Involvement of Cytochrome P450 in Organic-Solvent Tolerant Bacillus subtilis GRSW1-B1 in Vanillin Production via Ferulic Acid Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.1.1. Bacterial Strains
2.1.2. Growth Conditions
2.1.3. Plasmid Construction
2.2. Bioconversion of Ferulic Acid
2.2.1. Bioconversion of Ferulic Acid by B. subtilis GRSW1-B1
2.2.2. Potential Involvement of Bacillus P450s in the Production of Vanillin
2.3. Flask-Scale Two-Step Conversion of Ferulic Acid Using Resting Cells of Recombinant E. coli
2.4. One-Pot Conversion of Ferulic Acid Using Resting Cells of Recombinant E. coli Coexpressing PadC and CypD
2.5. Conversion of 4-Vinylguaiacol Using Resting Cells of Recombinant E. coli
2.6. Analytical Methods
2.6.1. High-Performance Liquid Chromatography
2.6.2. Gas Chromatography-Mass Spectrometry (GC-MS)
3. Results
3.1. Whole-Genome Sequencing of B. subtilis GRSW1-B1
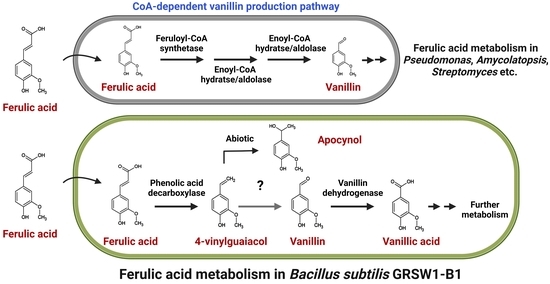
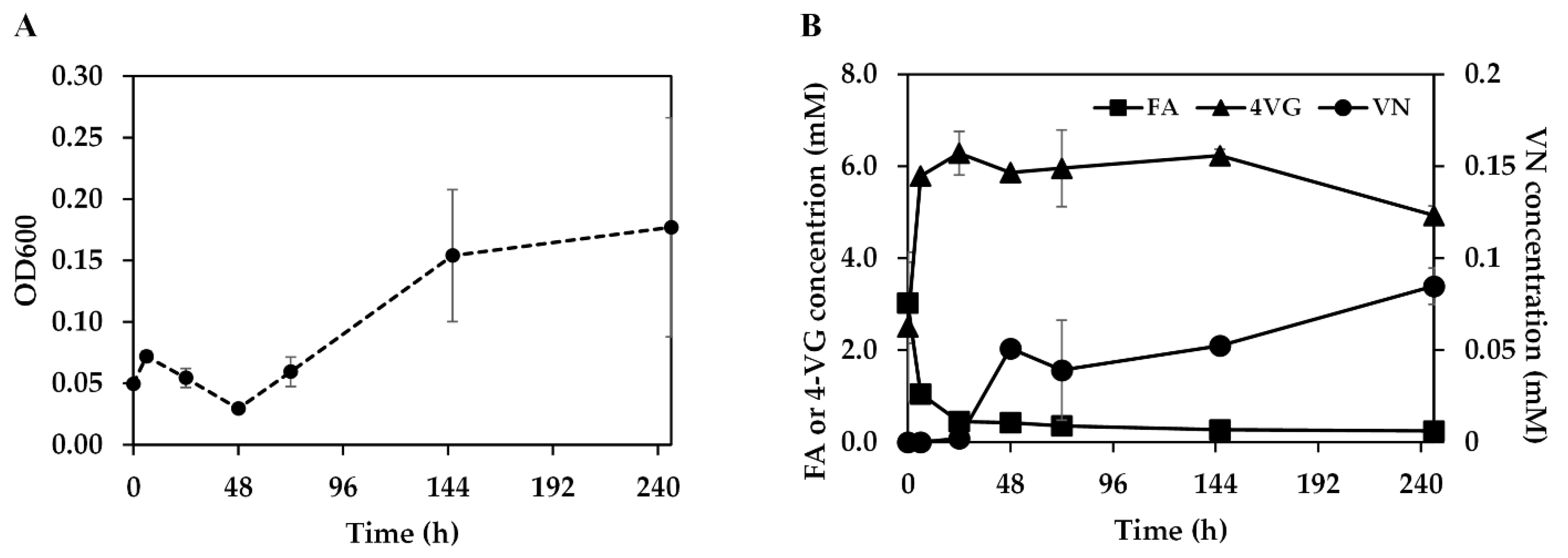
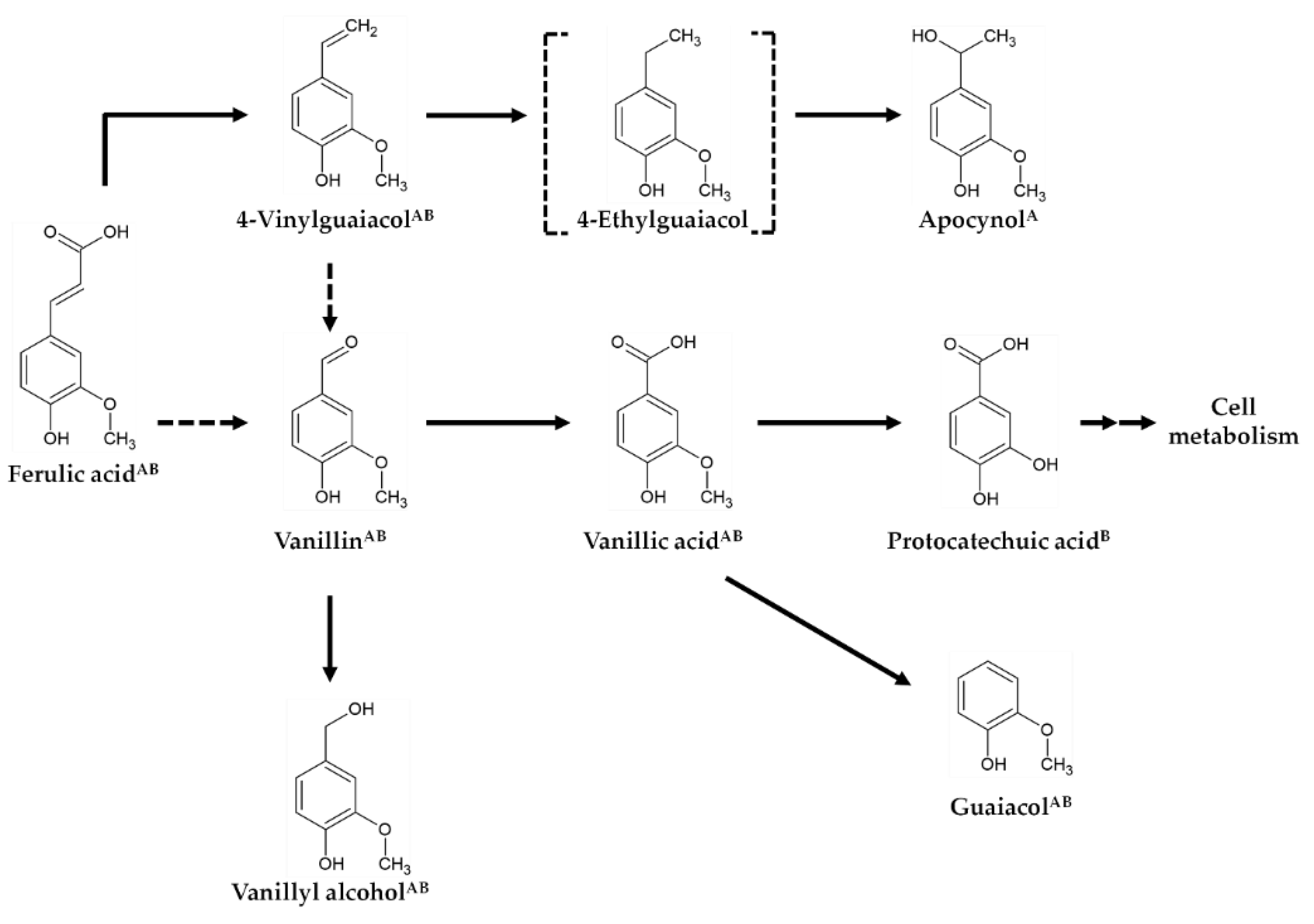
3.2. Ferulic Acid Metabolism by B. subtilis GRSW1-B1
3.3. Potential Involvement of Bacillus CypD in Vanillin Production during Ferulic Acid Metabolism
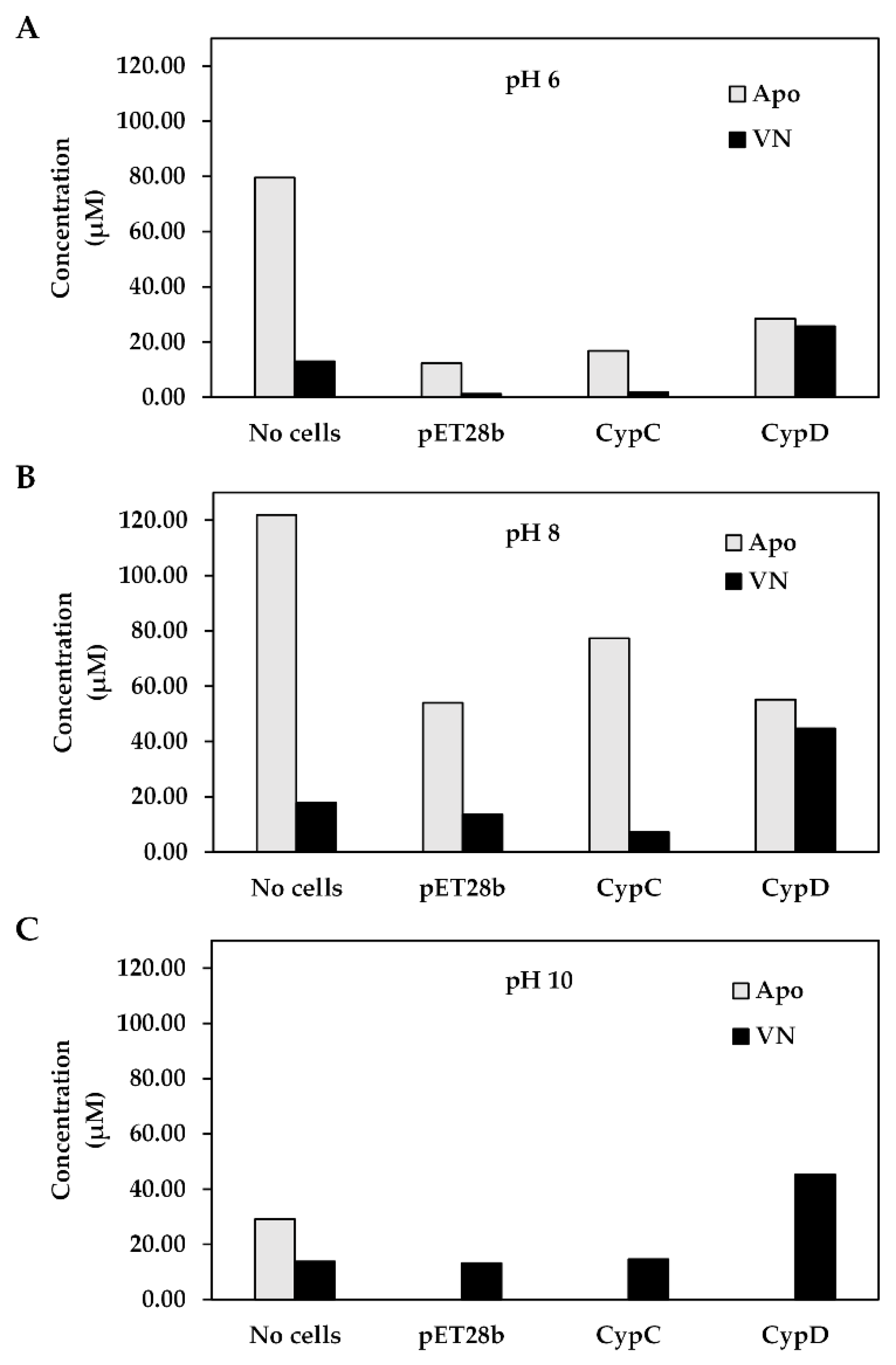
3.4. pH-Dependence of the Two-Step Vanillin Production Using Recombinant E. coli Harboring CypD
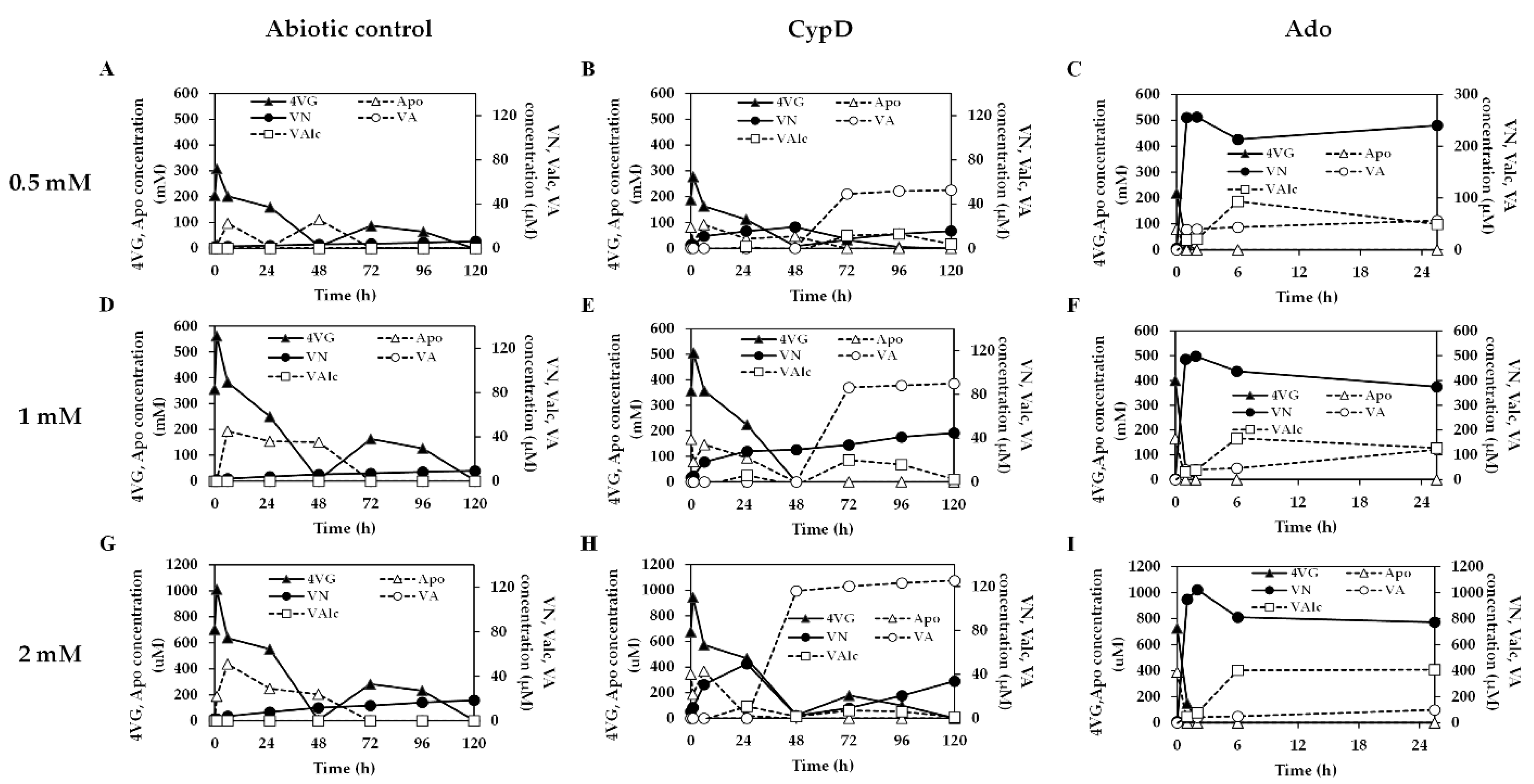
3.5. Conversion of Ferulic Acid to Vanillin via 4-Vinylguaiacol by Recombinant E. coli BL21(DE3) Coexpressing PadC and CypD
3.6. Vanillin Production from 4-Vinylguaiacol by Recombinant E. coli BL21(DE3) Expressing Ado or CypD
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muheim, A.; Lerch, K. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 1999, 51, 456–461. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E. Bioconversions of Ferulic Acid, an Hydroxycinnamic Acid. Crit. Rev. Microbiol. 2006, 32, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Achterholt, S.; Priefert, H.; Steinbüchel, A. Identification of Amycolatopsis sp. strain HR167 genes, involved in the bioconversion of ferulic acid to vanillin. Appl. Microbiol. Biotechnol. 2000, 54, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Plaggenborg, R.; Overhage, J.; Loos, A.; Archer, J.A.C.; Lessard, P.; Sinskey, A.J.; Steinbüchel, A.; Priefert, H. Potential of Rhodococcus strains for biotechnological vanillin production from ferulic acid and eugenol. Appl. Microbiol. Biotechnol. 2006, 72, 745–755. [Google Scholar] [CrossRef]
- Overhage, J.; Priefert, H.; Steinbüchel, A. Biochemical and Genetic Analyses of Ferulic Acid Catabolism in Pseudomonas sp. Strain HR199. Appl. Environ. Microbiol. 1999, 65, 4837–4847. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Li, C.; Kim, J.-E.; Lee, S.-H.; Yoon, J.-Y.; Choi, M.-S.; Seo, W.-T.; Yang, J.-K.; Kim, J.-Y.; Kim, S.-W. Production of Vanillin by Metabolically Engineered Escherichia coli. Biotechnol. Lett. 2005, 27, 1829–1832. [Google Scholar] [CrossRef]
- Barghini, P.; Di Gioia, D.; Fava, F.; Ruzzi, M. Vanillin production using metabolically engineered Escherichia coli under non-growing conditions. Microb. Cell Factories 2007, 6, 13. [Google Scholar] [CrossRef]
- Yang, W.; Tang, H.; Ni, J.; Wu, Q.; Hua, D.; Tao, F.; Xu, P. Characterization of Two Streptomyces Enzymes That Convert Ferulic Acid to Vanillin. PLoS ONE 2013, 8, e67339. [Google Scholar] [CrossRef]
- Chakraborty, D.; Selvam, A.; Kaur, B.; Wong, J.W.C.; Karthikeyan, O.P. Application of recombinant Pediococcus acidilactici BD16 (fcs +/ech +) for bioconversion of agrowaste to vanillin. Appl. Microbiol. Biotechnol. 2017, 101, 5615–5626. [Google Scholar] [CrossRef]
- Jang, S.; Gang, H.; Kim, B.-G.; Choi, K.-Y. FCS and ECH dependent production of phenolic aldehyde and melanin pigment from l-tyrosine in Escherichia coli. Enzym. Microb. Technol. 2018, 112, 59–64. [Google Scholar] [CrossRef]
- Chen, P.; Yan, L.; Wu, Z.; Li, S.; Bai, Z.; Yan, X.; Wang, N.; Liang, N.; Li, H. A microbial transformation using Bacillus subtilis B7-S to produce natural vanillin from ferulic acid. Sci. Rep. 2016, 6, 20400. [Google Scholar] [CrossRef]
- Paz, A.; Outeiriño, D.; de Souza Oliveira, R.P.; Domínguez, J.M. Fed-batch production of vanillin by Bacillus aryabhattai BA03. New Biotechnol. 2018, 40, 186–191. [Google Scholar] [CrossRef]
- Gou, J.; Guo, Y.; Liu, H.; Zhao, Y.; Zhu, R.; Dang, Y.; Liu, N.; Chen, M.; Chen, X. Process optimization of vanillin production by conversion of ferulic acid by Bacillus megaterium. J. Sci. Food Agric 2022, 102, 6047–6061. [Google Scholar] [CrossRef]
- van Dijl, J.M.; Hecker, M. Bacillus subtilis: From soil bacterium to super-secreting cell factory. Microb. Cell Factories 2013, 12, 3. [Google Scholar] [CrossRef]
- Apetroaie-Constantin, C.; Mikkola, R.; Andersson, M.A.; Teplova, V.; Suominen, I.; Johansson, T.; Salkinoja-Salonen, M. Bacillus subtilis and B. mojavensis strains connected to food poisoning produce the heat stable toxin amylosin. J. Appl. Microbiol. 2009, 106, 1976–1985. [Google Scholar] [CrossRef]
- Kataoka, N.; Tajima, T.; Kato, J.; Rachadech, W.; Vangnai, A.S. Development of butanol-tolerant Bacillus subtilis strain GRSW2-B1 as a potential bioproduction host. AMB Express 2011, 1, 10. [Google Scholar] [CrossRef]
- Mahipant, G.; Kato, J.; Kataoka, N.; Vangnai, A.S. An alternative genome-integrated method for undomesticated Bacillus subtilis and related species. J. Gen. Appl. Microbiol. 2019, 65, 96–105. [Google Scholar] [CrossRef]
- Wangrangsimagul, N.; Klinsakul, K.; Vangnai, A.S.; Wongkongkatep, J.; Inprakhon, P.; Honda, K.; Ohtake, H.; Kato, J.; Pongtharangkul, T. Bioproduction of vanillin using an organic solvent-tolerant Brevibacillus agri 13. Appl. Microbiol. Biotechnol. 2012, 93, 555–563. [Google Scholar] [CrossRef]
- Tang, J.; Shi, L.; Li, L.; Long, L.; Ding, S. Expression and characterization of a 9-cis-epoxycarotenoid dioxygenase from Serratia sp. ATCC 39006 capable of biotransforming isoeugenol and 4-vinylguaiacol to vanillin. Biotechnol. Rep. 2018, 18, e00253. [Google Scholar] [CrossRef]
- Angiuoli, S.V.; Gussman, A.; Klimke, W.; Cochrane, G.; Field, D.; Garrity, G.M.; Kodira, C.D.; Kyrpides, N.; Madupu, R.; Markowitz, V.; et al. Toward an Online Repository of Standard Operating Procedures (SOPs) for (Meta) genomic Annotation. OMICS A J. Integr. Biol. 2008, 12, 137–141. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Ikeda-Ohtsubo, W.; Nagata, Y.; Tsuda, M. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinform. 2008, 9, 376. [Google Scholar] [CrossRef]
- Cavin, J.-F.; Dartois, V.; Diviès, C. Gene cloning, transcriptional analysis, purification, and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl. Environ. Microbiol. 1998, 64, 1466–1471. [Google Scholar] [CrossRef]
- Hu, H.; Li, L.; Ding, S. An organic solvent-tolerant phenolic acid decarboxylase from Bacillus licheniformis for the efficient bioconversion of hydroxycinnamic acids to vinyl phenol derivatives. Appl. Microbiol. Biotechnol 2015, 99, 5071–5081. [Google Scholar] [CrossRef]
- Graf, N.; Wenzel, M.; Altenbuchner, J. Identification and characterization of the vanillin dehydrogenase YfmT in Bacillus subtilis 3NA. Appl. Microbiol. Biotechnol. 2016, 100, 3511–3521. [Google Scholar] [CrossRef]
- Furuya, T.; Miura, M.; Kino, K. A Coenzyme-Independent Decarboxylase/Oxygenase Cascade for the Efficient Synthesis of Vanillin. ChemBioChem 2014, 15, 2248–2254. [Google Scholar] [CrossRef]
- Furuya, T.; Miura, M.; Kuroiwa, M.; Kino, K. High-yield production of vanillin from ferulic acid by a coenzyme-independent decarboxylase/oxygenase two-stage process. New Biotechnol. 2015, 32, 335–339. [Google Scholar] [CrossRef]
- Ni, J.; Wu, Y.-T.; Tao, F.; Peng, Y.; Xu, P. A coenzyme-free biocatalyst for the value-added utilization of lignin-derived aromatics. J. Am. Chem. Soc. 2018, 140, 16001–16005. [Google Scholar] [CrossRef]
- Han, Z.; Long, L.; Ding, S. Expression and Characterization of Carotenoid Cleavage Oxygenases From Herbaspirillum seropedicae and Rhodobacteraceae bacterium Capable of Biotransforming Isoeugenol and 4-Vinylguaiacol to Vanillin. Front. Microbiol. 2015, 10, 1869. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, P.; Han, S.; Yan, H.; Ma, C. Metabolism of isoeugenol via isoeugenol-diol by a newly isolated strain of Bacillus subtilis HS8. Appl. Microbiol. Biotechnol. 2006, 73, 771–779. [Google Scholar] [CrossRef]
- Shimoni, E.; Ravid, U.; Shoham, Y. Isolation of a Bacillus sp. capable of transforming isoeugenol to vanillin. J. Biotechnol. 2000, 78, 1–9. [Google Scholar] [CrossRef]
- Hammer, S.C.; Kubik, G.; Watkins, E.; Huang, S.; Minges, H.; Arnold, F.H. Anti-Markovnikov alkene oxidation by metal-oxo-mediated enzyme catalysis. Science 2017, 358, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Fruetel, J.A.; Collins, J.R.; Camper, D.L.; Loew, G.H.; Ortiz de Montellano, P.R. Calculated and experimental absolute stereochemistry of the styrene and beta-methylstyrene epoxides formed by cytochrome P 450cam. J. Am. Chem. Soc. 1992, 114, 6987–6993. [Google Scholar] [CrossRef]
- Sun, C.; Hu, B.; Liu, Z. Efficient and ecofriendly options for the chemoselective oxidation of alkenes using manganese porphyrin and dioxygen. Chem. Eng. J. 2013, 232, 96–103. [Google Scholar] [CrossRef]
- Budde, M.; Maurer, S.C.; Schmid, R.D.; Urlacher, V.B. Cloning, expression and characterisation of CYP102A2, a self-sufficient P450 monooxygenase from Bacillus subtilis. Appl. Microbiol. Biotechnol. 2004, 66, 180–186. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, Q.; Horner, J.H.; Sheng, X.; Newcomb, M. Kinetics and activation parameters for oxidations of styrene by Compounds I from the cytochrome P450(BM-3) (CYP102A1) heme domain and from CYP119. Biochemistry 2009, 48, 9140–9146. [Google Scholar] [CrossRef][Green Version]
- Huang, W.C.; Cullis, P.M.; Raven, E.L.; Roberts, G.C.K. Control of the stereo-selectivity of styreneepoxidation by cytochrome P450 BM3 using structure-based mutagenesis. Metallomics 2011, 3, 410–416. [Google Scholar] [CrossRef]
- Rauch, M.C.R.; Tieves, F.; Paul, C.E.; Arends, I.; Alcalde, M.; Hollmann, F. Peroxygenase-Catalysed Epoxidation of Styrene Derivatives in Neat Reaction Media. ChemCatChem 2019, 11, 4519–4523. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Li, X.; Gu, W.; Huang, J.; Zhang, K.-Q. The metabolism of ferulic acid via 4-vinylguaiacol to vanillin by Enterobacter sp. Px6-4 isolated from Vanilla root. Process Biochem. 2008, 43, 1132–1137. [Google Scholar] [CrossRef]
- Maeda, M.; Tokashiki, M.; Tokashiki, M.; Uechi, K.; Ito, S.; Taira, T. Characterization and induction of phenolic acid decarboxylase from Aspergillus luchuensis. J. Biosci. Bioeng. 2018, 126, 162–168. [Google Scholar] [CrossRef]
- Mukai, N.; Masaki, K.; Fujii, T.; Iefuji, H. Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J. Biosci. Bioeng. 2014, 118, 50–55. [Google Scholar] [CrossRef]
- Vanbeneden, N.; Saison, D.; Delvaux, F.; Delvaux, F.R. Decrease of 4-vinylguaiacol during beer aging and formation of apocynol and vanillin in beer. J. Agric. Food Chem. 2008, 56, 11983–11988. [Google Scholar] [CrossRef]
- Hunter, W.J.; Manter, D.K.; van der Lelie, D. Biotransformation of Ferulic Acid to 4-Vinylguaiacol by Enterobacter soli and E. aerogenes. Curr. Microbiol. 2012, 65, 752–757. [Google Scholar] [CrossRef]
- Baqueiro-Peña, I.; Rodríguez-Serrano, G.; González-Zamora, E.; Augur, C.; Loera, O.; Saucedo-Castañeda, G. Biotransformation of ferulic acid to 4-vinylguaiacol by a wild and a diploid strain of Aspergillus niger. Bioresour. Technol. 2010, 101, 4721–4724. [Google Scholar] [CrossRef]
- Mathew, S.; Abraham, T.E.; Sudheesh, S. Rapid conversion of ferulic acid to 4-vinyl guaiacol and vanillin metabolites by Debaryomyces hansenii. J. Mol. Catal. B Enzym. 2007, 44, 48–52. [Google Scholar] [CrossRef]
- Shanker, K.S.; Kishore, K.H.; Kanjilal, S.; Misra, S.; Narayana Murty, U.S.; Prasad, R.B.N. Biotransformation of ferulic acid to acetovanillone using Rhizopus oryzae. Biocatal. Biotransform. 2007, 25, 109–112. [Google Scholar] [CrossRef]
- Ni, J.; Gao, Y.Y.; Tao, F.; Liu, H.Y.; Xu, P. Temperature-Directed Biocatalysis for the Sustainable Production of Aromatic Aldehydes or Alcohols. Angew. Chem. Int. Ed. 2018, 57, 1214–1217. [Google Scholar] [CrossRef]
- Labuda, I. Biotechnology of Vanillin: Vanillin from Microbial Sources. In Handbook of Vanilla Science and Technology; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 457–488. [Google Scholar]
- Yan, L.; Chen, P.; Zhang, S.; Li, S.; Yan, X.; Wang, N.; Liang, N.; Li, H. Biotransformation of ferulic acid to vanillin in the packed bed-stirred fermentors. Sci. Rep. 2016, 6, 34644. [Google Scholar] [CrossRef]
- Chen, P.; Li, S.; Yan, L.; Wang, N.; Yan, X.; Li, H. Draft Genome Sequence of Bacillus subtilis Type Strain B7-S, Which Converts Ferulic Acid to Vanillin. Genome Announc. 2014, 2, e00025-14. [Google Scholar] [CrossRef]
- Klaus, T.; Seifert, A.; Häbe, T.; Nestl, B.M.; Hauer, B. An Enzyme Cascade Synthesis of Vanillin. Catalysts 2019, 9, 252. [Google Scholar] [CrossRef]
- Nickerson, D.P.; Harford-Cross, C.F.; Fulcher, S.R.; Wong, L.-L. The catalytic activity of cytochrome P450cam towards styrene oxidation is increased by site-specific mutagenesis. FEBS Lett. 1997, 405, 153–156. [Google Scholar] [CrossRef]
- Zhou, Y.; Sekar, B.S.; Wu, S.; Li, Z. Benzoic acid production via cascade biotransformation and coupled fermentation-biotransformation. Biotechnol. Bioeng. 2020, 117, 2340–2350. [Google Scholar] [CrossRef]
- Zhu, D.; Xu, L.; Sethupathy, S.; Si, H.; Ahmad, F.; Zhang, R.; Zhang, W.; Yang, B.; Sun, J.-Z. Decoding lignin valorization pathways in the extremophilic Bacillus ligniniphilus L1 for vanillin biosynthesis. Green Chem. 2021, 23, 9554–9570. [Google Scholar] [CrossRef]


| Bacterial Strains | Details | Source |
|---|---|---|
| B. subtilis GRSW1-B1 | Solvent-tolerant bacterium | [16] |
| E. coli DH5α | ||
| E. coli BL21(DE3) | ||
| E. coli JM109(DE3) | ||
| Plasmids | Details | Source |
| pET28b | Expression vector pBR322 ori, kanamycin r | Novagen |
| pETDuet | Expression vector pBR322 ori, ampicillin r | Novagen |
| pCDFDuet | Expression vector CloDF13 ori; streptomycin r | Novagen |
| pETD-PadC | pETDuet harboring padC from B. subtilis GRSW1-B1 at MCS-2 | This study |
| pET28-CypC | pET28b harboring cypC from B. subtilis GRSW1-B1 | This study |
| pET28-CypD | pET28b harboring CYP102A2 (cypD) from B. subtilis GRSW1-B1 | This study |
| pCDFD-CypD | pCDFDuet harboring CYP102A2 (cypD) from B. subtilis GRSW1-B1 | This study |
| pCDFD-Ado | pCDFDuet harboring synthetic ado | This study |
| cypC-pET28 BamHI F | atgcGGATCCaATGAATGAGCAGATTCCACATG |
| cypC-pET28 XhoI R | atgcCTCGAGTTAACTTTTTCGTCTGATTCCG |
| cypD-pET28 BamHI F | atgcGGATCCaATGAAGGAAACAAGCCCGATTC |
| cypD-pET28 XhoI R | atgcCTCGAGCTATATCCCTGCCCAGACATC |
| padC_NdeI-F | atcgCATATGGAAAACTTTATCGGAAGC |
| padC-AatII-R | atcgGACGTCTTATAATCTTCCCGCGCGAATA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotchaplai, P.; Ninrat, J.; Mahipant, G.; Vangnai, A.S. Involvement of Cytochrome P450 in Organic-Solvent Tolerant Bacillus subtilis GRSW1-B1 in Vanillin Production via Ferulic Acid Metabolism. Fermentation 2022, 8, 508. https://doi.org/10.3390/fermentation8100508
Kotchaplai P, Ninrat J, Mahipant G, Vangnai AS. Involvement of Cytochrome P450 in Organic-Solvent Tolerant Bacillus subtilis GRSW1-B1 in Vanillin Production via Ferulic Acid Metabolism. Fermentation. 2022; 8(10):508. https://doi.org/10.3390/fermentation8100508
Chicago/Turabian StyleKotchaplai, Panaya, Jedsadakorn Ninrat, Gumpanat Mahipant, and Alisa S. Vangnai. 2022. "Involvement of Cytochrome P450 in Organic-Solvent Tolerant Bacillus subtilis GRSW1-B1 in Vanillin Production via Ferulic Acid Metabolism" Fermentation 8, no. 10: 508. https://doi.org/10.3390/fermentation8100508
APA StyleKotchaplai, P., Ninrat, J., Mahipant, G., & Vangnai, A. S. (2022). Involvement of Cytochrome P450 in Organic-Solvent Tolerant Bacillus subtilis GRSW1-B1 in Vanillin Production via Ferulic Acid Metabolism. Fermentation, 8(10), 508. https://doi.org/10.3390/fermentation8100508