Sexually-Driven Combinatorial Diversity in Native Saccharomyces Wine Yeasts
Abstract
1. Introduction
2. Results and Discussion
2.1. Genetic Diversity of Autochthonous S. cerevisiae Wine Yeasts at the Aljarafe Region
2.2. Sexually Driven Diversity of Microsatellite Patterns
2.3. Combinatorial Diversity of Phenotypic Characters of Oenological Interest
3. Materials and Methods
3.1. Strains and Culture Medium
3.2. Determination of Yeast Killer Genotype and Killer Activity
3.3. Microsatellites and Capillary Electrophoresis
3.4. Sporulation and Tetrad Dissection
3.5. Determination of Growth Rate
3.6. Microvinifications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martini, A.; Ciani, M.; Scorzetti, G. Direct Enumeration and Isolation of Wine Yeasts from Grape Surfaces. Am. J. Enol. Vitic. 1996, 47, 435–440. [Google Scholar]
- Fleet, G.H. Yeast Interactions and Wine Flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome Evolution across 1011 Saccharomyces Cerevisiae Isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Csoma, H.; Zakany, N.; Capece, A.; Romano, P.; Sipiczki, M. Biological Diversity of Saccharomyces Yeasts of Spontaneously Fermenting Wines in Four Wine Regions: Comparative Genotypic and Phenotypic Analysis. Int. J. Food Microbiol. 2010, 140, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Maio, S.D.; Genna, G.; Gandolfo, V.; Amore, G.; Ciaccio, M.; Oliva, D. Presence of Candida Zemplinina in Sicilian Musts and Selection of a Strain for Wine Mixed Fermentations. S. Afr. J. Enol. Vitic. 2012, 33, 80–87. [Google Scholar] [CrossRef]
- Legras, J.-L.; Karst, F. Optimisation of Interdelta Analysis for Saccharomyces Cerevisiae Strain Characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Basile, A.; De Pascale, F.; Bianca, F.; Rossi, A.; Frizzarin, M.; De Bernardini, N.; Bosaro, M.; Baldisseri, A.; Antoniali, P.; Lopreiato, R.; et al. Large-Scale Sequencing and Comparative Analysis of Oenological Saccharomyces Cerevisiae Strains Supported by Nanopore Refinement of Key Genomes. Food Microbiol. 2021, 97, 103753. [Google Scholar] [CrossRef]
- Cromie, G.A.; Hyma, K.E.; Ludlow, C.L.; Garmendia-Torres, C.; Gilbert, T.L.; May, P.; Huang, A.A.; Dudley, A.M.; Fay, J.C. Genomic Sequence Diversity and Population Structure of Saccharomyces Cerevisiae Assessed by RAD-Seq. G3 2013, 3, 2163–2171. [Google Scholar] [CrossRef]
- Ayoub, M.-J.; Legras, J.-L.; Abi-Nakhoul, P.; Nguyen, H.-V.; Saliba, R.; Gaillardin, C. Lebanon’s Native Oenological Saccharomyces Cerevisiae Flora: Assessment of Different Aspects of Genetic Diversity and Evaluation of Winemaking Potential. J. Fungi 2021, 7, 678. [Google Scholar] [CrossRef]
- Csoma, H.; Kállai, Z.; Antunovics, Z.; Czentye, K.; Sipiczki, M. Vinification without Saccharomyces: Interacting Osmotolerant and “Spoilage” Yeast Communities in Fermenting and Ageing Botrytised High-Sugar Wines (Tokaj Essence). Microorganisms 2020, 9, 19. [Google Scholar] [CrossRef]
- Raymond Eder, M.L.; Rosa, A.L. Non-Tandem Repeat Polymorphisms at Microsatellite Loci in Wine Yeast Species. Mol. Genet. Genom. 2020, 295, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Zabukovec, P.; Čadež, N.; Čuš, F. Isolation and Identification of Indigenous Wine Yeasts and Their Use in Alcoholic Fermentation. Food Technol. Biotechnol. 2020, 58, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Masneuf-Pomarede, I.; Bely, M.; Marullo, P.; Albertin, W. The Genetics of Non-Conventional Wine Yeasts: Current Knowledge and Future Challenges. Front. Microbiol. 2015, 6, 1563. [Google Scholar] [CrossRef] [PubMed]
- Vaudano, E.; Garcia-Moruno, E. Discrimination of Saccharomyces Cerevisiae Wine Strains Using Microsatellite Multiplex PCR and Band Pattern Analysis. Food Microbiol. 2008, 25, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Heard, G.M.; Fleet, G.H. Occurrence and Growth of Killer Yeasts during Wine Fermentation. Appl. Environ. Microbiol. 1987, 53, 2171–2174. [Google Scholar] [CrossRef] [PubMed]
- Pérez, F.; Ramírez, M.; Regodón, J.A. Influence of Killer Strains of Saccharomyces Cerevisiae on Wine Fermentation. Antonie Van Leeuwenhoek 2001, 79, 393–399. [Google Scholar] [CrossRef]
- Petering, J.E.; Symons, M.R.; Langridge, P.; Henschke, P.A. Determination of Killer Yeast Activity in Fermenting Grape Juice by Using a Marked Saccharomyces Wine Yeast Strain. Appl. Environ. Microbiol. 1991, 57, 3232–3236. [Google Scholar] [CrossRef]
- Quintero-Blanco, J.; Jimenez, J.; Garzón, A. A Simple Multiplex Reverse Transcription-PCR Method for the Diagnosis of L-A and M Totiviruses in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2022, 88, e0221321. [Google Scholar] [CrossRef]
- Fukuda, N. Crossbreeding of Yeasts Domesticated for Fermentation: Infertility Challenges. Int. J. Mol. Sci. 2020, 21, 7985. [Google Scholar] [CrossRef]
- Romano, P.; Soli, M.G.; Suzzi, G.; Zambonelli, C. Physiological Characteristics of Single Spore Cultures of Wine Yeasts for Industrial Purposes. Annali Di Microbiol. Ed Enzimol. 1988, 38, 123–129. [Google Scholar]
- Mortimer, R.K.; Romano, P.; Suzzi, G.; Polsinelli, M. Genome Renewal: A New Phenomenon Revealed from a Genetic Study of 43 Strains of Saccharomyces Cerevisiae Derived from Natural Fermentation of Grape Musts. Yeast 1994, 10, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Butler, G. Mating-Type Switching in Budding Yeasts, from Flip/Flop Inversion to Cassette Mechanisms. Microbiol. Mol. Biol. Rev. 2022, 86, e0000721. [Google Scholar] [CrossRef] [PubMed]
- Töpfer, R.; Trapp, O. A Cool Climate Perspective on Grapevine Breeding: Climate Change and Sustainability Are Driving Forces for Changing Varieties in a Traditional Market. Theor. Appl. Genet. 2022. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Wills, C. Abundant Microsatellite Polymorphism in Saccharomyces Cerevisiae, and the Different Distributions of Microsatellites in Eight Prokaryotes and S. Cerevisiae, Result from Strong Mutation Pressures and a Variety of Selective Forces. Proc. Natl. Acad. Sci. USA 1998, 95, 1647–1652. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Umek, L.; Zupan, B.; Schuller, D. Computational Approaches for the Genetic and Phenotypic Characterization of a Saccharomyces Cerevisiae Wine Yeast Collection. Yeast 2009, 26, 675–692. [Google Scholar] [CrossRef]
- Povhe Jemec, K.; Cadez, N.; Zagorc, T.; Bubic, V.; Zupec, A.; Raspor, P. Yeast Population Dynamics in Five Spontaneous Fermentations of Malvasia Must. Food Microbiol. 2001, 18, 247–259. [Google Scholar] [CrossRef]
- Santamaría, P.; Garijo, P.; López, R.; Tenorio, C.; Rosa Gutiérrez, A. Analysis of Yeast Population during Spontaneous Alcoholic Fermentation: Effect of the Age of the Cellar and the Practice of Inoculation. Int. J. Food Microbiol. 2005, 103, 49–56. [Google Scholar] [CrossRef]
- Torija, M.J.; Rozès, N.; Poblet, M.; Guillamón, J.M.; Mas, A. Yeast Population Dynamics in Spontaneous Fermentations: Comparison between Two Different Wine-Producing Areas over a Period of Three Years. Antonie Van Leeuwenhoek 2001, 79, 345–352. [Google Scholar] [CrossRef]
- Blanco, P.; Ramilo, A.; Cerdeira, M.; Orriols, I. Genetic Diversity of Wine Saccharomyces Cerevisiae Strains in an Experimental Winery from Galicia (NW Spain). Antonie Van Leeuwenhoek 2006, 89, 351–357. [Google Scholar] [CrossRef]
- Guillamón, J.M.; Cano, J.; Ramón, D.; Guarro, J. Molecular Differentiation of Keratinomyces (Trichophyton) Species. Antonie Van Leeuwenhoek 1996, 69, 223–227. [Google Scholar] [CrossRef]
- Martínez, C.; Gac, S.; Lavín, A.; Ganga, M. Genomic Characterization of Saccharomyces Cerevisiae Strains Isolated from Wine-Producing Areas in South America. J. Appl. Microbiol. 2004, 96, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Schuller, D.; Casal, M. The Genetic Structure of Fermentative Vineyard-Associated Saccharomyces Cerevisiae Populations Revealed by Microsatellite Analysis. Antonie Van Leeuwenhoek 2007, 91, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Querol, A.; Barrio, E.; Ramón, D. Population Dynamics of Natural Saccharomyces Strains during Wine Fermentation. Int. J. Food Microbiol. 1994, 21, 315–323. [Google Scholar] [CrossRef]
- Agnolucci, M.; Scarano, S.; Santoro, S.; Sassano, C.; Toffanin, A.; Nuti, M. Genetic and Phenotypic Diversity of Autochthonous Saccharomyces Spp. Strains Associated to Natural Fermentation of “Malvasia Delle Lipari”. Lett. Appl. Microbiol. 2007, 45, 657–662. [Google Scholar] [CrossRef]
- Versavaud, A.; Courcoux, P.; Roulland, C.; Dulau, L.; Hallet, J.N. Genetic Diversity and Geographical Distribution of Wild Saccharomyces Cerevisiae Strains from the Wine-Producing Area of Charentes, France. Appl. Environ. Microbiol. 1995, 61, 3521–3529. [Google Scholar] [CrossRef]
- Boynton, P.J. The Ecology of Killer Yeasts: Interference Competition in Natural Habitats. Yeast 2019, 36, 473–485. [Google Scholar] [CrossRef]
- Gutiérrez, A.R.; Epifanio, S.; Garijo, P.; López, R.; Santamaría, P. Killer Yeasts: Incidence in the Ecology of Spontaneous Fermentation. Am. J. Enol. Vitic. 2001, 52, 352–356. [Google Scholar]
- Maqueda, M.; Zamora, E.; Álvarez, M.L.; Ramírez, M. Characterization, Ecological Distribution, and Population Dynamics of Saccharomyces Sensu Stricto Killer Yeasts in the Spontaneous Grape Must Fermentations of Southwestern Spain. Appl. Environ. Microbiol. 2012, 78, 735–743. [Google Scholar] [CrossRef]
- Schmitt, M.J.; Breinig, F. Yeast Viral Killer Toxins: Lethality and Self-Protection. Nat. Rev. Microbiol. 2006, 4, 212–221. [Google Scholar] [CrossRef]
- Vuuren, H.J.J.V.; Jacobs, C.J. Killer Yeasts in the Wine Industry: A Review. Am. J. Enol. Vitic. 1992, 43, 119–128. [Google Scholar]
- Buskirk, S.W.; Rokes, A.B.; Lang, G.I. Adaptive Evolution of Nontransitive Fitness in Yeast. eLife 2020, 9, e62238. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Cousiño, N.; Maqueda, M.; Ambrona, J.; Zamora, E.; Esteban, R.; Ramírez, M. A New Wine Saccharomyces Cerevisiae Killer Toxin (Klus), Encoded by a Double-Stranded Rna Virus, with Broad Antifungal Activity Is Evolutionarily Related to a Chromosomal Host Gene. Appl. Environ. Microbiol. 2011, 77, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Fernández-González, M.; Úbeda, J.F.; Briones, A.I. Study of Saccharomyces Cerevisiae Wine Strains for Breeding through Fermentation Efficiency and Tetrad Analysis. Curr. Microbiol. 2015, 70, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Bakalinsky, A.T.; Snow, R. The Chromosomal Constitution of Wine Strains of Saccharomyces cerevisiae. Yeast 1990, 6, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Guijo, S.; Mauricio, J.C.; Salmon, J.M.; Ortega, J.M. Determination of the Relative Ploidy in Different Saccharomyces Cerevisiae Strains Used for Fermentation and “flor” Film Ageing of Dry Sherry-Type Wines. Yeast 1997, 13, 101–117. [Google Scholar] [CrossRef]
- Gallone, B.; Steensels, J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; Roncoroni, M.; Voordeckers, K.; Miraglia, L.; et al. Domestication and Divergence of Saccharomyces Cerevisiae Beer Yeasts. Cell 2016, 166, 1397–1410.e16. [Google Scholar] [CrossRef]
- Sipiczki, M.; Romano, P.; Capece, A.; Paraggio, M. Genetic Segregation of Natural Saccharomyces Cerevisiae Strains Derived from Spontaneous Fermentation of Aglianico Wine. J. Appl. Microbiol. 2004, 96, 1169–1175. [Google Scholar] [CrossRef]
- Timberlake, W.E.; Frizzell, M.A.; Richards, K.D.; Gardner, R.C. A New Yeast Genetic Resource for Analysis and Breeding. Yeast 2011, 28, 63–80. [Google Scholar] [CrossRef]
- Jiménez, J.; Benítez, T. Genetic Analysis of Highly Ethanol-Tolerant Wine Yeasts. Curr. Genet. 1987, 12, 421–428. [Google Scholar] [CrossRef]
- Koufopanou, V.; Hughes, J.; Bell, G.; Burt, A. The Spatial Scale of Genetic Differentiation in a Model Organism: The Wild Yeast Saccharomyces Paradoxus. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1941–1946. [Google Scholar] [CrossRef][Green Version]
- Kuehne, H.A.; Murphy, H.A.; Francis, C.A.; Sniegowski, P.D. Allopatric Divergence, Secondary Contact, and Genetic Isolation in Wild Yeast Populations. Curr. Biol. 2007, 17, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Magwene, P.M.; Kayıkçı, Ö.; Granek, J.A.; Reininga, J.M.; Scholl, Z.; Murray, D. Outcrossing, Mitotic Recombination, and Life-History Trade-Offs Shape Genome Evolution in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2011, 108, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Shields, D.C. Molecular Evidence for an Ancient Duplication of the Entire Yeast Genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Piskur, J.; Rozpedowska, E.; Polakova, S.; Merico, A.; Compagno, C. How Did Saccharomyces Evolve to Become a Good Brewer? Trends Genet. 2006, 22, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Piskur, J.; Langkjaer, R.B. Yeast Genome Sequencing: The Power of Comparative Genomics. Mol. Microbiol. 2004, 53, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Wolfe, K.H. Increased Glycolytic Flux as an Outcome of Whole-Genome Duplication in Yeast. Mol. Syst. Biol. 2007, 3, 129. [Google Scholar] [CrossRef]
- Hu, X.H.; Wang, M.H.; Tan, T.; Li, J.R.; Yang, H.; Leach, L.; Zhang, R.M.; Luo, Z.W. Genetic Dissection of Ethanol Tolerance in the Budding Yeast Saccharomyces cerevisiae. Genetics 2007, 175, 1479–1487. [Google Scholar] [CrossRef]
- Ibeas, J.I.; Jimenez, J. Mitochondrial DNA Loss Caused by Ethanol in Saccharomyces Flor Yeasts. Appl. Environ. Microbiol. 1997, 63, 7–12. [Google Scholar] [CrossRef]
- Birch, R.M.; Walker, G.M. Influence of Magnesium Ions on Heat Shock and Ethanol Stress Responses of Saccharomyces cerevisiae. Enzyme. Microb. Technol. 2000, 26, 678–687. [Google Scholar] [CrossRef]
- Aguilera, A.; Benítez, T. Role of Mitochondria in Ethanol Tolerance of Saccharomyces cerevisiae. Arch. Microbiol. 1985, 142, 389–392. [Google Scholar] [CrossRef]
- Jimenez, J.; Oballe, J. Ethanol-Hypersensitive and Ethanol-Dependent Cdc-Mutants in Schizosaccharomyces Pombe. Mol. Gen. Genet. 1994, 245, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Attfield, P.V. Stress Tolerance: The Key to Effective Strains of Industrial Baker’s Yeast. Nat. Biotechnol. 1997, 15, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Martí, E.; Zuzuarregui, A.; Gomar-Alba, M.; Gutiérrez, D.; Gil, C.; del Olmo, M. Molecular Response of Saccharomyces Cerevisiae Wine and Laboratory Strains to High Sugar Stress Conditions. Int. J. Food Microbiol. 2011, 145, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Mosedale, J.R.; Abernethy, K.E.; Smart, R.E.; Wilson, R.J.; Maclean, I.M.D. Climate Change Impacts and Adaptive Strategies: Lessons from the Grapevine. Glob. Chang. Biol. 2016, 22, 3814–3828. [Google Scholar] [CrossRef]
- Goddard, M.; Brakjovich, M.; Jun, C.; Sergeant, K. The Effect of Temperature on Yeast Species Composition during Ferment. Aust. N. Z. Grapegrow. Winemak. 2008, 533, 88–92. [Google Scholar]
- Williams, K.M.; Liu, P.; Fay, J.C. Evolution of Ecological Dominance of Yeast Species in High-Sugar Environments. Evolution 2015, 69, 2079–2093. [Google Scholar] [CrossRef]
- Alonso-Del-Real, J.; Contreras-Ruiz, A.; Castiglioni, G.L.; Barrio, E.; Querol, A. The Use of Mixed Populations of Saccharomyces Cerevisiae and S. Kudriavzevii to Reduce Ethanol Content in Wine: Limited Aeration, Inoculum Proportions, and Sequential Inoculation. Front. Microbiol. 2017, 8, 2087. [Google Scholar] [CrossRef]
- Henriques, D.; Alonso-Del-Real, J.; Querol, A.; Balsa-Canto, E. Saccharomyces Cerevisiae and S. Kudriavzevii Synthetic Wine Fermentation Performance Dissected by Predictive Modeling. Front. Microbiol. 2018, 9, 88. [Google Scholar] [CrossRef]
- Ruderfer, D.M.; Pratt, S.C.; Seidel, H.S.; Kruglyak, L. Population Genomic Analysis of Outcrossing and Recombination in Yeast. Nat. Genet. 2006, 38, 1077–1081. [Google Scholar] [CrossRef]
- Winston, F.; Dollard, C.; Ricupero-Hovasse, S.L. Construction of a Set of Convenient Saccharomyces Cerevisiae Strains That Are Isogenic to S288C. Yeast 1995, 11, 53–55. [Google Scholar] [CrossRef]
- Morin, A.; Moores, A.W.; Sacher, M. Dissection of Saccharomyces Cerevisiae Asci. J. Vis. Exp. 2009, 27, e1146. [Google Scholar]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth Rates Made Easy. Mol. Biol. Evol. 2014, 31, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Cumming, G.; Fidler, F.; Vaux, D.L. Error Bars in Experimental Biology. J. Cell Biol. 2007, 177, 7–11. [Google Scholar] [CrossRef] [PubMed]
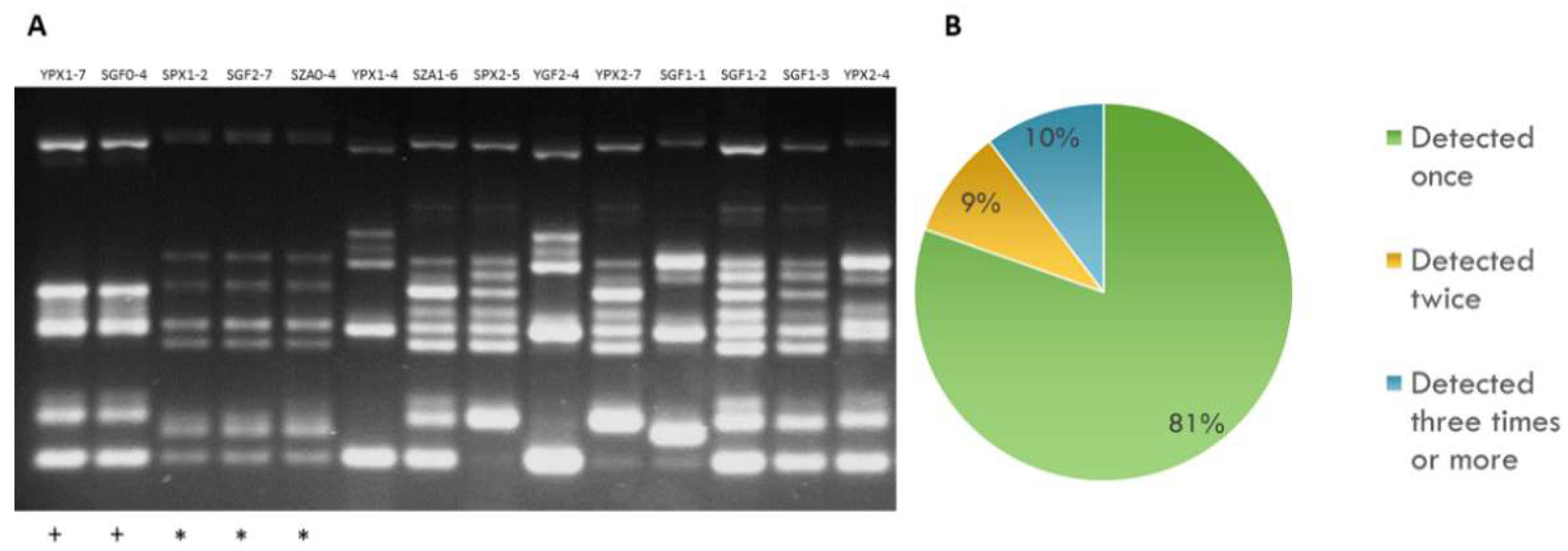
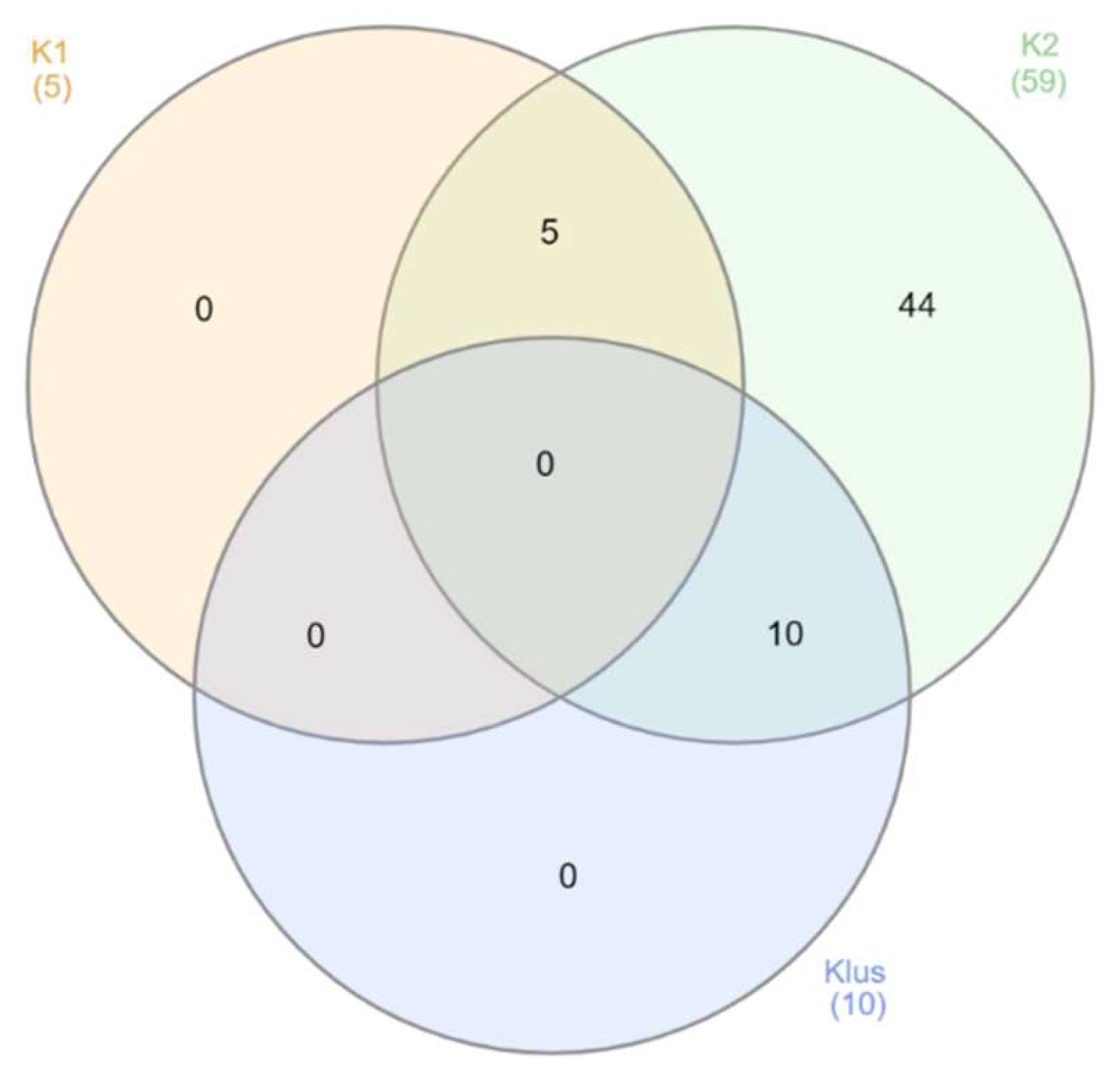
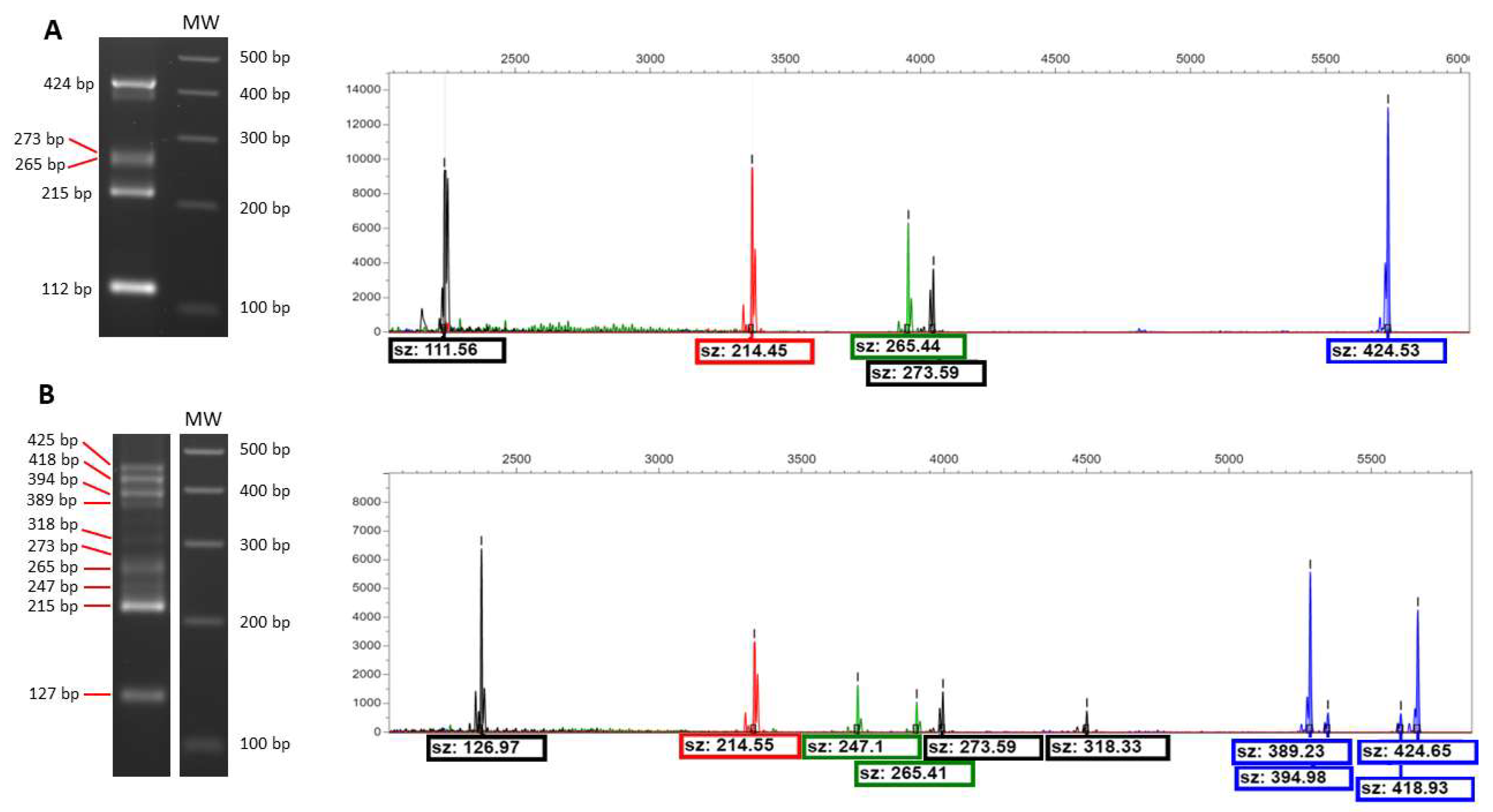

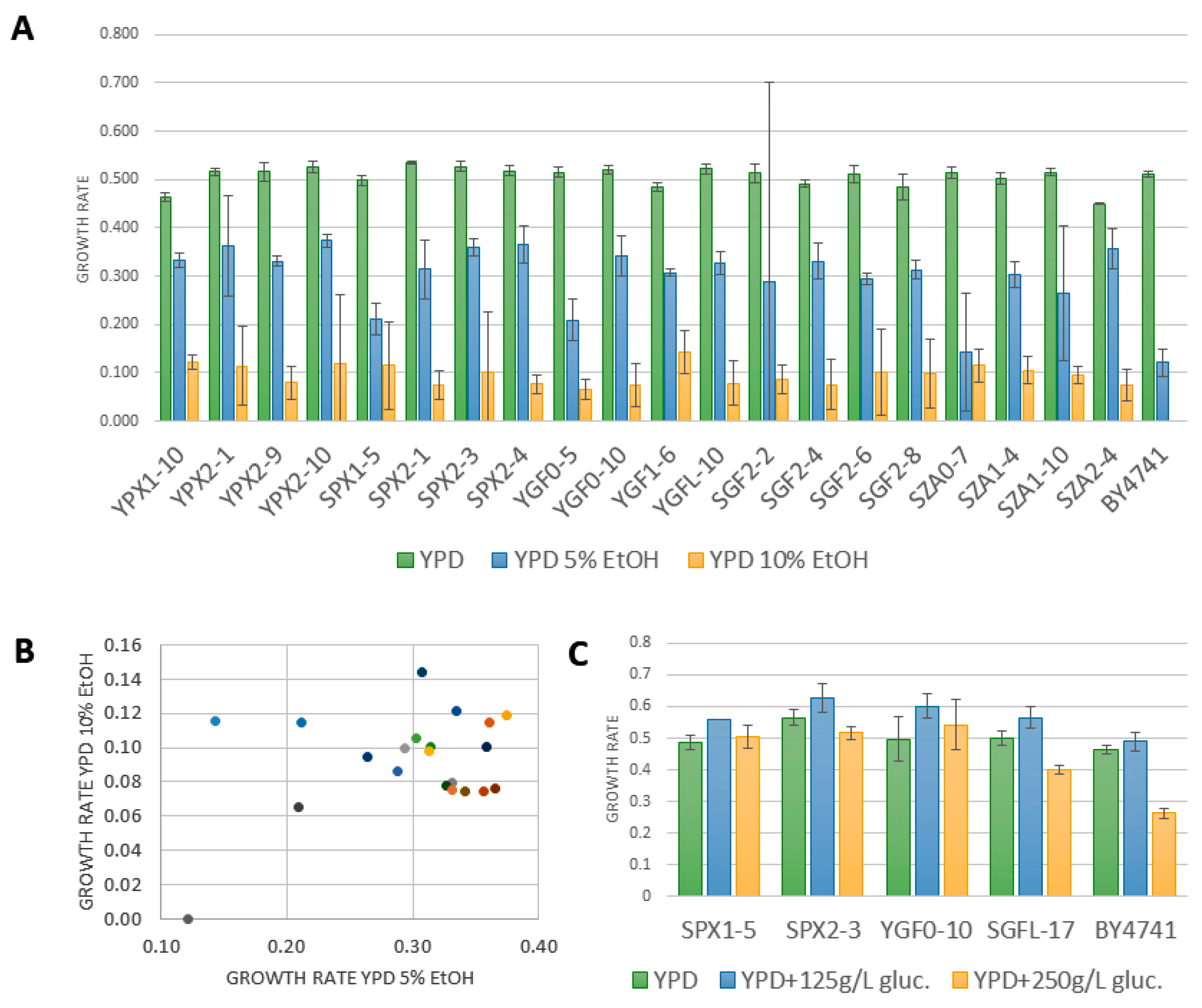
| YPX | SPX | YGF | SGF | SZA | |
|---|---|---|---|---|---|
| T0 | 2/10 | 2/10 | 1/10 | 1/10 | 5/10 |
| T1 | 4/10 | 5/10 | 1/10 | 5/10 | 1/10 |
| T2 | 4/10 | 4/10 | 1/10 | 1/10 | 3/10 |
| Overall different strains | 26/30 | 24/30 | 11/30 | 18/30 | 25/30 |
| Killer yeasts/total different strains | 9/26 | 7/24 | 2/11 | 5/18 | 9/25 |
| Strain | 4:0 Tetrads | 3:1 Tetrads | 2:2 Tetrads | 1:3 Tetrads | Sexual Cycle | Killer Factor | |
|---|---|---|---|---|---|---|---|
| Simple patterns | YPX2-1 | 8 | 0 | 0 | 0 | Homothallic | + |
| YPX2-9 | 0 | 0 | 4 | 4 | Heterozygous | - | |
| SPX2-4 | 8 | 0 | 0 | 0 | Homothallic | + | |
| SPX1-5 | 7 | 1 | 0 | 0 | Homothallic | - | |
| YGF0-5 | 7 | 1 | 0 | 0 | Homothallic | + | |
| YGF0-10 | 8 | 0 | 0 | 0 | Homothallic | + | |
| SGF2-8 | 7 | 1 | 0 | 0 | Homothallic | - | |
| SGF2-4 | 7 | 1 | 0 | 0 | Homothallic | + | |
| SZA0-7 | 5 | 3 | 0 | 0 | Homothallic | + | |
| SZA1-10 | 8 | 0 | 0 | 0 | Homothallic | - | |
| Complex patterns | YPX1-10 | 7 | 1 | 0 | 0 | Homothallic | + |
| YPX2-10 | 2 | 3 | 3 | 0 | Heterothallic | - | |
| SPX2-1 | 7 | 1 | 0 | 0 | Homothallic | + | |
| SPX2-3 | 7 | 1 | 0 | 0 | Homothallic | - | |
| YGF1-6 | 8 | 0 | 0 | 0 | Homothallic | + | |
| YGF2-10 | 0 | 0 | 0 | 0 | Non-sporulating | - | |
| SGF2-2 | 0 | 0 | 0 | 0 | Non-sporulating | - | |
| SGF2-6 | 7 | 1 | 0 | 0 | Heterozygous | + | |
| SZA1-4 | 0 | 2 | 6 | 0 | Heterozygous | + | |
| SZA2-4 | 0 | 0 | 7 | 1 | Heterothallic | - |
| Name | Sequence | Locus | Fluorochrome |
|---|---|---|---|
| SCY F | GGTGACTCTAACGGCAGAGTGG | SCYOR267C | 6-FAM |
| SCY R | GGATCTACTTGCAGTATACGGG | ||
| SCP F | CCCTTTTAAGGAAGAGCAAGCC | SCPTSY7 | NED |
| SCP R | CCACTCTCAGCTTATTGGGG | ||
| SC8 F | CTGCTCAACTTGTGATGGGTTTTGG | SC8132X | VIC |
| SC8 R | CCTCGTTACTATCGTCTTCATCTTGC | ||
| SCAAT F | TGGGAGGAGGGAAATGGACAG | ScAAT3 | PET |
| SCAAT R | TTCAGTTACCCGCACAATCTA | ||
| C5 F | TGACACAATAGCAATGGCCTTCA | C5 | NED |
| C5 R | GCAAGCGACTAGAACAACAATCACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintero-Blanco, J.; Delodi, E.; Garzón, A.; Jimenez, J. Sexually-Driven Combinatorial Diversity in Native Saccharomyces Wine Yeasts. Fermentation 2022, 8, 569. https://doi.org/10.3390/fermentation8100569
Quintero-Blanco J, Delodi E, Garzón A, Jimenez J. Sexually-Driven Combinatorial Diversity in Native Saccharomyces Wine Yeasts. Fermentation. 2022; 8(10):569. https://doi.org/10.3390/fermentation8100569
Chicago/Turabian StyleQuintero-Blanco, Juan, Eugenia Delodi, Andrés Garzón, and Juan Jimenez. 2022. "Sexually-Driven Combinatorial Diversity in Native Saccharomyces Wine Yeasts" Fermentation 8, no. 10: 569. https://doi.org/10.3390/fermentation8100569
APA StyleQuintero-Blanco, J., Delodi, E., Garzón, A., & Jimenez, J. (2022). Sexually-Driven Combinatorial Diversity in Native Saccharomyces Wine Yeasts. Fermentation, 8(10), 569. https://doi.org/10.3390/fermentation8100569





