisen, J.A.; Nelson, K.E.; Paulsen, I.T.; Heidelberg, J.F.; Wu, M.; Dodson, R.J.; Deboy, R.; Gwinn, M.L.; Nelson, W.C.; Haft, D.H.; et al. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 2002, 99, 9509–9514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuge, K.; Sato, Y.; Kobayashi, Y.; Gondo, M.; Hasebe, M.; Togashi, T.; Tomita, M.; Itaya, M. Method of preparing an equimolar DNA mixture for one-step DNA assembly of over 50 fragments. Sci. Rep. 2015, 5, 10655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.; Tabita, F.R. Both subunits of ATP-citrate lyase from Chlorobium tepidum contribute to catalytic activity. J. Bacteriol. 2006, 188, 6544–6552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawers, R.G.; Ballantine, S.P.; Boxer, D.H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: Evidence for a third isoenzyme. J. Bacteriol. 1985, 164, 1324–1331. [Google Scholar] [CrossRef] [Green Version]
- Wilbur, K.M.; Anderson, N.G. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 1948, 176, 147–154. [Google Scholar] [CrossRef]
- Jensen, E.L.; Clement, R.; Kosta, A.; Maberly, S.C.; Gontero, B. A new widespread subclass of carbonic anhydrase in marine phytoplankton. Isme J. 2019, 13, 2094–2106. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.-J.; Shin, J.-S.; Li, S.-Y. The catalytic role of RuBisCO for in situ CO2 recycling in Escherichia coli. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Li, Y.H.; Ou-Yang, F.Y.; Yang, C.H.; Li, S.Y. The coupling of glycolysis and the Rubisco-based pathway through the non-oxidative pentose phosphate pathway to achieve low carbon dioxide emission fermentation. Bioresour. Technol. 2015, 187, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.Y.; Li, S.Y. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour. Technol. 2013, 150, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Hirayama, A.; Ishikawa, T.; Nakamura, S.; Shimizu, K.; Ueno, Y.; Tomita, M.; Soga, T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. Biosyst. 2008, 4, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Chin, W.C.; Lin, K.H.; Liu, C.C.; Tsuge, K.; Huang, C.C. Improved n-butanol production via co-expression of membrane-targeted tilapia metallothionein and the clostridial metabolic pathway in Escherichia coli. BMC Biotechnol. 2017, 17, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511. Available online: https://www.nature.com/articles/nbt.1621#supplementary-information (accessed on 16 June 2021). [CrossRef] [PubMed] [Green Version]

| Enzyme Activity (nmol/min/mg Protein) | |||
|---|---|---|---|
| Strain | ATP-Dependent Citrate Lyase a | α-Ketoglutarate Oxidoreductase | Fumarate Reductase |
| J-KAFS b | 82 ± 5 | 6.5 ± 0.6 | 142 ± 11 |
| J-vector control c | 14 ± 3 | <0.01 | 30 ± 2 |
| Gene Name | Description | Log2 Ratio (J-KAFS/J-Vector Control) a,b | |
|---|---|---|---|
| Heterologous expression (Chlorobaculum tepidum genes cloned into Escherichia coli) | |||
| aclA (CT1088) | ATP-citrate lyase α subunit | 22.2 | |
| aclB (CT1089) | ATP-citrate lyase β subunit | 23.8 | |
| frdC (CT2040) | succinate dehydrogenase/fumarate reductase, cytochrome b subunit | 21.6 | |
| frdB (CT2041) | succinate dehydrogenase/fumarate reductase, iron-sulfur subunit | 22 | |
| frdA (CT2042) | succinate dehydrogenase/fumarate reductase, flavoprotein subunit | 21.6 | |
| korB (CT0162) | 2-oxoglutarate ferredoxin oxidoreductase subunit β | 25.1 | |
| korA (CT0163) | α-oxoglutarate ferredoxin oxidoreductase subunit α | 17.1 | |
| sdhB (CT2266) | succinate dehydrogenase/fumarate reductase iron-sulfur subunit | 16.6 | |
| sdhA (CT2267) | succinate dehydrogenase/fumarate reductase, flavoprotein subunit | 17.9 | |
| sdhC (CT2268) | succinate dehydrogenase/fumarate reductase, cytochrome b subunit | 20.2 | |
| E. coli genes | |||
| Pyrimidine synthesis | |||
| carA | carbamoyl-phosphate synthase small subunit | −2.7 | |
| carB | carbamoyl-phosphate synthase large chain | −2.6 | |
| pyrI | aspartate carbamoyltransferase regulatory subunit | −3.4 | |
| pyrB | aspartate carbamoyltransferase catalytic subunit | −3.8 | |
| upp | uracil phosphoribosyltransferase | −2.4 | |
| ybbY | uracil/xanthine transporter | −3.1 | |
| Others | |||
| lacI | lac repressor | −4.9 | |
| yhdV | lipoprotein | −1.8 | |
| bfd | bacterioferritin-associated ferredoxin | −2.0 | |
| ygaV | transcriptional regulator | −2.1 | |
| yjjZ | DUF1435 domain-containing protein | −2.2 | |
| proU | tRNA-Pro(UGG) | −24.2 | |
| Strain | J-Vector Control | J-KAFS |
|---|---|---|
| Carbon | Amount (mmol/L) | |
| Consumed carbon a | 31.14 | 51.91 |
| Consumed glucose b | 5.18 ± 0.54 | 8.65 ± 0.51 |
| Produced carbon c | 25.68 | 47.3 |
| Biomass carbon | 1.18 | 2.89 |
| CO2 release d | 0.70 ± 0.26 | −1.08 ± 0.56 |
| Fermentation product carbon | 23.79 | 45.49 |
| Pyruvate | 0.04 ± 0.01 | 0.31 ± 0.07 |
| Lactate | 0.8 ± 0.03 | 1.88 ± 0.36 |
| Formate | 7.59 ± 0.91 | 13.88 ± 0.73 |
| Acetate | 3.52 ± 0.58 | 5.49 ± 0.53 |
| Ethanol | 3.28 ± 0.31 | 6.99 ± 0.75 |
| Succinate | 0.02 ± 0.02 | 0.02 ± 0.27 |
| Fermentation product per biomass carbon | 20.15 | 15.74 |
| Biomass formation percentage e | 3.7% | 5.5% |
| Carbon balance f | 82.46% | 91.11% |
| Gene Name | Description | Expression log2 Ratio |
|---|---|---|
| (GenBank CDS Number) | (J-KAFS/Vector Control) a,b | |
| Heterologous expression (Chlorobaculum tepidum genes cloned into Escherichia coli) | ||
| aclA (CT1088) | ATP-citrate lyase α subunit | 19.1 |
| aclB (CT1089) | ATP-citrate lyase β subunit | 21 |
| frdC (CT2040) | Succinate dehydrogenase/fumarate reductase, cytochrome b subunit | 18.8 |
| frdB (CT2041) | Succinate dehydrogenase/fumarate reductase, iron-sulfur subunit | 19 |
| frdA (CT2042) | Succinate dehydrogenase/fumarate reductase, flavoprotein subunit | 18.5 |
| korB (CT0162) | 2-Oxoglutarate ferredoxin oxidoreductase subunit β | 23 |
| korA (CT0163) | α-oxoglutarate ferredoxin oxidoreductase subunit α | 26.4 |
| sdhB (CT2266) | Succinate dehydrogenase/fumarate reductase iron-sulfur subunit | 24.3 |
| sdhA (CT2267) | Succinate dehydrogenase/fumarate reductase, flavoprotein subunit | 19 |
| sdhC (CT2268) | Succinate dehydrogenase/fumarate reductase, cytochrome b subunit | 18.3 |
| E. coli genes | ||
| Chemotaxis | ||
| aer | Fused signal transducer for aerotaxis sensory component and methyl-accepting chemotaxis component | 2.5 |
| cheA | Fused chemotactic sensory histidine kinase (soluble) in two-component regulatory system with CheB and CheY | 3.2 |
| cheB | Fused chemotaxis regulator and protein-glutamate methylesterase in two-component regulatory system with CheA | 2.5 |
| cheR | Chemotaxis regulator | 2.6 |
| cheW | Purine-binding chemotaxis protein | 3.1 |
| cheY | Chemotaxis regulator transmitting signal to flagellar motor component | 2.6 |
| tap | Methyl-accepting protein IV | 3.3 |
| tar | Methyl-accepting chemotaxis protein II | 3.3 |
| trg | Methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | 2.7 |
| tsr | Methyl-accepting chemotaxis protein I, serine sensor receptor | 2.3 |
| Flagellar assembly | ||
| flgK | Flagellar hook-filament junction protein 1 | 3.1 |
| flgL | Flagellar hook-filament junction protein | 2.7 |
| flgM | Anti-sigma factor for FliA (sigma 28) | 3 |
| flgN | Export chaperone for FlgK and FlgL | 2.6 |
| fliA | RNA polymerase, sigma 28 (sigma F) factor | 2.7 |
| fliC | Flagellar filament structural protein | 3.3 |
| fliD | Flagellar filament capping protein | 3.2 |
| fliS | Flagellar protein potentiates polymerization | 3 |
| fliT | Predicted chaperone | 2.9 |
| fliZ | Predicted regulator of FliA activity | 2.5 |
| flxA | Hypothetical protein | 2.8 |
| motA | Proton conductor component of flagella motor | 3.3 |
| motB | Protein that enables flagellar motor rotation | 3.4 |
| ycgR | Protein involved in flagellar function | 3.1 |
| yhjH | EAL domain-containing protein involved in flagellar function | 3.5 |
| Pyrimidine synthesis | ||
| carA | Carbamoyl-phosphate synthetase small subunit, glutamine amidotransferase | −2.2 |
| carB | Carbamoyl-phosphate synthase large subunit | −2 |
| pyrB | Aspartate carbamoyltransferase, catalytic subunit | −2.6 |
| Stress resistance | ||
| gadA | Glutamate decarboxylase A, PLP-dependent | 2.9 |
| gadB | Glutamate decarboxylase B, PLP-dependent | 3 |
| gadC | Predicted glutamate:gamma-aminobutyric acid antiporter | 2.6 |
| hdeA | Stress response protein acid-resistance protein | 3.6 |
| hdeB | Acid-resistance protein | 3.9 |
| hdeD | Acid-resistance membrane protein | 3.2 |
| yjbJ | Predicted stress response protein | 2.2 |
| Metabolite | Concentration (pmol/O.D./mL) | |||||
|---|---|---|---|---|---|---|
| J-Vector Control | J-KAFS | |||||
| 1 h | 6 h | 24 h | 1 h | 6 h | 24 h | |
| Adenylate energy charge a | 0.403 | 0.447 | 0.354 | 0.411 | 0.448 | 0.319 |
| ATP | 53 | 81 | 38 | 58 | 95 | 40 |
| ADP | 87 | 95 | 55 | 104 | 138 | 84 |
| AMP | 99 | 111 | 92 | 105 | 133 | 133 |
| Strain b | Changes in Carbonic Anhydrase Activity (WAU/mL) a |
|---|---|
| J-KAFS | −0.81 |
| J-vector control | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lo, S.-C.; Chiang, E.-P.I.; Yang, Y.-T.; Li, S.-Y.; Peng, J.-H.; Tsai, S.-Y.; Wu, D.-Y.; Yu, C.-H.; Huang, C.-H.; Su, T.-T.; et al. Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli. Fermentation 2021, 7, 98. https://doi.org/10.3390/fermentation7020098
Lo S-C, Chiang E-PI, Yang Y-T, Li S-Y, Peng J-H, Tsai S-Y, Wu D-Y, Yu C-H, Huang C-H, Su T-T, et al. Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli. Fermentation. 2021; 7(2):98. https://doi.org/10.3390/fermentation7020098
Chicago/Turabian StyleLo, Shou-Chen, En-Pei Isabel Chiang, Ya-Tang Yang, Si-Yu Li, Jian-Hau Peng, Shang-Yieng Tsai, Dong-Yan Wu, Chia-Hua Yu, Chu-Han Huang, Tien-Tsai Su, and et al. 2021. "Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli" Fermentation 7, no. 2: 98. https://doi.org/10.3390/fermentation7020098
APA StyleLo, S.-C., Chiang, E.-P. I., Yang, Y.-T., Li, S.-Y., Peng, J.-H., Tsai, S.-Y., Wu, D.-Y., Yu, C.-H., Huang, C.-H., Su, T.-T., Tsuge, K., & Huang, C.-C. (2021). Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli. Fermentation, 7(2), 98. https://doi.org/10.3390/fermentation7020098







