Kocuria Strains from Unique Radon Spring Water from Jachymov Spa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Growth at Different Temperatures
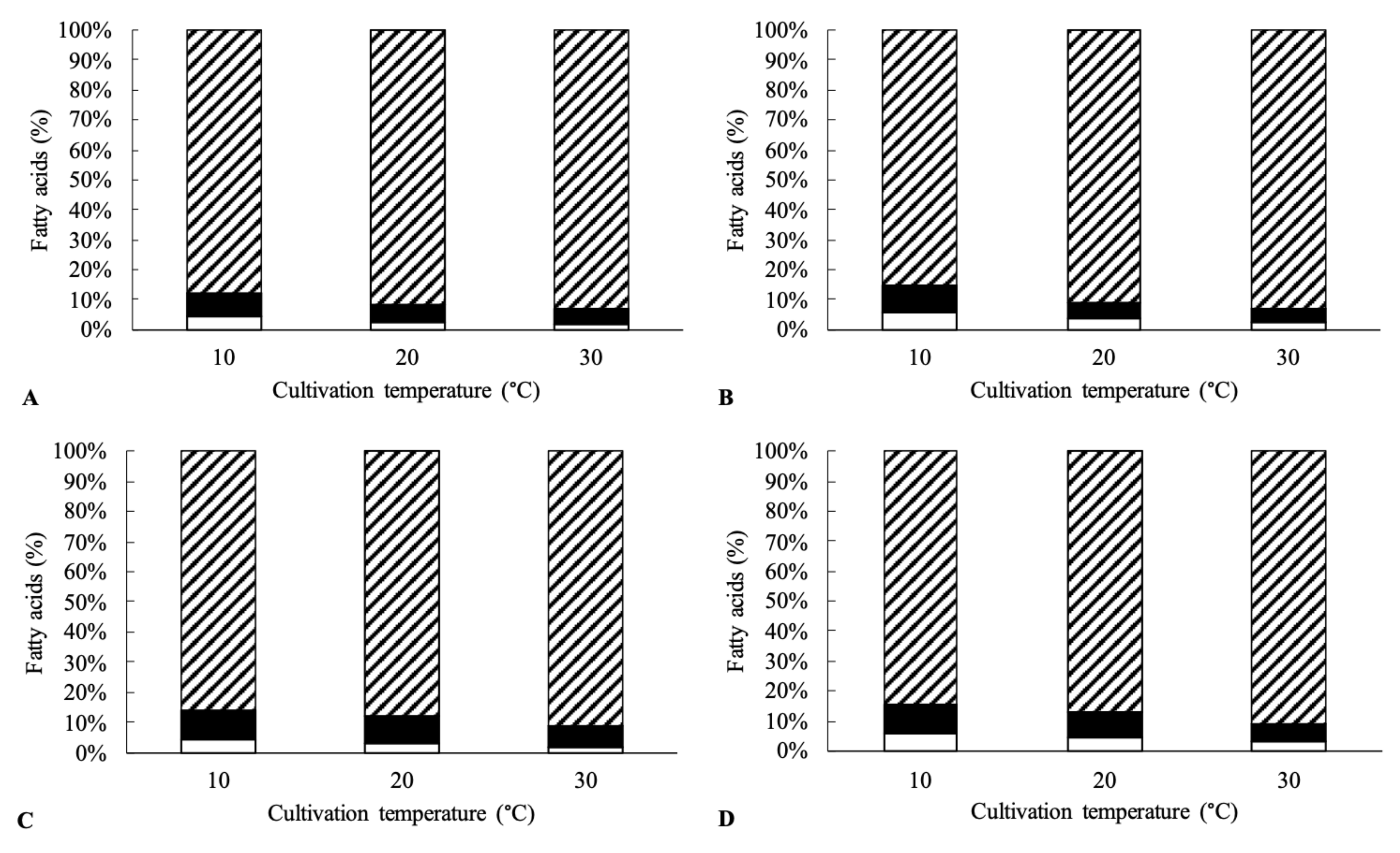
2.3. Isolation of Lipids and Fatty Acid Methyl Ester (FAME) Characterization
2.4. Carbon Source Utilization Characterization
2.5. Determination of Antioxidant Production
2.6. Determination of Siderophore Production
2.7. Determination of UV-C Resistance
2.8. Determination of Tolerance to Hydrogen Peroxide (H2O2)
2.9. Determination of Desiccation Tolerance
2.10. Statistical Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Griebler, C.; Lueders, T. Microbial biodiversity in groundwater ecosystems. Freshw. Biol. 2009, 54, 649–677. [Google Scholar] [CrossRef]
- Krejbichova, Z. Radioactivity of mineral waters in Bohemia. Czech. J. Phys. 1999, 49, 127–132. [Google Scholar] [CrossRef]
- Asgarani, E.; Soudi, M.R.; Borzooee, F.; Dabbagh, R. Radio-resistance in psychrotrophic Kocuria sp ASB 107 isolated from Ab-e-Siah radioactive spring. J. Environ. Radioact. 2012, 113, 171–176. [Google Scholar] [CrossRef]
- Gholami, M.; Etemadifar, Z.; Bouzari, M. Isolation a new strain of Kocuria rosea capable of tolerating extreme conditions. J. Environ. Radioact. 2015, 144, 113–119. [Google Scholar] [CrossRef]
- Bernal, C.; Cairo, J.; Coello, N. Purification and characterization of a novel exocellular keratinase from Kocuria rosea. Enzym. Microb. Technol. 2006, 38, 49–54. [Google Scholar] [CrossRef]
- Goswami, D.; Pithwa, S.; Dhandhukia, P.; Thakker, J.N. Delineating Kocuria turfanensis 2M4 as a credible PGPR: A novel IAA-producing bacteria isolated from saline desert. J. Plant Interact. 2014, 9, 566–576. [Google Scholar] [CrossRef]
- Nesheli, M.A.; Asgarani, E.; Dabbagh, R. Biosorption potential of Cr(VI) by Kocuria sp ASB107, a radio-resistant bacterium isolated from Ramsar, Iran. Chem. Ecol. 2018, 34, 163–176. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Koch, C.; Gvozdiak, O.; Schumann, P. Taxonomic Dissection Of The Genus Micrococcus-Kocuria Gen-Nov, Nesterenkonia Gen-Nov, Kytococcus Gen-Nov, Dermacoccus Gen-Nov, And Micrococcus Cohn 1872 Gen Emend. Int. J. Syst. Bacteriol. 1995, 45, 682–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastager, S.G.; Tang, S.K.; Srinivasan, K.; Lee, J.C.; Li, W.J. Kocuria indica sp nov., isolated from a sediment sample. Int. J. Syst. Evol. Microbiol. 2014, 64, 869–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, M.S.; Wang, E.J.; Zimmermann, S.; Boutin, S.; Wagner, H.; Wink, M. Kocuria tytonicola, new bacteria from the preen glands of American barn owls (Tyto furcata). Syst. Appl. Microbiol. 2019, 42, 198–204. [Google Scholar] [CrossRef]
- Pridmore, D.; Rekhif, N.; Pittet, A.C.; Suri, B.; Mollet, B. Variacin, a new lanthionine-containing bacteriocin produced by Micrococcus varians: Comparison to lacticin 481 of Lactococcus lactis. Appl. Environ. Microbiol. 1996, 62, 1799–1802. [Google Scholar] [CrossRef] [Green Version]
- Sarafin, Y.; Birdilla, M.; Donio, S.; Velmurugan, S.; Michaelbabu, M.; Citarasu, T. Kocuria marina BS-15 a biosurfactant producing halophilic bacteria isolated from solar salt works in India. Saudi J. Biol. Sci. 2014, 21, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Budel, B.; Karsten, U.; GarciaPichel, F. Ultraviolet-absorbing scytonemin and mycosporine-like amino acid derivatives in exposed, rock-inhabiting cyanobacterial lichens. Oecologia 1997, 112, 165–172. [Google Scholar] [CrossRef]
- Shick, J.M.; Dunlap, W.C. Mycosporine-like amino acids and related gadusols: Biosynthesis, accumulation, and UV-protective functions in aquatic organisms. Annu. Rev. Physiol. 2002, 64, 223–262. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, W.C.; Shick, J.M. Ultraviolet radiation-absorbing mycosporine-like amino acids in coral reef organisms: A biochemical and environmental perspective. J. Phycol. 1998, 34, 418–430. [Google Scholar] [CrossRef]
- Trincone, A. Potential biocatalysts originating from sea environments. J. Mol. Catal. B-Enzym. 2010, 66, 241–256. [Google Scholar] [CrossRef]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulzele, R.; DeSa, E.; Yadav, A.; Shouche, Y.; Bhadekar, R. Characterization of Novel Extracellular Protease Produced By Marine Bacterial Isolate from the Indian Ocean. Braz. J. Microbiol. 2011, 42, 1364–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, B.; Xu, Z.J.; Sun, Z.T.; Lin, J.; Hua, Y.J. Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim. Biophys. Acta-Gen. Subj. 2007, 1770, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Tristan, S.; Parra-Saldivar, R.; Iqbal, H.M.N.; Carrillo-Nieves, D. Bioinspired biomolecules: Mycosporine-like amino acids and scytonemin from Lyngbya sp. with UV-protection potentialities. J. Photochem. Photobiol. B-Biol. 2019, 201, 11. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Eichman, C.; Jackson, J.R.; Mattern, M.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharmacol. Exp. Ther. 2002, 303, 858–866. [Google Scholar] [CrossRef]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: Correlation between stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef]
- Haidar, B.; Ferdous, M.; Fatema, B.; Ferdous, A.S.; Islam, M.R.; Khan, H. Population diversity of bacterial endophytes from jute (Corchorus olitorius) and evaluation of their potential role as bioinoculants. Microbiol. Res. 2018, 208, 43–53. [Google Scholar] [CrossRef]
- Retamal-Morales, G.; Mehnert, M.; Schwabe, R.; Tischler, D.; Zapata, C.; Chavez, R.; Schlomann, M.; Levican, G. Detection of arsenic-binding siderophores in arsenic-tolerating Actinobacteria by a modified CAS assay. Ecotoxicol. Environ. Saf. 2018, 157, 176–181. [Google Scholar] [CrossRef]
- Samanta, A.K.; Chaudhuri, S.; Dutta, D. Antioxidant efficacy of carotenoid extract from bacterial strain Kocuria marina DAGII. Mater. Today-Proc. 2016, 3, 3427–3433. [Google Scholar] [CrossRef]
- Mal, S.A.; Ibrahim, G.S.; Al Khalaf, M.I.; Al-Hejin, A.M.; Bataweel, N.M.; Abu-Zaid, M. Production and Partial Characterization of Yellow Pigment Produced by Kocuria flava Isolate and Testing its Antioxidant and Antimicrobial activity. Int. J. Life Sci. Pharma Res. 2020, 10, L58–L66. [Google Scholar] [CrossRef]
- Kumar, C.G.; Sujitha, P. Kocuran, an exopolysaccharide isolated from Kocuria rosea strain BS-1 and evaluation of its in vitro immunosuppression activities. Enzym. Microb. Technol. 2014, 55, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Palomo, S.; Gonzalez, I.; de la Cruz, M.; Martin, J.; Tormo, J.R.; Anderson, M.; Hill, R.T.; Vicente, F.; Reyes, F.; Genilloud, O. Sponge-Derived Kocuria and Micrococcus spp. as Sources of the New Thiazolyl Peptide Antibiotic Kocurin. Mar. Drugs 2013, 11, 1071–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezanka, T.; Gharwalova, L.; Novakova, G.; Kolouchova, I.; Uhlik, O.; Sigler, K. Kocuria Bacterial Isolates from Radioactive Springs of Jachymov spa (Joachimsthal) as Sources of Polyunsaturated Fatty Acids. Lipids 2019, 54, 177–187. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Rezanka, T.; Rozentsvet, O.A. Lipid-Composition of 3 Macrophytes from the Caspian Sea. Phytochemistry 1993, 33, 1015–1019. [Google Scholar] [CrossRef]
- Vancura, A.; Rezanka, T.; Marslalek, J.; Melzoch, K.; Basarova, G.; Kristan, V. Metabolism of L-Threonine and Fatty-Acids and Tylosin Biosynthesis in Streptomyces-Fradiae. FEMS Microbiol. Lett. 1988, 49, 411–415. [Google Scholar] [CrossRef]
- Garland, J.L.; Mills, A.L. Classification and Characterization of Heterotrophic Microbial Communities on the Basis of Patterns of Community-Level Sole-Carbon-Source Utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef] [Green Version]
- Schwyn, B.; Neilands, J.B. Universal Chemical-Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Chick, H. An investigation of the laws of disinfection. J. Hyg. 1908, 8, 92–158. [Google Scholar] [CrossRef] [Green Version]
- Stiefel, P.; Schmidt-Emrich, S.; Maniura-Weber, K.; Ren, Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015, 15, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.B.; Nedashkovskaya, O.I.; Mikhailov, V.V.; Han, S.K.; Kim, K.O.; Rhee, M.S.; Bae, K.S. Kocuria marina sp. nov., a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2004, 54, 1617–1620. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1015. [Google Scholar] [CrossRef] [PubMed]
- Russell, N.J.; Nichols, D.S. Polyunsaturated fatty acids in marine bacteria—A dogma rewritten. Microbiology 1999, 145, 767–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Close, D.; Nelson, W.; Bernhard, W. DNA Damage by the Direct Effect of Ionizing Radiation: Products Produced by Two Sequential One-Electron Oxidations. J. Phys. Chem. A 2013, 117, 12608–12615. [Google Scholar] [CrossRef]
- Pavlopoulou, A.; Savva, G.D.; Louka, M.; Bagos, P.G.; Vorgias, C.E.; Michalopoulos, I.; Georgakilas, A.G. Unraveling the mechanisms of extreme radioresistance in prokaryotes: Lessons from nature. Mutat. Res.-Rev. Mutat. Res. 2016, 767, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Leapman, R.D.; Lai, B.; Ravel, B.; Li, S.M.W.; Kemner, K.M.; et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS. Biol. 2007, 5, 769–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rich, T.; Allen, R.L.; Wyllie, A.H. Defying death after DNA damage. Nature 2000, 407, 777–783. [Google Scholar] [CrossRef]
- Rivas, M.; Rojas, E.; Calaf, G.M.; Barberan, M.; Liberman, C.; Correa, M.D. Association between non-melanoma and melanoma skin cancer rates, vitamin D and latitude. Oncol. Lett. 2017, 13, 3787–3792. [Google Scholar] [CrossRef] [PubMed]
- Gabani, P.; Singh, O.V. Radiation-resistant extremophiles and their potential in biotechnology and therapeutics. Appl. Microbiol. Biotechnol. 2013, 97, 993–1004. [Google Scholar] [CrossRef]
- Ben Ghorbal, S.K.; Chatti, A.; Sethom, M.M.; Maalej, L.; Mihoub, M.; Kefacha, S.; Feki, M.; Landoulsi, A.; Hassen, A. Changes in Membrane Fatty Acid Composition of Pseudomonas aeruginosa in Response to UV-C Radiations. Curr. Microbiol. 2013, 67, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Venkateswaran, A.; Hess, M.; Omelchenko, M.V.; Kostandarithes, H.M.; Makarova, K.S.; et al. Accumulation of Mn(II) in, Deinococcus radiodurans facilitates gamma-radiation resistance. Science 2004, 306, 1025–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cain, T.J.; Smith, A.T. Ferric iron reductases and their contribution to unicellular ferrous iron uptake. J. Inorg. Biochem. 2021, 218, 9. [Google Scholar] [CrossRef]
- Yoshida, K.; Hashimoto, M.; Hori, R.; Adachi, T.; Okuyama, H.; Orikasa, Y.; Nagamine, T.; Shimizu, S.; Ueno, A.; Morita, N. Bacterial Long-Chain Polyunsaturated Fatty Acids: Their Biosynthetic Genes, Functions, and Practical Use. Mar. Drugs 2016, 14, 23. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Li, S.-M.W.; Gaidamakova, E.K.; Matrosova, V.Y.; Zhai, M.; Sulloway, H.M.; Scholten, J.C.; Brown, M.G.; Balkwill, D.L.; Daly, M.J. Protein oxidation: Key to bacterial desiccation resistance? ISME J. 2008, 2, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Wang, L.L.; Chen, Y.J.; Long, Y. Optimization of staining with SYTO 9/propidium iodide: Interplay, kinetics and impact on Brevibacillus brevis. Biotechniques 2020, 69, 89. [Google Scholar] [CrossRef]
- Stocks, S.M. Mechanism and use of the commercially available viability stain, BacLight. Cytom. Part A 2004, 61A, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.; Burghardt, J.; Pradella, S.; Schumann, P.; Stackebrandt, E.; Marialigeti, K. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia). Int. J. Syst. Bacteriol. 1999, 49, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Roman-Ponce, B.; Wang, D.; Vasquez-Murrieta, M.S.; Chen, W.F.; Estrada-de los Santos, P.; Sui, X.H.; Wang, E.T. Kocuria arsenatis sp nov., an arsenic-resistant endophytic actinobacterium associated with Prosopis laegivata grown on high-arsenic-polluted mine tailing. Int. J. Syst. Evol. Microbiol. 2016, 66, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Shibata, C.; Tamura, T.; Nurkanto, A.; Ratnakomala, S.; Lisdiyanti, P.; Suzuki, K. Kocuria pelophila sp nov., an actinobacterium isolated from the rhizosphere of a mangrove. Int. J. Syst. Evol. Microbiol. 2016, 66, 3276–3280. [Google Scholar] [CrossRef]
- Singh, O.V.; Gabani, P. Extremophiles: Radiation resistance microbial reserves and therapeutic implications. J. Appl. Microbiol. 2011, 110, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Grace, K.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. Scytonemin—A marine natural product inhibitor of kinases key in hyperproliferative inflammatory diseases. Inflamm. Res. 2002, 51, 112–114. [Google Scholar] [CrossRef] [PubMed]
- Lemee, L.; Peuchant, E.; Clerc, M.; Brunner, M.; Pfander, H. Deinoxanthin: A new carotenoid isolated from Deinococcus radiodurans. Tetrahedron 1997, 53, 919–926. [Google Scholar] [CrossRef]
- Shahmohammadi, H.R.; Asgarani, E.; Terato, H.; Saito, T.; Ohyama, Y.; Gekko, K.; Yamamoto, O.; Ide, H. Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents. J. Radiat. Res. 1998, 39, 251–262. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; Li, J.L.; Pang, R.J.; Dai, S.; Li, T.; Weng, Y.L.; Jin, Y.; Hua, Y.J. Gold Nanoparticles Biosynthesized and Functionalized Using a Hydroxylated Tetraterpenoid Trigger Gene Expression Changes and Apoptosis in Cancer Cells. ACS Appl. Mater. Interfaces 2018, 10, 37353–37363. [Google Scholar] [CrossRef]
- Santhanam, R.; Rong, X.Y.; Huang, Y.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M. Streptomyces bullii sp nov., isolated from a hyper-arid Atacama Desert soil. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 367–373. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Harrison, W.T.A.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; et al. Diverse Metabolic Profiles of a Streptomyces Strain Isolated from a Hyper-arid Environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Beese, P.; Ohlendorf, B.; Erhard, A.; Zinecker, H.; Dorador, C.; Imhoff, J.F. Abenquines A-D: Aminoquinone derivatives produced by Streptomyces sp strain DB634. J. Antibiot. 2011, 64, 763–768. [Google Scholar] [CrossRef] [PubMed]
 E. coli CCM 4517,
E. coli CCM 4517,  D. radiodurans CCM 1700T,
D. radiodurans CCM 1700T,  isolate 101,
isolate 101,  isolate 208,
isolate 208,  isolate 301,
isolate 301,  isolate 401.
isolate 401.
 E. coli CCM 4517,
E. coli CCM 4517,  D. radiodurans CCM 1700T,
D. radiodurans CCM 1700T,  isolate 101,
isolate 101,  isolate 208,
isolate 208,  isolate 301,
isolate 301,  isolate 401.
isolate 401.



| Temperature (°C) | Kocuria sp. 101 | Kocuria sp. 208 | Kocuria sp. 301 | Kocuria sp. 401 | |
|---|---|---|---|---|---|
| 5 | Specific growth rate (h−1) OD600 max | 0.013 ± 0.005 | - | 0.013 ± 0.002 | - |
| 10 | Specific growth rate (h−1) | 0.021 ± 0.004 | - | 0.020 ± 0.005 | - |
| OD600 max | 13.5 | 10.8 | |||
| 20 | Specific growth rate (h−1) | 0.070 ± 0.010 | 0.068 ± 0.009 | 0.074 ± 0.002 | 0.083 ± 0.006 |
| OD600 max | 18.9 | 15.3 | 15.5 | 12.6 | |
| 30 | Specific growth rate (h−1) | 0.066 ± 0.008 | 0.080 ± 0.012 | 0.072 ± 0.009 | 0.085 ± 0.013 |
| OD600 max | 19.1 | 14.9 | 15.7 | 12.6 | |
| 40 | Specific growth rate (h−1) | 0.042 ± 0.007 | 0.064 ± 0.011 | 0.053 ± 0.002 | 0.061 ± 0.005 |
| OD600 max | 6.8 | 7.5 | 6.0 | 4.9 | |
| 45 | Specific growth rate (h−1) | 0.021 ± 0.006 | 0.031 ± 0.007 | 0.025 ± 0.002 | 0.020 ± 0.008 |
| OD600 max | 6.3 | 6.2 | 5.7 | 4.0 |
| Siderophore Production | Antioxidant Activity (mgAA/L) | C20:4 ω-6 (% of Total Fatty Acid) | C20:5 ω-3 (% of Total Fatty Acid) | |
|---|---|---|---|---|
| Kocuria sp. 101 | + | 4.1 ± 1.3 | 0.31 ± 0.09 | 2.21 ± 0.12 |
| Kocuria sp. 208 | + | 4.2 ± 0.3 | 1.01 ± 0.13 | 2.11 ± 0.17 |
| Kocuria sp. 301 | + | 8.9 ± 0.6 | 1.21 ± 0.17 | 3.71 ± 0.13 |
| Kocuria sp. 401 | + | 5.4 ± 0.4 | 1.11 ± 0.08 | 3.31 ± 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timkina, E.; Drábová, L.; Palyzová, A.; Řezanka, T.; Maťátková, O.; Kolouchová, I. Kocuria Strains from Unique Radon Spring Water from Jachymov Spa. Fermentation 2022, 8, 35. https://doi.org/10.3390/fermentation8010035
Timkina E, Drábová L, Palyzová A, Řezanka T, Maťátková O, Kolouchová I. Kocuria Strains from Unique Radon Spring Water from Jachymov Spa. Fermentation. 2022; 8(1):35. https://doi.org/10.3390/fermentation8010035
Chicago/Turabian StyleTimkina, Elizaveta, Lucie Drábová, Andrea Palyzová, Tomáš Řezanka, Olga Maťátková, and Irena Kolouchová. 2022. "Kocuria Strains from Unique Radon Spring Water from Jachymov Spa" Fermentation 8, no. 1: 35. https://doi.org/10.3390/fermentation8010035
APA StyleTimkina, E., Drábová, L., Palyzová, A., Řezanka, T., Maťátková, O., & Kolouchová, I. (2022). Kocuria Strains from Unique Radon Spring Water from Jachymov Spa. Fermentation, 8(1), 35. https://doi.org/10.3390/fermentation8010035






