Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose
Abstract
:1. Introduction
2. Production of Organic Acids through Consolidated Bioprocessing (CBP) of Lignocellulosic Biomass
2.1. C3 Organic Acids
2.1.1. Lactic Acid (LA)
2.1.2. 3-Hydroxypropionic Acid (3-HP)
2.2. C4 Organic Acids: Fumaric Acid (FA), Malic Acid (MA), and Succinic Acid (SA)
2.2.1. Fumaric Acid (FA)
2.2.2. Malic Acid (MA)
2.2.3. Succinic Acid (SA)
2.3. C5 Organic Acids
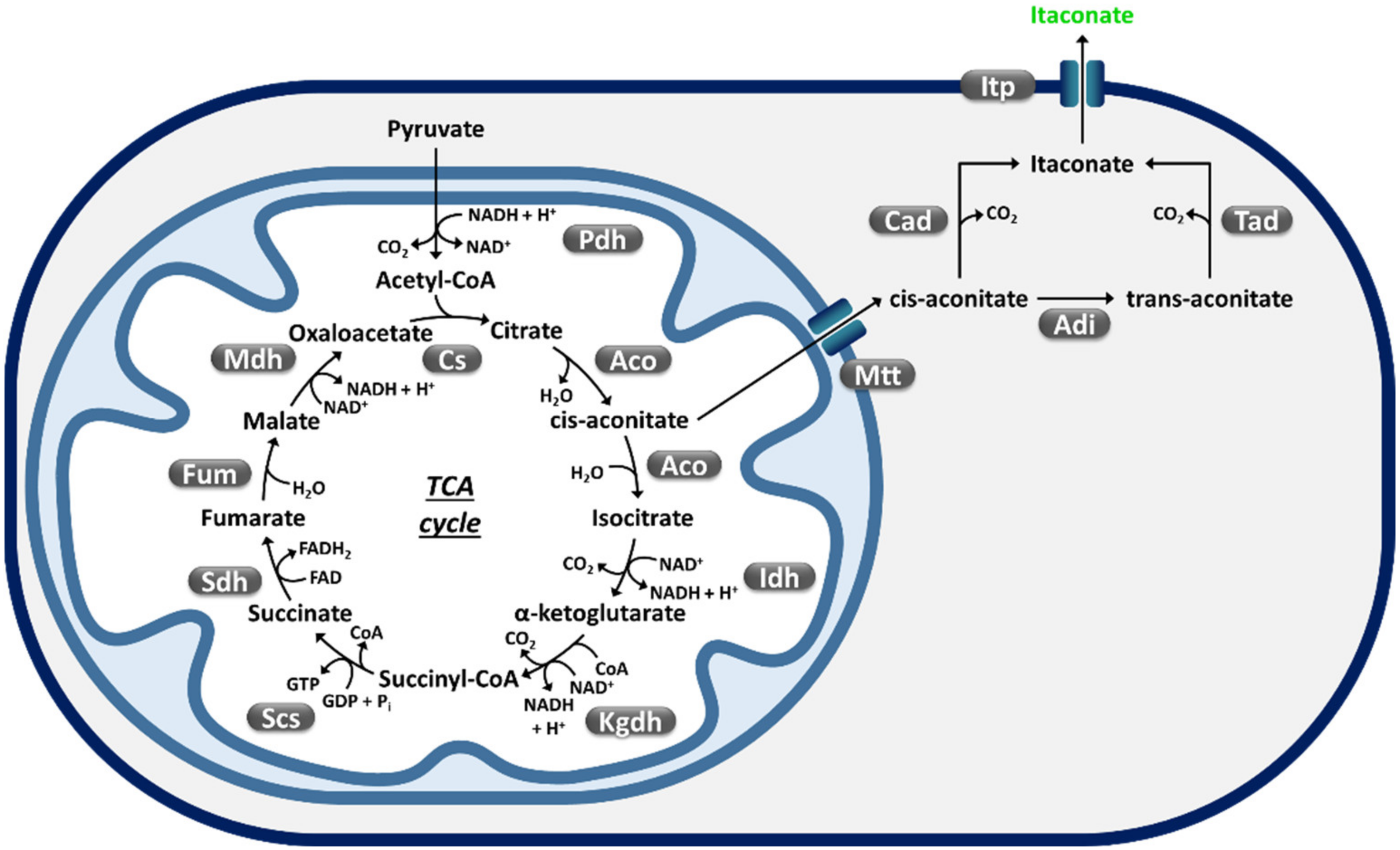
Itaconic Acid (IA)
2.4. C6 Organic Acids
Adipic Acid (AA), cis,cis-Muconic Acid (CCM), Glucaric Acid (GA)
3. Improvement of Acid Tolerance
4. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Sims, R.E.H.; Mabee, W.; Saddler, J.N.; Taylor, M. An overview of second generation biofuel technologies. Bioresour. Technol. 2010, 101, 1570–1580. [Google Scholar] [CrossRef]
- Gray, K.A.; Zhao, L.; Emptage, M. Bioethanol. Curr. Opin. Chem. Biol. 2006, 10, 141–146. [Google Scholar] [CrossRef]
- Nuss, P.; Gardner, K.H. Attributional life cycle assessment (ALCA) of polyitaconic acid production from northeast US softwood biomass. Int. J. Life Cycle Assess. 2013, 18, 603–612. [Google Scholar] [CrossRef]
- International Sugar Organization. International Sugar Organization Daily Sugar Prices. Available online: https://www.isosugar.org/prices.php (accessed on 27 July 2021).
- Mazzoli, R.; Olson, D.G. Clostridium thermocellum: A microbial platform for high-value chemical production from lignocellulose. Adv. Appl. Microbiol. 2020, 113, 111–161. [Google Scholar]
- Lynd, L.R. The grand challenge of cellulosic biofuels. Nat. Biotechnol. 2017, 35, 912–915. [Google Scholar] [CrossRef] [Green Version]
- Mazzoli, R. Metabolic engineering strategies for consolidated production of lactic acid from lignocellulosic biomass. Biotechnol. Appl. Biochem. 2020, 67, 61–72. [Google Scholar] [CrossRef]
- Francois, J.M.; Alkim, C.; Morin, N. Engineering microbial pathways for production of bio-based chemicals from lignocellulosic sugars: Current status and perspectives. Biotechnol. Biofuels 2020, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Gong, M.; Lv, X.; Huang, Z.; Gu, Y.; Li, J.; Du, G.; Liu, L. Current advance in biological production of short-chain organic acid. Appl. Microbiol. Biotechnol. 2020, 104, 9109–9124. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value Added Chemicals from Biomass, Volume I: Results of Screening for Potential Candidates from Sugars and Synthesis Gas; US Department of Energy: Washington, DC, USA, 2004. [Google Scholar] [CrossRef] [Green Version]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “top 10” revisited. Green Chem. 2010, 12, 539–555. [Google Scholar] [CrossRef]
- Sauer, M.; Porro, D.; Mattanovich, D.; Branduardi, P. Microbial production of organic acids: Expanding the markets. Trends Biotechnol. 2008, 26, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Precedence Research Biofuels Market Size Worth Around US$307.01 billion by 2030. Available online: https://www.globenewswire.com/news-release/2021/01/22/2162581/0/en/Biofuels-Market-Size-Worth-Around-US-307-01-Billion-by-2030.html (accessed on 10 August 2021).
- MarketStudy Global Adipic Acid Market Research Report 2020—Market Study Report. Available online: https://www.marketstudyreport.com/reports/global-adipic-acid-market-research-report-2020?gclid=Cj0KCQjw6s2IBhCnARIsAP8RfAgKz8fqb3GzGMqUAeSsabN9dUFoteDvrE6hHowwfb6McXQ2nvLwEYsaAhRrEALw_wcB (accessed on 11 August 2021).
- European Commission. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions. For a European Industrial Renaissance (COM (2014) 14 Final). Brussels 2014-01-22. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:52014DC0014&from=ET (accessed on 9 October 2021).
- Klotz, S.; Kuenz, A.; Prüße, U. Nutritional requirements and the impact of yeast extract on the d-lactic acid production by Sporolactobacillus inulinus. Green Chem. 2017, 19, 4633–4641. [Google Scholar] [CrossRef]
- Biddy, M.J.; Scarlata, C.J.; Kinchin, C.M. Chemicals from Biomass: A Market Assessment of Bioproducts with Near-Term Potential; NREL: Golden, CO, USA, 2016. [Google Scholar] [CrossRef] [Green Version]
- MarketsandMarkets. Markets and Markets Lactic Acid Market Size & Share | Industry Growth, Trends & Forecasts | COVID-19 Impact Analysis. Available online: https://www.marketsandmarkets.com/Market-Reports/polylacticacid-387.html?gclid=Cj0KCQjw6s2IBhCnARIsAP8RfAgISU79DUg8my_rRKY3ly98GZIdnJuh6gogfiiwScOqI9j0iRsPWmIaAvs-EALw_wcB (accessed on 11 August 2021).
- De Fouchécour, F.; Sánchez-Castañeda, A.K.; Saulou-Bérion, C.; Spinnler, H.É. Process engineering for microbial production of 3-hydroxypropionic acid. Biotechnol. Adv. 2018, 36, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Tian, P. Biosynthesis pathways and strategies for improving 3-hydroxypropionic acid production in bacteria. World J. Microbiol. Biotechnol. 2021, 37, 117. [Google Scholar] [CrossRef]
- Mordor Intelligence. Mordor Intelligence Fumaric Acid Market Size, Trends & Forecast | Global Industry Report 2021 to 2026. Available online: https://www.mordorintelligence.com/industry-reports/fumaric-acid-market (accessed on 10 August 2021).
- IMARC Fumaric Acid Market Size, Price Trends, Research Report 2020—Radiant Insights. Available online: https://www.imarcgroup.com/fumaric-acid-market (accessed on 11 August 2021).
- Skoog, E.; Shin, J.H.; Saez-Jimenez, V.; Mapelli, V.; Olsson, L. Biobased adipic acid—The challenge of developing the production host. Biotechnol. Adv. 2018, 36, 2248–2263. [Google Scholar] [CrossRef]
- PharmaCompass. Fumaric Acid | Price | per kg | USD. Available online: https://www.pharmacompass.com/price/fumaric-acid (accessed on 9 October 2021).
- Imarc Global Malic Acid Market Size, Share, Trends & Forecast 2021–2026. Available online: https://www.imarcgroup.com/malic-acid-technical-material-market-report (accessed on 11 August 2021).
- 360 Market Updates. Global Malic Acid Market Research Report 2020—Market Study Report. 2020. Available online: https://www.marketstudyreport.com/reports/global-malic-acid-market-research-report-2020?gclid=CjwKCAjwx8iIBhBwEiwA2quaq97ZpW (accessed on 10 August 2021).
- Mondala, A.H. Direct fungal fermentation of lignocellulosic biomass into itaconic, fumaric, and malic acids: Current and future prospects. J. Ind. Microbiol. Biotechnol. 2015, 42, 487–506. [Google Scholar] [CrossRef] [PubMed]
- PharmaCompass. Malic Acid | Price | per kg | USD. Available online: https://www.pharmacompass.com/price/malic-acid (accessed on 11 October 2021).
- MarketsandMarkets. Markets and Markets Succinic Acid Market by Type, End-Use Industry & Geography | COVID-19 Impact Analysis. Available online: https://www.marketsandmarkets.com/Market-Reports/succinic-acid-market-402.html (accessed on 11 August 2021).
- PharmaCompass. Succinic Acid | Price | per kg | USD. Available online: https://www.pharmacompass.com/price/succinic-acid (accessed on 9 October 2021).
- MarketWatch. Market Watch Itaconic Acid Market Research Report 2021—Industry Size and Share by Global Business Growth, Revenue Sales Estimation and Key Trends with Future Demand Status Forecast to 2027. Available online: https://www.marketwatch.com/press-release/itaconic-acid-market-research-report-2021---industry-size-and-share-by-global-business-growth-revenue-sales-estimation-and-key-trends-with-future-demand-status-forecast-to-2027-2021-08-08 (accessed on 11 August 2021).
- De Carvalho, J.C.; Magalhães, A.I.; Soccol, C.R. Biobased itaconic acid market and research trends-is it really a promising chemical? Chim. Oggi/Chem. Today 2018, 36, 56–58. [Google Scholar]
- Klement, T.; Büchs, J. Itaconic acid—A biotechnological process in change. Bioresour. Technol. 2013, 135, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Schlembach, I.; Tehrani, H.H.; Blank, L.M.; Büchs, J.; Wierckx, N.; Regestein, L.; Rosenbaum, M.A. Consolidated bioprocessing of cellulose to itaconic acid by a co-culture of Trichoderma reesei and Ustilago maydis. Biotechnol. Biofuels 2020, 13, 207. [Google Scholar] [CrossRef]
- PharmaCompass. Adipic Acid | Price | per kg | USD. Available online: https://www.pharmacompass.com/price/adipic-acid (accessed on 9 October 2021).
- IMARC Global Muconic Acid Market to Reach US$ 119.4 Million by 2024. Available online: https://www.imarcgroup.com/global-muconic-acid-market (accessed on 11 August 2021).
- Intrado Globe Newswire Global Glucaric Acid Market Is Anticipated to Reach USD 1.41 Billion by 2028: Fior Markets. Available online: https://www.globenewswire.com/news-release/2021/05/26/2236067/0/en/Global-Glucaric-Acid-Market-Is-Anticipated-to-Reach-USD-1-41-billion-by-2028-Fior-Markets.html (accessed on 11 August 2021).
- Tarraran, L.; Mazzoli, R. Alternative strategies for lignocellulose fermentation through lactic acid bacteria: The state of the art and perspectives. FEMS Microbiol. Lett. 2018, 365, fny126. [Google Scholar] [CrossRef]
- Lynd, L.R.; Laser, M.S.; Bransby, D.; Dale, B.E.; Davison, B.; Hamilton, R.; Himmel, M.; Keller, M.; McMillan, J.D.; Sheehan, J.; et al. How biotech can transform biofuels. Nat. Biotechnol. 2008, 26, 169–172. [Google Scholar] [CrossRef]
- Lynd, L.R.; Van Zyl, W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotechnol. 2005, 16, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Shahab, R.L.; Luterbacher, J.S.; Brethauer, S.; Studer, M.H. Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal-bacterial consortium. Biotechnol. Bioeng. 2018, 115, 1207–1215. [Google Scholar] [CrossRef]
- Minty, J.J.; Singer, M.E.; Scholz, S.A.; Bae, C.H.; Ahn, J.H.; Foster, C.E.; Liao, J.C.; Lin, X.N. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc. Natl. Acad. Sci. USA 2013, 110, 14592–14597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholz, S.A.; Graves, I.; Minty, J.J.; Lin, X.N. Production of cellulosic organic acids via synthetic fungal consortia. Biotechnol. Bioeng. 2018, 115, 1096–1100. [Google Scholar] [CrossRef]
- Shahab, R.L.; Brethauer, S.; Davey, M.P.; Smith, A.G.; Vignolini, S.; Luterbacher, J.S.; Studer, M.H. A heterogeneous microbial consortium producing short-chain fatty acids from lignocellulose. Science 2020, 369. [Google Scholar] [CrossRef]
- Gandini, C.; Tarraran, L.; Kalemasi, D.; Pessione, E.; Mazzoli, R. Recombinant Lactococcus lactis for efficient conversion of cellodextrins into L-lactic acid. Biotechnol. Bioeng. 2017, 114, 2807–2817. [Google Scholar] [CrossRef]
- Stern, J.; Moraïs, S.; Ben-David, Y.; Salama, R.; Shamshoum, M.; Lamed, R.; Shoham, Y.; Bayer, E.A.; Mizrahi, I. Assembly of synthetic functional cellulosomal structures onto the cell surface of Lactobacillus plantarum, a potent member of the gut microbiome. Appl. Environ. Microbiol. 2018, 84, e00282-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davison, S.A.; den Haan, R.; van Zyl, W.H. Exploiting strain diversity and rational engineering strategies to enhance recombinant cellulase secretion by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2020, 104, 5163–5184. [Google Scholar] [CrossRef]
- Wen, Z.; Ledesma-Amaro, R.; Lu, M.; Jin, M.; Yang, S. Metabolic engineering of Clostridium cellulovorans to improve butanol production by consolidated bioprocessing. ACS Synth. Biol. 2020, 9, 304–315. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, R.; Zhou, J.; He, A.; Xu, J.; Xin, F.; Zhang, W.; Ma, J.; Jiang, M.; Dong, W. Recent advances of biofuels and biochemicals production from sustainable resources using co-cultivation systems. Biotechnol. Biofuels 2019, 12, 155. [Google Scholar] [CrossRef] [Green Version]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.C.; Wang, H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016, 31, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 739. [Google Scholar] [CrossRef] [Green Version]
- Alper, H.; Stephanopoulos, G. Engineering for biofuels: Exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 2009, 7, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.P.; Robin, J.; Duquesne, S.; O’Donohue, M.J.; Marty, A.; Bordes, F. Developing cellulolytic Yarrowia lipolytica as a platform for the production of valuable products in consolidated bioprocessing of cellulose. Biotechnol. Biofuels 2018, 11, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarraran, L.; Gandini, C.; Luganini, A.; Mazzoli, R. Cell-surface binding domains from Clostridium cellulovorans can be used for surface display of cellulosomal scaffoldins in Lactococcus lactis. Biotechnol. J. 2021, 16, e2100064. [Google Scholar] [CrossRef]
- Olson, D.G.; Lynd, L.R. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol. 2012, 510, 317–330. [Google Scholar]
- Walker, J.E.; Lanahan, A.A.; Zheng, T.; Toruno, C.; Lynd, L.R.; Cameron, J.C.; Olson, D.G.; Eckert, C.A. Development of both type I–B and type II CRISPR/Cas genome editing systems in the cellulolytic bacterium Clostridium thermocellum. Metab. Eng. Commun. 2020, 10, e00116. [Google Scholar] [CrossRef] [PubMed]
- Hon, S.; Tian, L.; Zheng, T.; Cui, J.; Lynd, L.R.; Olson, D.G. Methods for metabolic engineering of Thermoanaerobacterium saccharolyticum. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2096, pp. 21–43. [Google Scholar]
- Yang, X.; Xu, M.; Yang, S.T. Metabolic and process engineering of Clostridium cellulovorans for biofuel production from cellulose. Metab. Eng. 2015, 32, 39–48. [Google Scholar] [CrossRef]
- Chung, D.; Cha, M.; Guss, A.M.; Westpheling, J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc. Natl. Acad. Sci. USA 2014, 111, 8931–8936. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lin, L.; Sun, T.; Xu, J.; Ji, J.; Liu, Q.; Tian, C. Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophila. Metab. Eng. 2019, 61, 416–426. [Google Scholar] [CrossRef]
- Mazzoli, R.; Lamberti, C.; Pessione, E. Engineering new metabolic capabilities in bacteria: Lessons from recombinant cellulolytic strategies. Trends Biotechnol. 2012, 30, 111–119. [Google Scholar] [CrossRef]
- Lambertz, C.; Garvey, M.; Klinger, J.; Heesel, D.; Klose, H.; Fischer, R.; Commandeur, U. Challenges and advances in the heterologous expression of cellulolytic enzymes: A review. Biotechnol. Biofuels 2014, 7, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Den Haan, R.; van Rensburg, E.; Rose, S.H.; Görgens, J.F.; van Zyl, W.H. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr. Opin. Biotechnol. 2015, 33, 32–38. [Google Scholar] [CrossRef]
- Anandharaj, M.; Lin, Y.J.; Rani, R.P.; Nadendla, E.K.; Ho, M.C.; Huang, C.C.; Cheng, J.F.; Chang, J.J.; Li, W.H. Constructing a yeast to express the largest cellulosome complex on the cell surface. Proc. Natl. Acad. Sci. USA 2020, 117, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Auer, L.; Lazuka, A.; Sillam-Dussès, D.; Miambi, E.; O’Donohue, M.; Hernandez-Raquet, G. Uncovering the potential of termite gut microbiome for lignocellulose bioconversion in anaerobic batch bioreactors. Front. Microbiol. 2017, 8, 2623. [Google Scholar] [CrossRef] [Green Version]
- Rajeswari, G.; Jacob, S.; Chandel, A.K.; Kumar, V. Unlocking the potential of insect and ruminant host symbionts for recycling of lignocellulosic carbon with a biorefinery approach: A review. Microb. Cell Fact. 2021, 20, 107. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tang, Y.; Gao, H.; Zhang, W.; Jiang, Y.; Xin, F.; Jiang, M. Challenges and future perspectives of promising biotechnologies for lignocellulosic biorefinery. Molecules 2021, 26, 5411. [Google Scholar] [CrossRef]
- Luo, Z.; Yu, S.; Zeng, W.; Zhou, J. Comparative analysis of the chemical and biochemical synthesis of keto acids. Biotechnol. Adv. 2021, 47, 107706. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, R.O.; Godan, T.K.; Sindhu, R.; Pandey, A.; Binod, P. Bioengineering advancements, innovations and challenges on green synthesis of 2, 5-furan dicarboxylic acid. Bioengineered 2020, 11, 19–38. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lin, M.; Xu, M.; Yang, S.T. Anaerobic fermentation for production of carboxylic acids as bulk chemicals from renewable biomass. In Advances in Biochemical Engineering/Biotechnology; Springer-Verlag: Berlin, Germany, 2016; Volume 156, pp. 323–363. [Google Scholar]
- Warnecke, T.; Gill, R.T. Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb. Cell Fact. 2005, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Upadhyaya, B.P.; DeVeaux, L.C.; Christopher, L.P. Metabolic engineering as a tool for enhanced lactic acid production. Trends Biotechnol. 2014, 32, 637–644. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Sonomoto, K. Opportunities to overcome the current limitations and challenges for efficient microbial production of optically pure lactic acid. J. Biotechnol. 2016, 236, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Straathof, A.J.J. The Proportion of Downstream Costs in Fermentative Production Processes. In Comprehensive Biotechnology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 2, pp. 811–814. ISBN 9780080885049. [Google Scholar]
- López-Garzón, C.S.; Straathof, A.J.J. Recovery of carboxylic acids produced by fermentation. Biotechnol. Adv. 2014, 32, 873–904. [Google Scholar] [CrossRef] [PubMed]
- Murali, N.; Srinivas, K.; Ahring, B.K. Biochemical production and separation of carboxylic acids for biorefinery applications. Fermentation 2017, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, R.A.; Komesu, A.; Vaz Rossell, C.E.; Maciel Filho, R. Challenges and opportunities in lactic acid bioprocess design—From economic to production aspects. Biochem. Eng. J. 2018, 133, 219–239. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: Overview and limits. J. Biotechnol. 2011, 156, 286–301. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 2013, 31, 877–902. [Google Scholar] [CrossRef] [PubMed]
- Jem, K.J.; van der Pol, J.F.; de Vos, S. Microbial Lactic Acid, Its Polymer Poly(lactic acid), and Their Industrial Applications. In Plastics from Bacteria; Springer: Berlin/Heidelberg, Germany, 2010; pp. 323–346. [Google Scholar]
- Poudel, P.; Tashiro, Y.; Sakai, K. New application of Bacillus strains for optically pure L-lactic acid production: General overview and future prospects. Biosci. Biotechnol. Biochem. 2016, 80, 642–654. [Google Scholar] [CrossRef]
- Okano, K.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: Recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 2010, 85, 413–423. [Google Scholar] [CrossRef]
- Mazzoli, R.; Bosco, F.; Mizrahi, I.; Bayer, E.A.; Pessione, E. Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 2014, 32, 1216–1236. [Google Scholar] [CrossRef] [PubMed]
- Scheirlinck, T.; Mahillon, J.; Joos, H.; Dhaese, P.; Michiels, F. Integration and expression of α-amylase and endoglucanase genes in the Lactobacillus plantarum chromosome. Appl. Environ. Microbiol. 1989, 55, 2130–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, E.E.; Gilbert, H.J.; Hazlewood, G.P.; Huckle, J.; Laurie, J.I.; Mann, S.P. Expression of a Clostridium thermocellum endoglucanase gene in Lactobacillus plantarum. Appl. Environ. Microbiol. 1989, 55, 2095–2097. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, Y.; Moraïs, S.; Bayer, E.A.; Mizrahi, I. Rapid adaptation for fibre degradation by changes in plasmid stoichiometry within Lactobacillus plantarum at the synthetic community level. Microb. Biotechnol. 2020, 13, 1748–1764. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.L. Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr. Opin. Biotechnol. 1995, 6, 517–522. [Google Scholar] [CrossRef]
- Fu, L.L.; Xu, Z.R.; Li, W.F.; Shuai, J.B.; Lu, P.; Hu, C.X. Protein secretion pathways in Bacillus subtilis: Implication for optimization of heterologous protein secretion. Biotechnol. Adv. 2007, 25, 1–12. [Google Scholar]
- Chang, J.J.; Anandharaj, M.; Ho, C.Y.; Tsuge, K.; Tsai, T.Y.; Ke, H.M.; Lin, Y.J.; Ha Tran, M.D.; Li, W.H.; Huang, C.C. Biomimetic strategy for constructing Clostridium thermocellum cellulosomal operons in Bacillus subtilis. Biotechnol. Biofuels 2018, 11, 157. [Google Scholar] [CrossRef] [Green Version]
- Lo, J.; Zheng, T.; Hon, S.; Olson, D.G.; Lynd, L.R. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J. Bacteriol. 2015, 197, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Mazzoli, R.; Olson, D.G.; Lynd, L.R. Construction of lactic acid overproducing Clostridium thermocellum through enhancement of lactate dehydrogenase expression. Enzyme Microb. Technol. 2020, 141, 109645. [Google Scholar] [CrossRef]
- Williams-Rhaesa, A.M.; Awuku, N.K.; Lipscomb, G.L.; Poole, F.L.; Rubinstein, G.M.; Conway, J.M.; Kelly, R.M.; Adams, M.W.W. Native xylose-inducible promoter expands the genetic tools for the biomass-degrading, extremely thermophilic bacterium Caldicellulosiruptor bescii. Extremophiles 2018, 22, 629–638. [Google Scholar] [CrossRef]
- Yao, S.; Mikkelsen, M.J. Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J. Mol. Microbiol. Biotechnol. 2010, 19, 123–133. [Google Scholar] [CrossRef]
- Zhou, J.; Olson, D.G.; Lanahan, A.A.; Tian, L.; Murphy, S.J.L.; Lo, J.; Lynd, L.R. Physiological roles of pyruvate ferredoxin oxidoreductase and pyruvate formate-lyase in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biotechnol. Biofuels 2015, 8, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandiwad, A.; Guseva, A.; Lynd, L. Metabolic Engineering of Thermoanaerobacterium thermosaccharolyticum for Increased n-Butanol Production. Adv. Microbiol. 2013, 3, 46–51. [Google Scholar] [CrossRef] [Green Version]
- Sander, K.; Chung, D.; Hyatt, D.; Westpheling, J.; Klingeman, D.M.; Rodriguez, M.; Engle, N.L.; Tschaplinski, T.J.; Davison, B.H.; Brown, S.D. Rex in Caldicellulosiruptor bescii: Novel regulon members and its effect on the production of ethanol and overflow metabolites. Microbiologyopen 2019, 8, e00639. [Google Scholar] [CrossRef] [PubMed]
- Ravcheev, D.A.; Li, X.; Latif, H.; Zengler, K.; Leyn, S.A.; Korostelev, Y.D.; Kazakov, A.E.; Novichkov, P.S.; Osterman, A.L.; Rodionov, D.A. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor rex. J. Bacteriol. 2012, 194, 1145–1157. [Google Scholar] [CrossRef]
- Lo, J.; Olson, D.G.; Murphy, S.J.L.; Tian, L.; Hon, S.; Lanahan, A.; Guss, A.M.; Lynd, L.R. Engineering electron metabolism to increase ethanol production in Clostridium thermocellum. Metab. Eng. 2017, 39, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Mazzoli, R.; Olson, D.G.; Concu, A.M.; Holwerda, E.K.; Lynd, L.R. In vivo evolution of lactic acid hyper-tolerant Clostridium thermocellum. N. Biotechnol. 2021. Minor revisions. [Google Scholar]
- Bryant, F.O. Characterization of the fructose 1,6-bisphosphate-activated, l(+lactate dehydrogenase from Thermoanaerobacter ethanolicus. J. Enzyme Inhib. Med. Chem. 1991, 5, 235–248. [Google Scholar] [CrossRef]
- Willquist, K.; van Niel, E.W.J. Lactate formation in Caldicellulosiruptor saccharolyticus is regulated by the energy carriers pyrophosphate and ATP. Metab. Eng. 2010, 12, 282–290. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y.; Zhang, X. Metabolic engineering of a laboratory-evolved Thermobifida fusca muC strain for malic acid production on cellulose and minimal treated lignocellulosic biomass. Biotechnol. Prog. 2016, 32, 14–20. [Google Scholar] [CrossRef]
- Lu, J.; Lv, Y.; Jiang, Y.; Wu, M.; Xu, B.; Zhang, W.; Zhou, J.; Dong, W.; Xin, F.; Jiang, M. Consolidated Bioprocessing of Hemicellulose-Enriched Lignocellulose to Succinic Acid through a Microbial Cocultivation System. ACS Sustain. Chem. Eng. 2020, 8, 9035–9045. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, T.; Zhao, M.; Wang, Z.; Zhao, X. Engineering Escherichia coli for succinate production from hemicellulose via consolidated bioprocessing. Microb. Cell Fact. 2012, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Geiser, E.; Reindl, M.; Blank, L.M.; Feldbrügge, M.; Wierckx, N.; Schipper, K. Activating intrinsic carbohydrate-active enzymes of the smut fungus Ustilago maydis for the degradation of plant cell wall components. Appl. Environ. Microbiol. 2016, 82, 5174–5185. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Chen, S.; Fang, H. Consolidated bioprocessing of lignocellulosic biomass to itaconic acid by metabolically engineering Neurospora crassa. Appl. Microbiol. Biotechnol. 2018, 102, 9577–9584. [Google Scholar] [CrossRef]
- Elmore, J.R.; Dexter, G.N.; Salvachúa, D.; Martinez-Baird, J.; Hatmaker, E.A.; Huenemann, J.D.; Klingeman, D.M.; Peabody, G.L.; Peterson, D.J.; Singer, C.; et al. Production of itaconic acid from alkali pretreated lignin by dynamic two stage bioconversion. Nat. Commun. 2021, 12, 2261. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J. Appl. Microbiol. 2015, 119, 1057–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvachúa, D.; Johnson, C.W.; Singer, C.A.; Rohrer, H.; Peterson, D.J.; Black, B.A.; Knapp, A.; Beckham, G.T. Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 2018, 20, 5007–5019. [Google Scholar] [CrossRef]
- Kohlstedt, M.; Starck, S.; Barton, N.; Stolzenberger, J.; Selzer, M.; Mehlmann, K.; Schneider, R.; Pleissner, D.; Rinkel, J.; Dickschat, J.S.; et al. From lignin to nylon: Cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 2018, 47, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Kuhl, M.; Kohlstedt, M.; Starck, S.; Wittmann, C. Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb. Cell Fact. 2018, 17, 115. [Google Scholar] [CrossRef]
- Li, C.; Lin, X.; Ling, X.; Li, S.; Fang, H. Consolidated bioprocessing of lignocellulose for production of glucaric acid by an artificial microbial consortium. Biotechnol. Biofuels 2021, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Mika, L.T.; Cséfalvay, E.; Németh, Á. Catalytic Conversion of Carbohydrates to Initial Platform Chemicals: Chemistry and Sustainability. Chem. Rev. 2018, 118, 505–613. [Google Scholar] [CrossRef]
- Jiang, X.R.; Yan, X.; Yu, L.P.; Liu, X.Y.; Chen, G.Q. Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis. Nat. Commun. 2021, 12, 1513. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, J.; Wu, Y.; Wu, W.; Zhang, Y.; Liu, D. Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab. Eng. 2017, 39, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.Y.; Lee, J.W.; Min, W.K.; Park, Y.C.; Seo, J.H. Simultaneous conversion of glucose and xylose to 3-hydroxypropionic acid in engineered Escherichia coli by modulation of sugar transport and glycerol synthesis. Bioresour. Technol. 2015, 198, 709–716. [Google Scholar] [CrossRef]
- Kildegaard, K.R.; Wang, Z.; Chen, Y.; Nielsen, J.; Borodina, I. Production of 3-hydroxypropionic acid from glucose and xylose by metabolically engineered Saccharomyces cerevisiae. Metab. Eng. Commun. 2015, 2, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Jang, Y.S.; Lee, S.Y. Production of succinic acid by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2016, 42, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Zhou, H.; Zhang, S.; Gu, H.; Yang, Q.; Zhang, W.; Dong, W.; Ma, J.; Fang, Y.; Jiang, M.; et al. Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresour. Technol. 2018, 258, 345–353. [Google Scholar] [CrossRef]
- Mazière, A.; Prinsen, P.; García, A.; Luque, R.; Len, C. A review of progress in (bio)catalytic routes from/to renewable succinic acid. Biofuels Bioprod. Biorefining 2017, 11, 908–931. [Google Scholar] [CrossRef]
- Beauprez, J.J.; De Mey, M.; Soetaert, W.K. Microbial succinic acid production: Natural versus metabolic engineered producers. Process Biochem. 2010, 45, 1103–1114. [Google Scholar] [CrossRef]
- Zelle, R.M.; De Hulster, E.; Van Winden, W.A.; De Waard, P.; Dijkema, C.; Winkler, A.A.; Geertman, J.M.A.; Van Dijken, J.P.; Pronk, J.T.; Van Maris, A.J.A. Malic acid production by Saccharomyces cerevisiae: Engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 2008, 74, 2766–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roa Engel, C.A.; Straathof, A.J.J.; Zijlmans, T.W.; Van Gulik, W.M.; Van Der Wielen, L.A.M. Fumaric acid production by fermentation. Appl. Microbiol. Biotechnol. 2008, 78, 379–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Lübeck, M.; Ahring, B.K.; Lübeck, P.S. Enhanced succinic acid production in Aspergillus saccharolyticus by heterologous expression of fumarate reductase from Trypanosoma brucei. Appl. Microbiol. Biotechnol. 2016, 100, 1799–1809. [Google Scholar] [CrossRef]
- Goldberg, I.; Rokem, J.S.; Pines, O. Organic acids: Old metabolites, new themes. J. Chem. Technol. Biotechnol. 2006, 81, 1601–1611. [Google Scholar] [CrossRef]
- Mrowietz, U.; Christophers, E.; Altmeyer, P. Treatment of psoriasis with fumaric acid esters: Results of a prospective multicentre study. Br. J. Dermatol. 1998, 138, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Rietzschel, I.; Pawlak, F.; Hoffmann, K. Antipsoriatic effect of fumaric acid derivatives: Results of a multicenter double-blind study in 100 patients. J. Am. Acad. Dermatol. 1994, 30, 977–981. [Google Scholar] [CrossRef]
- McGinn, S.M.; Beauchemin, K.A.; Coates, T.; Colombatto, D. Methane emissions from beef cattle: Effects of monensin, sunflower oil, enzymes, yeast, and fumaric acid. J. Anim. Sci. 2004, 82, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.H.; Bright, M.M.; Scott, W.E. Mechanism of fumaric acid accumulation in Rhizopus nigricans. J. Bacteriol. 1967, 93, 600–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, R.A.; Moyer, A.J.; Smith, M.L.; Kelley, S.E. Production of Fumaric Acid by Rhizopus arrhizus. Appl. Microbiol. 1959, 7, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Kenealy, W.; Zaady, E.; Du Preez, J.C. Biochemical aspects of fumaric acid accumulation by Rhizopus arrhizus. Appl. Environ. Microbiol. 1986, 52, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Juhász, T.; Szengyel, Z.; Réczey, K.; Siika-Aho, M.; Viikari, L. Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem. 2005, 40, 3519–3525. [Google Scholar] [CrossRef]
- Gu, S.; Li, J.; Chen, B.; Sun, T.; Liu, Q.; Xiao, D.; Tian, C. Metabolic engineering of the thermophilic filamentous fungus Myceliophthora thermophila to produce fumaric acid. Biotechnol. Biofuels 2018, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Gross, R.A.; Kalra, B. Biodegradable polymers for the environment. Science 2002, 297, 803–807. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.; Zhou, Y.; Lin, M.; Wei, P.; Yang, S.T. Polymalic acid fermentation by Aureobasidium pullulans for malic acid production from soybean hull and soy molasses: Fermentation kinetics and economic analysis. Bioresour. Technol. 2017, 223, 166–174. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, Y.; Luo, H.; Cheng, X.; Zhang, R.; Teng, W. A comparative evaluation of different types of microbial electrolysis desalination cells for malic acid production. Bioresour. Technol. 2015, 198, 87–93. [Google Scholar] [CrossRef]
- Battat, E.; Peleg, Y.; Bercovitz, A.; Rokem, J.S.; Goldberg, I. Optimization of L-malic acid production by Aspergillus flavus in a stirred fermentor. Biotechnol. Bioeng. 1991, 37, 1108–1116. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, Y.; Cao, W.; Liu, H. Improved Production of Malic Acid in Aspergillus Niger by Abolishing Citric Acid Accumulation and Enhancing Glycolytic Flux. ACS Synth. Biol. 2020, 9, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Fong, S.S. Laboratory evolution and multi-platform genome re-sequencing of the cellulolytic actinobacterium Thermobifida fusca. J. Biol. Chem. 2011, 286, 39958–39966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Li, J.; Gao, H.; Zhou, D.; Xu, H.; Cong, Y.; Zhang, W.; Xin, F.; Jiang, M. Recent progress on bio-succinic acid production from lignocellulosic biomass. World J. Microbiol. Biotechnol. 2021, 37, 16. [Google Scholar] [CrossRef]
- Andersson, C.; Hodge, D.; Berglund, K.A.; Rova, U. Effect of different carbon sources on the production of succinic acid using metabolically engineered Escherichia coli. Biotechnol. Prog. 2007, 23, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Alcantara, J.; Mondala, A.; Hughey, L.; Shields, S. Direct succinic acid production from minimally pretreated biomass using sequential solid-state and slurry fermentation with mixed fungal cultures. Fermentation 2017, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Runtsch, M.C.; O’Neill, L.A.J. Pseudomonas Persists by Feeding off Itaconate. Cell Metab. 2020, 31, 1045–1047. [Google Scholar] [CrossRef]
- Zhao, M.; Lu, X.; Zong, H.; Li, J.; Zhuge, B. Itaconic acid production in microorganisms. Biotechnol. Lett. 2018, 40, 455–464. [Google Scholar] [CrossRef]
- Kuenz, A.; Krull, S. Biotechnological production of itaconic acid—things you have to know. Appl. Microbiol. Biotechnol. 2018, 102, 3901–3914. [Google Scholar] [CrossRef]
- Geiser, E.; Przybilla, S.K.; Friedrich, A.; Buckel, W.; Wierckx, N.; Blank, L.M.; Bölker, M. Ustilago maydis produces itaconic acid via the unusual intermediate trans-aconitate. Microb. Biotechnol. 2016, 9, 116–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bafana, R.; Pandey, R.A. New approaches for itaconic acid production: Bottlenecks and possible remedies. Crit. Rev. Biotechnol. 2018, 38, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Krull, S.; Hevekerl, A.; Kuenz, A.; Prüße, U. Process development of itaconic acid production by a natural wild type strain of Aspergillus terreus to reach industrially relevant final titers. Appl. Microbiol. Biotechnol. 2017, 101, 4063–4072. [Google Scholar] [CrossRef] [PubMed]
- Okabe, M.; Lies, D.; Kanamasa, S.; Park, E.Y. Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl. Microbiol. Biotechnol. 2009, 84, 597–606. [Google Scholar] [CrossRef] [Green Version]
- Karaffa, L.; Díaz, R.; Papp, B.; Fekete, E.; Sándor, E.; Kubicek, C.P. A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl. Microbiol. Biotechnol. 2015, 99, 7937–7944. [Google Scholar] [CrossRef] [Green Version]
- Soccol, C.R.; Vandenberghe, L.P.S.; Rodrigues, C.; Pandey, A. New perspectives for citric acid production and application. Food Technol. Biotechnol. 2006, 44, 141–149. [Google Scholar]
- Cano-Canchola, C.; Acevedo, L.; Ponce-Noyola, P.; Flores-Martínez, A.; Flores-Carreón, A.; Leal-Morales, C.A. Induction of lytic enzymes by the interaction of Ustilago maydis with zea mays tissues. Fungal Genet. Biol. 2000, 29, 145–151. [Google Scholar] [CrossRef]
- Mueller, O.; Kahmann, R.; Aguilar, G.; Trejo-Aguilar, B.; Wu, A.; de Vries, R.P. The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 2008, 45, S63–S70. [Google Scholar] [CrossRef]
- Couturier, M.; Navarro, D.; Olivé, C.; Chevret, D.; Haon, M.; Favel, A.; Lesage-Meessen, L.; Henrissat, B.; Coutinho, P.M.; Berrin, J.G. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genom. 2012, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Regestein, L.; Klement, T.; Grande, P.; Kreyenschulte, D.; Heyman, B.; Maßmann, T.; Eggert, A.; Sengpiel, R.; Wang, Y.; Wierckx, N.; et al. From beech wood to itaconic acid: Case study on biorefinery process integration. Biotechnol. Biofuels 2018, 11, 279. [Google Scholar] [CrossRef]
- Kerssemakers, A.A.J.; Doménech, P.; Cassano, M.; Yamakawa, C.K.; Dragone, G.; Mussatto, S.I. Production of itaconic acid from cellulose pulp: Feedstock feasibility and process strategies for an efficient microbial performance. Energies 2020, 13, 1654. [Google Scholar] [CrossRef] [Green Version]
- Stoffels, P.; Müller, M.J.; Stachurski, S.; Terfrüchte, M.; Schröder, S.; Ihling, N.; Wierckx, N.; Feldbrügge, M.; Schipper, K.; Büchs, J. Complementing the intrinsic repertoire of Ustilago maydis for degradation of the pectin backbone polygalacturonic acid. J. Biotechnol. 2020, 307, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Van der Straat, L.; Vernooij, M.; Lammers, M.; van den Berg, W.; Schonewille, T.; Cordewener, J.; van der Meer, I.; Koops, A.; De Graaff, L.H. Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger. Microb. Cell Fact. 2014, 13, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, A.H.; Van Gerven, R.; Overkamp, K.M.; Lübeck, P.S.; Taşplnar, H.; Türker, M.; Punt, P.J. Metabolic engineering with ATP-citrate lyase and nitrogen source supplementation improves itaconic acid production in Aspergillus niger. Biotechnol. Biofuels 2019, 12, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, H.; Ma, Q.; Wei, D.; Wang, F. Metabolic engineering of an industrial Aspergillus niger strain for itaconic acid production. 3 Biotech 2020, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- McFadden, B.A.; Purohit, S. Itaconate, an isocitrate lyase directed inhibitor in Pseudomonas indigofera. J. Bacteriol. 1977, 131, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, I.A.; Filatova, L.V.; Ivanovsky, R.N. Inhibition of acetate and propionate assimilation by itaconate via propionyl-CoA carboxylase in isocitrate lyase-negative purple bacterium Rhodospirillum rubrum. FEMS Microbiol. Lett. 2002, 216, 49–54. [Google Scholar] [CrossRef]
- Harder, B.J.; Bettenbrock, K.; Klamt, S. Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab. Eng. 2016, 38, 29–37. [Google Scholar] [CrossRef]
- Otten, A.; Brocker, M.; Bott, M. Metabolic engineering of Corynebacterium glutamicum for the production of itaconate. Metab. Eng. 2015, 30, 156–165. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef] [PubMed]
- Hosseinpour Tehrani, H.; Becker, J.; Bator, I.; Saur, K.; Meyer, S.; Rodrigues Lóia, A.C.; Blank, L.M.; Wierckx, N. Integrated strain- And process design enable production of 220 g L-1 itaconic acid with Ustilago maydis. Biotechnol. Biofuels 2019, 12, 263. [Google Scholar] [CrossRef]
- Choi, S.; Lee, H.N.; Park, E.; Lee, S.J.; Kim, E.S. Recent advances in microbial production of cis,cis-muconic acid. Biomolecules 2020, 10, 1238. [Google Scholar] [CrossRef] [PubMed]
- De Jong, E.; Higson, A.; Walsh, P.; Wellisch, M. Task 42 Biobased Chemicals—Value Added Products from Biorefineries; IEA Bioenergy: London, UK, 2011; Available online: https://www.ieabioenergy.com/wp-content/uploads/2013/10/Task-42-Biobased-Chemicals-value-added-products-from-biorefineries.pdf (accessed on 9 October 2021).
- Polen, T.; Spelberg, M.; Bott, M. Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 2013, 167, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Aryapratama, R.; Janssen, M. Prospective life cycle assessment of bio-based adipic acid production from forest residues. J. Clean. Prod. 2017, 164, 434–443. [Google Scholar] [CrossRef] [Green Version]
- Diamond, G.M.; Murphy, V.; Boussie, T.R. Application of High Throughput Experimentation to the Production of Commodity Chemicals from Renewable Feedstocks. In Modern Applications of High Throughput R&D in Heterogeneous Catalysis; Bentham Science Publishers: Sharjah, United Arab Emirates, 2014; pp. 288–309. [Google Scholar]
- Yang, J.; Wei, Y.; Li, G.; Zhou, S.; Deng, Y. Computer-aided engineering of adipyl-CoA synthetase for enhancing adipic acid synthesis. Biotechnol. Lett. 2020, 42, 2693–2701. [Google Scholar] [CrossRef]
- Weber, C.; Brückner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef] [Green Version]
- Mazzoli, R.; Pessione, E.; Giuffrida, M.G.; Fattori, P.; Barello, C.; Giunta, C.; Lindley, N.D. Degradation of aromatic compounds by Acinetobacter radioresistens S13: Growth characteristics on single substrates and mixtures. Arch. Microbiol. 2007, 188, 55–68. [Google Scholar] [CrossRef]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Schweigert, N.; Zehnder, A.J.B.; Eggen, R.I.L. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 2001, 3, 81–91. [Google Scholar] [CrossRef]
- Park, W.; Jeon, C.O.; Cadillo, H.; DeRito, C.; Madsen, E.L. Survival of naphthalene-degrading Pseudomonas putida NCIB 9816-4 in naphthalene-amended soils: Toxicity of naphthalene and its metabolites. Appl. Microbiol. Biotechnol. 2004, 64, 429–435. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef]
- Linger, J.G.; Vardon, D.R.; Guarnieri, M.T.; Karp, E.M.; Hunsinger, G.B.; Franden, M.A.; Johnson, C.W.; Chupka, G.; Strathmann, T.J.; Pienkos, P.T.; et al. Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. USA 2014, 111, 12013–12018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvachúa, D.; Karp, E.M.; Nimlos, C.T.; Vardon, D.R.; Beckham, G.T. Towards lignin consolidated bioprocessing: Simultaneous lignin depolymerization and product generation by bacteria. Green Chem. 2015, 17, 4951–4967. [Google Scholar] [CrossRef]
- Kaneko, A.; Ishii, Y.; Kirimura, K. High-yield production of cis,cis-muconic acid from catechol in aqueous solution by biocatalyst. Chem. Lett. 2011, 40, 381–383. [Google Scholar] [CrossRef]
- Beckham, G.T.; Johnson, C.W.; Karp, E.M.; Salvachúa, D.; Vardon, D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016, 42, 40–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, E.M.; Nimlos, C.T.; Deutch, S.; Salvachúa, D.; Cywar, R.M.; Beckham, G.T. Quantification of acidic compounds in complex biomass-derived streams. Green Chem. 2016, 18, 4750–4760. [Google Scholar] [CrossRef]
- Johnson, C.W.; Abraham, P.E.; Linger, J.G.; Khanna, P.; Hettich, R.L.; Beckham, G.T. Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab. Eng. Commun. 2017, 5, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.C.; Khusnutdinova, A.N.; Flick, R.; Kim, T.; Bornscheuer, U.T.; Yakunin, A.F.; Mahadevan, R. Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem. Sci. 2017, 8, 1406–1413. [Google Scholar] [CrossRef] [Green Version]
- Raj, K.; Partow, S.; Correia, K.; Khusnutdinova, A.N.; Yakunin, A.F.; Mahadevan, R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. Commun. 2018, 6, 28–32. [Google Scholar] [CrossRef]
- Sun, J.; Raza, M.; Sun, X.; Yuan, Q. Biosynthesis of adipic acid via microaerobic hydrogenation of cis,cis-muconic acid by oxygen-sensitive enoate reductase. J. Biotechnol. 2018, 280, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.S.; Yoon, S.H.; Lanza, A.M.; Roy-Mayhew, J.D.; Prather, K.L.J. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitham, J.M.; Moon, J.W.; Rodriguez, M.; Engle, N.L.; Klingeman, D.M.; Rydzak, T.; Abel, M.M.; Tschaplinski, T.J.; Guss, A.M.; Brown, S.D. Clostridium thermocellum LL1210 pH homeostasis mechanisms informed by transcriptomics and metabolomics. Biotechnol. Biofuels 2018, 11, 98. [Google Scholar] [CrossRef]
- Ju, S.Y.; Kim, J.H.; Lee, P.C. Long-term adaptive evolution of Leuconostoc mesenteroides for enhancement of lactic acid tolerance and production. Biotechnol. Biofuels 2016, 9, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boontawan, P.; Kanchanathawee, S.; Boontawan, A. Extractive fermentation of l-(+)-lactic acid by Pediococcus pentosaceus using electrodeionization (EDI) technique. Biochem. Eng. J. 2011, 54, 192–199. [Google Scholar] [CrossRef]
- Sandström, A.G.; Almqvist, H.; Portugal-Nunes, D.; Neves, D.; Lidén, G.; Gorwa-Grauslund, M.F. Saccharomyces cerevisiae: A potential host for carboxylic acid production from lignocellulosic feedstock? Appl. Microbiol. Biotechnol. 2014, 98, 7299–7318. [Google Scholar] [CrossRef]
- Valli, M.; Sauer, M.; Branduardi, P.; Borth, N.; Porro, D.; Mattanovich, D. Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl. Environ. Microbiol. 2006, 72, 5492–5499. [Google Scholar] [CrossRef] [Green Version]
- Bellasio, M.; Mattanovich, D.; Sauer, M.; Marx, H. Organic acids from lignocellulose: Candida lignohabitans as a new microbial cell factory. J. Ind. Microbiol. Biotechnol. 2015, 42, 681–691. [Google Scholar] [CrossRef]
- Chen, Y.; Nielsen, J. Biobased organic acids production by metabolically engineered microorganisms. Curr. Opin. Biotechnol. 2016, 37, 165–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Usai, G.; Cirrincione, S.; Re, A.; Manfredi, M.; Pagnani, A.; Pessione, E.; Mazzoli, R. Clostridium cellulovorans metabolism of cellulose as studied by comparative proteomic approach. J. Proteom. 2020, 216, 103667. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Spike, T.; Klingeman, D.M.; Rodriguez, M.; Bremer, V.R.; Brown, S.D. Generation and Characterization of Acid Tolerant Fibrobacter succinogenes S85. Sci. Rep. 2017, 7, 2277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef] [Green Version]
- Cǎlinescu, O.; Paulino, C.; Kühlbrandt, W.; Fendler, K. Keeping it simple, transport mechanism and pH regulation in Na +/H+ exchangers. J. Biol. Chem. 2014, 289, 13168–13176. [Google Scholar] [CrossRef] [Green Version]
- Macpherson, N.; Shabala, L.; Rooney, H.; Jarman, M.G.; Davies, J.M. Plasma membrane H+ and K+ transporters are involved in the weak-acid preservative response of disprate food spoilage yeasts. Microbiology 2005, 151, 1995–2003. [Google Scholar] [CrossRef] [Green Version]
- Miller, E.F.; Maier, R.J. Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J. Bacteriol. 2014, 196, 3074–3081. [Google Scholar] [CrossRef] [Green Version]
- Laroute, V.; Yasaro, C.; Narin, W.; Mazzoli, R.; Pessione, E.; Cocaign-Bousquet, M.; Loubière, P. GABA production in Lactococcus lactis Is enhanced by Arginine and co-addition of malate. Front. Microbiol. 2016, 7, 1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, P.; Usai, G.; Re, A.; Manfredi, M.; Mannino, G.; Bertea, C.M.; Pessione, E.; Mazzoli, R. Clostridium cellulovorans Proteomic Responses to Butanol Stress. Front. Microbiol. 2021, 12, 1881. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Du, G.; Chen, J. Heterologous expression of Lactobacillus casei RecO improved the multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 during salt stress. Bioresour. Technol. 2013, 143, 238–241. [Google Scholar] [CrossRef]
- Peetermans, A.; Foulquié-Moreno, M.R.; Thevelein, J.M. Mechanisms underlying lactic acid tolerance and its influence on lactic acid production in Saccharomyces cerevisiae. Microb. Cell 2021, 8, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Jarboe, L.R.; Royce, L.A.; Liu, P. Understanding biocatalyst inhibition by carboxylic acids. Front. Microbiol. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narendranath, N.V.; Thomas, K.C.; Ingledew, W.M. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J. Ind. Microbiol. Biotechnol. 2001, 26, 171–177. [Google Scholar] [CrossRef]
- Wilbanks, B.; Trinh, C.T. Comprehensive characterization of toxicity of fermentative metabolites on microbial growth. Biotechnol. Biofuels 2017, 10, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patnaik, R.; Louie, S.; Gavrilovic, V.; Perry, K.; Stemmer, W.P.C.; Ryan, C.M.; Del Cardayré, S. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 2002, 20, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Pei, X.; Yu, L.; Feng, Y. Genome-shuffling improved acid tolerance and l-lactic acid volumetric productivity in Lactobacillus rhamnosus. J. Biotechnol. 2007, 129, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, Y.; Tang, Y.; Wang, X.; Chen, Y.; Shen, W.; Zhan, Y.; Gao, J.; Wu, B.; He, M.; et al. Development and characterization of acidic-pH-tolerant mutants of Zymomonas mobilis through adaptation and next-generation sequencing-based genome resequencing and RNA-Seq. Biotechnol. Biofuels 2020, 13, 144. [Google Scholar] [CrossRef] [PubMed]
- Si, T.; Chao, R.; Min, Y.; Wu, Y.; Ren, W.; Zhao, H. Automated multiplex genome-scale engineering in yeast. Nat. Commun. 2017, 8, 15187. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, H.; Yu, J.; Chen, X.; Liu, L. Med15B regulates acid stress response and tolerance in Candida glabrata by altering membrane lipid composition. Appl. Environ. Microbiol. 2017, 83, e01128-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Yang, J.; Yang, P.; Wu, Z.; Zhang, J.; Du, G. Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Microb. Cell Fact. 2019, 18, 136. [Google Scholar] [CrossRef]
- Wen, Z.; Ledesma-Amaro, R.; Lu, M.; Jiang, Y.; Gao, S.; Jin, M.; Yang, S. Combined evolutionary engineering and genetic manipulation improve low pH tolerance and butanol production in a synthetic microbial Clostridium community. Biotechnol. Bioeng. 2020, 117, 2008–2022. [Google Scholar] [CrossRef]
- Trip, H.; Mulder, N.L.; Lolkema, J.S. Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524. J. Biol. Chem. 2012, 287, 11195–11204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Caiyin, Q.; Feng, W.; Zhao, X.; Qiao, B.; Zhao, G.; Qiao, J. Enhance nisin yield via improving acid-tolerant capability of Lactococcus lactis F44. Sci. Rep. 2016, 6, 27973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Sakamoto, T.; Sugiyama, M.; Ishida, N.; Kambe, H.; Obata, S.; Kaneko, Y.; Takahashi, H.; Harashima, S. Disruption of multiple genes whose deletion causes lactic-acid resistance improves lactic-acid resistance and productivity in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2013, 115, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, J.; Bai, X.; Chang, Y.; Mou, J.; Song, J.; Wang, M. Improving the acetic acid tolerance and fermentation of Acetobacter pasteurianus by nucleotide excision repair protein UvrA. Appl. Microbiol. Biotechnol. 2018, 102, 6493–6502. [Google Scholar] [CrossRef]
- Abdullah-Al-Mahin; Sugimoto, S.; Higashi, C.; Matsumoto, S.; Sonomoto, K. Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl. Environ. Microbiol. 2010, 76, 4277–4285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Jiang, L.; Zhu, L.; Xu, Q.; Xu, X.; Huang, H. Tailoring of global transcription sigma D factor by random mutagenesis to improve Escherichia coli tolerance towards low-pHs. J. Biotechnol. 2016, 224, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schellhorn, H.E. Elucidating the function of the RpoS regulon. Future Microbiol. 2014, 9, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.M.D.; Benjamin, W.H.; Swords, W.E.; Foster, J.W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 1996, 178, 2572–2579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli *. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [Green Version]
- Waterman, S.R.; Small, P.L.C. Identification of σ(S)-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 1996, 21, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, J.; Yan, X.; Yang, J.; Li, X.; Chen, P.; Yang, X. Engineering of the Small Noncoding RNA (sRNA) DsrA Together with the sRNA Chaperone Hfq Enhances the Acid Tolerance of Escherichia coli. Appl. Environ. Microbiol. 2021, 87, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, B.; Qin, H.; Liu, P.; Qin, Y.; Duan, G.; Hu, G.; He, M. Genome shuffling enhances stress tolerance of Zymomonas mobilis to two inhibitors. Biotechnol. Biofuels 2019, 12, 288. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Luo, Q.; Liu, J.; Chen, X.; Liu, L. Enhanced pyruvate production in Candida glabrata by overexpressing the CgAMD1 gene to improve acid tolerance. Biotechnol. Lett. 2018, 40, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fu, R.Y.; Hugenholtz, J.; Li, Y.; Chen, J. Glutathione protects Lactococcus lactis against acid stress. Appl. Environ. Microbiol. 2007, 73, 5268–5275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.L.; Cardoso, F.S.; Bohn, A.; Neves, A.R.; Santos, H. Engineering trehalose synthesis in Lactococcus lactis for improved stress tolerance. Appl. Environ. Microbiol. 2011, 77, 4189–4199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Tan, J.; Zhang, L.; Gu, X.; Xu, W.; Guo, X.; Luo, Y. Increase of stress resistance in Lactococcus lactis via a novel food-grade vector expressing a shsp gene from Streptococcus thermophilus. Braz. J. Microbiol. 2012, 43, 1157–1164. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.G.; Liu, X.Y.; He, X.P.; Guo, X.N.; Lu, Y.; Zhang, B. run Improvement of acetic acid tolerance and fermentation performance of Saccharomyces cerevisiae by disruption of the FPS1 aquaglyceroporin gene. Biotechnol. Lett. 2011, 33, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Holzwarth, G.; Penner, M.H.; Patton-Vogt, J.; Bakalinsky, A.T. Overexpression of acetyl-CoA synthetase in Saccharomyces cerevisiae increases acetic acid tolerance. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Luan, Y.; Wang, Q.; Liang, Q.; Qi, Q. Construction of cellulose-utilizing Escherichia coli based on a secretable cellulase. Microb. Cell Fact. 2015, 14, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokinsky, G.; Peralta-Yahya, P.P.; George, A.; Holmes, B.M.; Steen, E.J.; Dietrich, J.; Lee, T.S.; Tullman-Ercek, D.; Voigt, C.A.; Simmons, B.A.; et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 19949–19954. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.J.; Ho, F.J.; Ho, C.Y.; Wu, Y.C.; Hou, Y.H.; Huang, C.C.; Shih, M.C.; Li, W.H. Assembling a cellulase cocktail and a cellodextrin transporter into a yeast host for CBP ethanol production. Biotechnol. Biofuels 2013, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Wang, J.; Wang, S.; Shen, Y.; Petranovic, D.; Hou, J.; Bao, X. Efficient yeast surface-display of novel complex synthetic cellulosomes. Microb. Cell Fact. 2018, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.P.; Duquesne, S.; Bozonnet, S.; Cioci, G.; Nicaud, J.M.; Marty, A.; O’Donohue, M.J. Conferring cellulose-degrading ability to Yarrowia lipolytica to facilitate a consolidated bioprocessing approach. Biotechnol. Biofuels 2017, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Gambacorta, F.V.; Dietrich, J.J.; Yan, Q.; Pfleger, B.F. Rewiring yeast metabolism to synthesize products beyond ethanol. Curr. Opin. Chem. Biol. 2020, 59, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zheng, T.; Ye, X.; Xin, F.; Zhang, W.; Dong, W.; Ma, J.; Jiang, M. Metabolic engineering of Escherichia coli for l-malate production anaerobically. Microb. Cell Fact. 2020, 19, 165. [Google Scholar] [CrossRef]
- Martinez, I.; Gao, H.; Bennett, G.N.; San, K.Y. High yield production of four-carbon dicarboxylic acids by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2018, 45, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.T.; Kunjapur, A.M. Deployment of Engineered Microbes: Contributions to the Bioeconomy and Considerations for Biosecurity. Heal. Secur. 2020, 18, 278–296. [Google Scholar] [CrossRef]




| N° of C Atoms | Organic Acid | Price (US$/kg) | Market Size (US$ Million) | Annual Production (k Tons) | CAGR | Application Field | Currently Exploited Feedstocks | References |
|---|---|---|---|---|---|---|---|---|
| C3 | LA | 1.30–4.0 | 1100 | ≈300 | 12.8% | Biodegradable polymers, food and beverages, personal care/cosmetics, pharmaceuticals | corn, sugarcane | [16,17,18] |
| 3-HP | n.a. | n.a. | n.a. | n.a. | Food/feed additive/preservative, building block for plastic polymers, acrylic acid precursor | Glucose, glycerol, 1,3-propanediol | [19,20] | |
| C4 | FA | 1.4 | 661 | 225 | 5.5% | Food/beverage additive, rosin paper sizing, unsaturated polyester resin, alkyd resin, personal care/cosmetics | petroleum | [21,22,23,24] |
| MA | 2.0–4.4 | 296 | 70 | 1.2% | Food/beverage additive, personal care/cosmetics, pharmaceuticals, biodegradable polymers | petroleum | [23,25,26,27,28] | |
| SA | 6.4 | 132 | 76 | 6.8% | Production of 1,4-butanediol and polymers, lubricants, pigments, personal care, food colorants | petroleum | [23,29,30] | |
| C5 | IA | 1.5–2.0 | 98 | 41 | 3.9% | Plasticizer, lubricating oil additive, adhesives, sealants, finishing agents, paints, coatings | glucose | [23,31,32,33,34] |
| C6 | AA | 1.0 | 5200 | 3000 | 4.4% | Nylon 6,6, polyurethanes, adipic esters | petroleum | [14,23,35] |
| CCM | n.a. | 79.6 | n.a. | 7% | Bio-plastics, agrochemicals, pharmaceuticals, new functional resins, food additives, production of bulk chemicals | glucose, petroleum/lignin derived aromatics | [23,36] | |
| GA | n.a. | 750 | n.a. | 8.1% | Detergents, polymer synthesis, corrosion inhibitors, pharmaceuticals, food additives/health supplements | glucose | [23,37] |
| N° of C Atoms | Product | Feedstock | CBP Strategy | Type of Fermentation | Microbial Strains | T (g/L) | Y (g/g) [%M] | P (g/L/h) | pH regulation | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| C3 | LA | Avicel | MC | Batch | Trichoderma reesei + Lactobacillus pentosus | 34.7 | 0.62 [62.4%] | 0.16 | 5.0 | [41] |
| Whole-slurry beech wood | MC | Fed-Batch | T. reesei + L. pentosus | 19.8 | 0.78 [85.2%] | 0.10 | 5.0 | [41] | ||
| Cellodextrin mixture (DP2-10) | RCS | Batch | a Lactococcus lactis P32blgA:engD | 3.42 | 0.89 [89%] | 0.31 | No | [45] | ||
| Cellobiose | NCS | Batch | b Clostridium thermocellum ΔadhE ldhS161R | 7.9 | 0.9 [85%] | 0.2 | No | [99] | ||
| C4 | FA | Microcrystalline cellulose | MC | Batch | T. reesei + Rhizopus delemar | 6.87 | 0.17 [13%] | 31.8 × 10−3 | No | [43] |
| Alkaline pretreated corn stover | MC | Batch | T. reesei + R. delemar | 0.69 | 0.05 [≈4.6%] | ≈3.5 × 10−3 | No | [43] | ||
| MA | Avicel | NCS | Batch | c Thermobifida fusca muC ΔcelR::PcdxCgpyk | 62.8 | 0.63 [42.2%] | 0.51 | n.a. | [102] | |
| Milled corn stover | NCS | Batch | c T. fusca muC ΔcelR::PcdxCgpyk | 21.5 | 0.43 [≈62%] | 0.18 | n.a. | [102] | ||
| Avicel | NCS | Fed-batch | d Myceliophthora thermophila JG207 (PtrpChph PtefAomae PAngpdAAopyc) | 181 | e 1.1 [n.a.] | ≈0.85 | 6.0 | [60] | ||
| Pulverized corn cob | NCS | Fed-batch | d M. thermophila JG207 (PtrpChph PtefAomae PAngpdAAopyc) | 105 | e 0.4 [n.a.] | ≈0.66 | 6.0 | [60] | ||
| Avicel | NCS | Batch | f M. thermophila JG574 (JG207 PMtgpdAglt-1 PgpdAppc PgpdAmdh Δldh Δpdc Δpck::Ppdcca PldhbicA) | 83.3 | 1.11 [75%] | ≈0.43 | 6.0 | [60] | ||
| SA | Xylan | MC | Batch | Thermoanaerobacterium thermosaccharolyticum + Actinobacillus succinogenes | 32.5 | 0.39 [≈49.6%] | ≈0.14 | 6.5–6.8 | [103] | |
| Corn cob | MC | Batch | T. thermosaccharolyticum + A. succinogenes | 12.5 | 0.16 [≈19.7%] | ≈0.065 | 6.5–6.8 | [103] | ||
| Xylan + xylose | - | Batch | gEscherichia coli ∆ldh ∆pflB ∆ptsG PtrcosmY-xynC-A:osmY-xyloA:osmY-abf2 Ptrcpyc PdsbAdsbA | 14.4 | 0.37 [≈47%] | 0.12 | No | [104] | ||
| C5 | IA | Cellobiose | RCS | Batch | h Ustilago maydis Pomabgl1 | 5.2 | 0.1 [14%] | ≈0.035 | No | [105] |
| Avicel | NCS | Batch | i Neurospora crassa FGSC 9720 Pccg-1cad1 | 20.4 × 10−3 | 1.02 × 10−3 [0.14%] | 4.3 × 10−4 | No | [106] | ||
| α-cellulose | MC | Fed-batch | j T. reesei RUT-C30 + U. maydis Δcyp3 ΔPria1::Petef Δfuz7 PetefmttA | 34 | 0.16 [22%] | 0.07 | ≈6–7.5 | [34] | ||
| k Depolymerized corn stover lignin | - | Batch | l Pseudomonas putida JE90 PT7:tad1:adi1 (trans) PurtAT7pol PcatlysY ΔphaC1ZC2 icdGTG:idhGTG | 1.43 | m 0.79 [99%] | 0.03 | No | [107] | ||
| C6 | AA | Avicel | - | Batch | T. fusca | 0.06 | 0.012 [1.61%] | 1.40 × 10−3 | n.a. | [108] |
| Milled corncob | - | Batch | T. fusca | 0.22 | 0.011 [1.47%] | 3.14 × 10−3 | n.a. | [108] | ||
| CCM | k Depolymerized corn stover lignin | - | Fed-batch | n P. putida KT2440 ΔcatRBC::PtaccatA ΔpcaHG::Ptac aroY:ecdB:ecdD Δcrc | 3.7 | m About 100% | 0.06 | 7 | [109] | |
| o Depolymerized softwood lignin | - | Fed-batch | p P. putida KT2440 ΔcatBC ΔendA-1 ΔendA-2 PcatcatA:catA2 PGRO*dmpKLMNOP | 13 | m About 100% | 0.24 | 7.0 | [110] | ||
| o Depolymerized softwood lignin | - | Fed-batch | q Corynebacterium glutamicum ΔcatB PtufcatA | 1.8 | m About 100% | 0.067 | 7.0 | [111] | ||
| GA | Avicel | MC | Batch | r T. reesei RUT-C30 + Saccharomyces cerevisiae INVSc1 ∆opi1 Pgpdmiox4 Ptefudh | 0.54 | 0.036 [≈3.1%] | 2.14 × 10−4 | No | [112] | |
| Steam-exploded corn stover | MC | Batch | r T. reesei RUT-C30 + S. cerevisiae INVSc1 ∆opi1 Pgpdmiox4 Ptefudh | 0.45 | 0.030 [n.a.] | 1.80 × 10−4 | No | [112] |
| Strategy | Microorganism | RME Gene Modification (Protein Function Encoded) | Improved Phenotype | Reference |
|---|---|---|---|---|
| ALE | Clostridium thermocellum | - | 2.7-fold higher LA tolerance (from 15 to 35 g/L) of strain LL1111, 50–85% faster growth of strain LL345 with 45 g/L LA | [99] |
| Leuconostoc mesenteroides | - | 2-fold higher growth rate with 70 g/L LA * | [190] | |
| Zymomonas mobilis | - | 2.5–3.5 fold higher biomass production at pH 3.5 | [212] | |
| GS | Lactobacillus sp. | - | pH limit for growth lowered from 4.2 to 3.8 * | [210] |
| Z. mobilis | - | ≈2-fold higher biomass production with 7 g/L acetic acid | [228] | |
| MAGE | Saccharomyces cerevisiae | - | Ability to growth with 11 g/L acetic acid | [213] |
| RME | Acetobacter pasteurianus | Overexpression of Lactobacillus casei uvrA (DNA repair protein) | 2-fold higher survival after 60 min with 60 g/L acetic acid | [220] |
| Candida glabrata | Overexpression of amd1 (AMP deaminase) | 59% increased biomass and 51% increased cell viability at pH 4 | [229] | |
| Escherichia coli | Overexpression of dsrA (sRNA) and hfq (sRNA chaperone) | About 50% improved biomass production at pH 4.5, about 100-fold higher survival at pH 2.5 | [227] | |
| Lactococcus lactis | Overexpression of E. coli gshA (γ-glutamyl cysteine synthetase) and gshB (glutathione synthetase) | 15-fold-higher survival after 30 min at pH 2.5; 18-fold higher survival after 10 h at pH 4.0 | [230] | |
| L. lactis | Overexpression of E. coli dnaK chaperone | 2.6-fold shorter generation time and 1.4-fold higher maximum biomass in medium supplemented with 5 g/L LA * | [221] | |
| L. lactis | Overexpression of lactococcal trePP (trehalose 6-phosphate phosphorylase) pgmB (β-phosphoglucomutase), and Propionibacterium freudenreichii otsBPf (trehalose 6-phosphate phosphatase) | 7-fold increased survival at pH 3.0 | [231] | |
| L. lactis | Overexpression of Streptococcus thermophilus shsp (small heat shock protein) | 6-fold higher survival after 4 h at pH 3.0 | [232] | |
| L. lactis | Overexpression of S. thermophilus hdcAPB (histidine decarboxylase operon) | pH limit for survival lowered from 3.5 to 3.0 | [217] | |
| L. lactis | Overexpression of Lactobacillus casei recO (DNA repair protein) | 22% increased biomass production in medium supplemented with LA, pH 5.0* | [205] | |
| L. lactis | Overexpression of hdeAB (protein chaperones), murG (peptidoglycan biosynthesis), and ldh (lactate dehydrogenase) | pH limit for survival lowered from 4.6 to 4.2 | [218] | |
| S. cerevisiae | ∆fps1 (acquaglyceroporin involved in acetic acid transport) | Ability to grow in medium with up to 6 g/L acetic acid instead of 4.5 g/L | [233] | |
| S. cerevisiae | ∆dse2 (daughter cell-specific secreted protein with similarity to glucanases), ∆scw11 (cell wall protein with similarity to glucanases), ∆eaf3 (involved in gene transcription), ∆sed1 (cell surface glycoprotein) | ≈2-fold shorter lag phase and 40-fold higher biomass production with 60 g/L LA* | [219] | |
| S. cerevisiae | Overexpression of acs2 (acetyl-coenzyme A synthetase) | 2-fold shorter lag phase, 25% increase in growth rate, 5.5 fold higher biomass with 8.4 g/L acetic acid | [234] | |
| RAISE | E. coli | Mutant rpoS (global regulator sigma D factor) | Increased biomass production at pH 4.15 (+ 75%), 3.62 (+ 50%), 3.17 (+15%) | [222] |
| ALE + RME | Clostridium cellulovorans | ∆Clocel_0798, ∆Clocel_2169 (cell wall lyase genes possibly involved in cell autolysis), overexpression of Clostridium beijerinckii augA (agmatine deiminase, agmatine → N-carbamoylputrescine + NH4+) | pH limit for growth lowered from ≈6.0 to 5.5 | [216] |
| CM + ALE | Fibrobacter succinogenes | - | pH limit for growth lowered from 6.10 to 5.65 | [198] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazzoli, R. Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose. Fermentation 2021, 7, 248. https://doi.org/10.3390/fermentation7040248
Mazzoli R. Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose. Fermentation. 2021; 7(4):248. https://doi.org/10.3390/fermentation7040248
Chicago/Turabian StyleMazzoli, Roberto. 2021. "Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose" Fermentation 7, no. 4: 248. https://doi.org/10.3390/fermentation7040248
APA StyleMazzoli, R. (2021). Current Progress in Production of Building-Block Organic Acids by Consolidated Bioprocessing of Lignocellulose. Fermentation, 7(4), 248. https://doi.org/10.3390/fermentation7040248






