Temperature and pH Profiling of Extracellular Amylase from Antarctic and Arctic Soil Microfungi
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. Crude Enzyme Production
2.3. Enzyme Activity Assessment
2.4. Partial Purification
2.5. Enzyme Activity and Temperature and pH Optimization
2.6. Visualization of Partially Purified α-Amylases
3. Results
3.1. Specific Activity of Crude Amylases
3.2. Specific Activity of the Partially Purified Enzymes
3.3. Temperature and pH Optimization
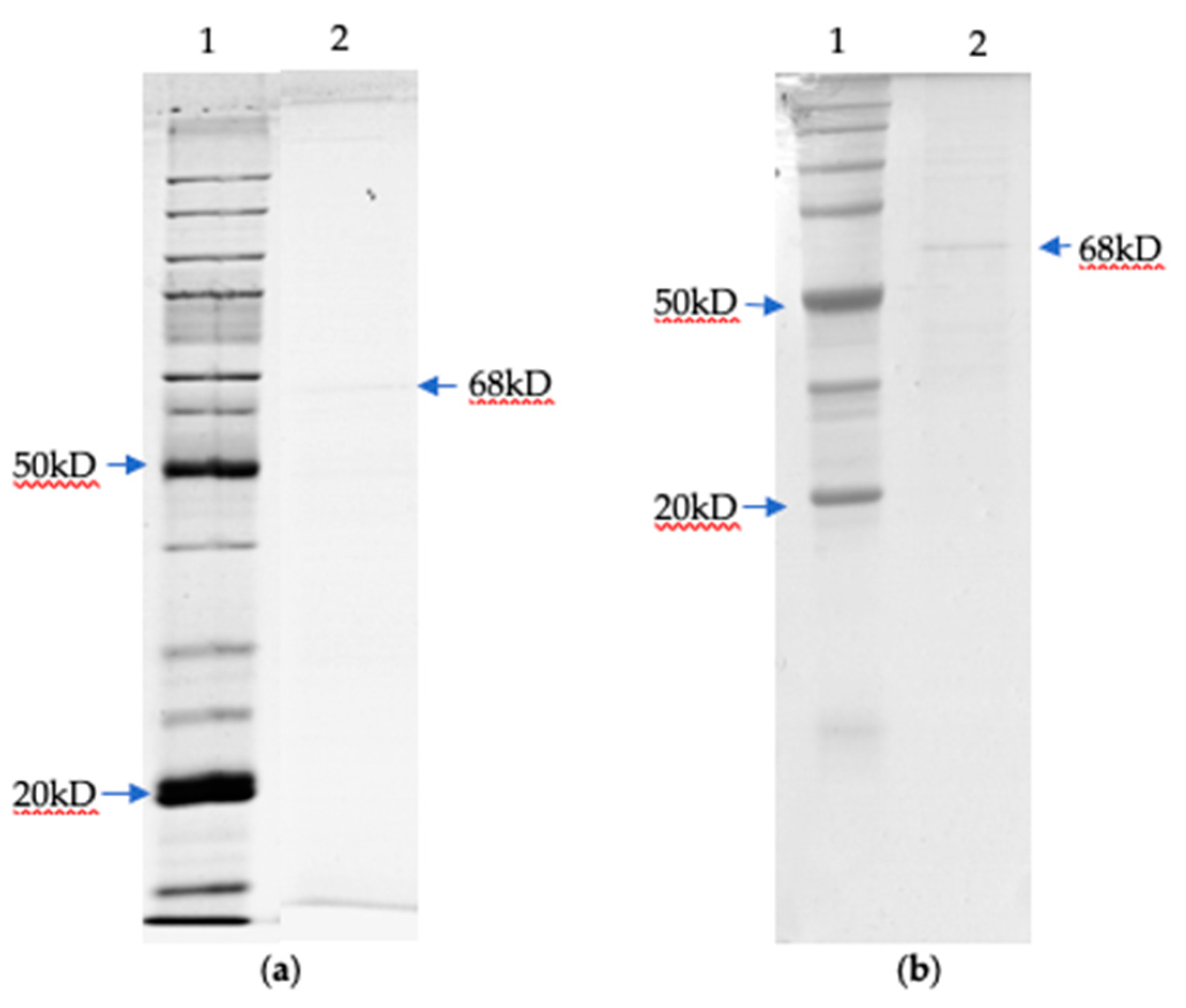
3.4. Determination of Molecular Weight by SDS PAGE
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruisi, S.; Barreca, D.; Selbmann, L.; Zucconi, L.; Onofri, S. Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 2007, 6, 127–141. [Google Scholar] [CrossRef]
- Tian, Y.; Li, Y.L.; Zhao, F.C. Secondary Metabolites from Polar Organisms. Mar. Drugs 2017, 15, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, S.; Gu, X.; Li, J.; Lin, X. Biosynthesis, characterization and antibacterial activity of silver nanoparticles by the Arctic anti-oxidative bacterium Paracoccus sp. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1488–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colby, G.A.; Ruuskanen, M.O.; St. Pierre, K.A.; St. Louis, V.L.; Poulain, A.J.; Aris-Brosou, S. Warming Climate Is Reducing the Diversity of Dominant Microbes in the Largest High Arctic Lake. Front. Microbiol. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F. Metagenomics: A gateway to drug discovery. In Advances in Biological Science Research; Meena, S.N., Naik, M.M., Eds.; Academic Press: Goa, India, 2019; pp. 453–468. [Google Scholar]
- Zucconi, L.; Canini, F.; Temporiti, M.E.; Tosi, S. Extracellular Enzymes and Bioactive Compounds from Antarctic Terrestrial Fungi for Bioprospecting. Int. J. Environ. Res. Public Health 2020, 17, 6459. [Google Scholar] [CrossRef] [PubMed]
- Arenz, B.E.; Blanchette, R.A.; Farrell, R.L. Fungal Diversity in Antarctic Soils. In Antarctic Terrestrial Microbiology; Cowan, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 35–53. [Google Scholar]
- Walton, D.W.H. Cellulose decomposition and its relationship to nutrient cycling at South Georgia. In Antarctic Nutrient Cycling and Food Webs, Proceedings of the 4th SCAR Symposium on Antarctic Biology, Wilderness, South Africa, 12–16 September 1983; Siegfried, W.R., Condy, P.R., Laws, R.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1985; pp. 192–199. [Google Scholar]
- Crowther, T.W.; Van Den Hoogen, J.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef]
- Singh, S.M.; Singh, S.K.; Yadav, L.S.; Singh, P.N.; Ravindra, R. Filamentous soil fungi from Ny-Ålesund, Spitsbergen, and screening for extracellular enzymes. Polar Biol. 2012, 65, 45–55. [Google Scholar] [CrossRef]
- Abneuf, M.A.; Krishnan, A.; Gonzalez-Aravena, M.; Pang, K.L.; Convey, P.; Mohamad-Fauzi, N.; Rizman-Idid, M.; Alias, S.A. Antimicrobial activity of microfungi from maritime Antarctic soil. Czech Polar Rep. 2016, 6, 141–154. [Google Scholar] [CrossRef]
- Irawan, B.; Afandi, A.; Hadi, S. Effects of saprophytic microfungi application on soil fertility based on their decomposition properties. J. Appl. Biol. Sci. 2017, 11, 15–19. [Google Scholar]
- IPCC. Climate Change: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Rajendra, K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland, 2014; 151p. [Google Scholar]
- Carrasco, M.; Villarreal, P.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Screening and characterization of amylase and cellulase activities in psychrotolerant yeasts. BMC Microbiol. 2016, 16, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Duarte, A.F.; Dayo-Owoyemi Nobre, F.S.; Pagnocca, F.C.; Chaud, L.S.; Pessoa, A.; Felipe, M.D.G.A.; Sette, L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles 2013, 17, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.M.; Fitzgerald, G.F.; Sinderen, D.V. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 2006, 72, 5289–5296. [Google Scholar] [CrossRef] [Green Version]
- Offen, W.A.; Viksoe-Nielsen, A.; Borchert, T.V.; Wilson, K.S.; Davies, G. Three-dimensional structure of a variant ‘Termamyl-like’ Geobacillus stearothermophilus α-amylase at 1.9 Å resolution. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 66–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yu, J.; Li, F.; Peng, H.; Zhang, X.; Xiao, Y.; He, C. Crystal structure of a raw-starch-degrading bacterial α-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13. Sci. Rep. 2017, 7, 44067. [Google Scholar] [CrossRef] [PubMed]
- Goto, C.E.; Barbosa, E.P.; Kistner, L.C.; Moreira, F.G.; Lenartovicz, V.; Peralta, R.M. Production of amylase by Aspergillus fumigatus utilizing alpha-methyl-D-glycoside, a synthetic analogue of maltose, as substrate. FEMS Microbiol. Lett. 1998, 167, 139–143. [Google Scholar] [PubMed] [Green Version]
- Bin, G.; Laisu, X.; Youfang, D.; Yanquan, L. Screening of alpha amylase high-producing strains from Bacillus subtilis. J. Zhejiang 1999, 23, 88–92. [Google Scholar]
- Singh, S.; Singh, S.; Bali, V.; Sharma, L.; Mangla, J. Production of Fungal Amylases Using Cheap, Readily Available Agriresidues, for Potential Application in Textile Industry. Biomed. Res. Int. 2014, 2014, 215748. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Mangla, J.; Singh, S. Evaluation of Aspergillus fumigatus NTCC1222 as a source of enzymes for detergent industry. Res. Environ. Sustain. 2021, 5, 1–5. [Google Scholar] [CrossRef]
- Naili, B.; Sahnoun, M.; Bejar, S.; Kammoun, R. Optimization of Submerged Aspergillus oryzae S2 α-Amylase Production. Food Sci. Biotechnol. 2016, 25, 185–192. [Google Scholar] [CrossRef]
- Abdullah, R.; ul-Haq, I. Purification and characterisation of a-amylase produced by mutant strain of Aspergillus oryzae EMS-18. Nat. Prod. Res. 2014, 29, 710–716. [Google Scholar] [CrossRef]
- Kamaraj, M.; Subramaniam, D. Optimization of Experimental Variables for the Production of α- Amylase by Aspergillus Oryzae Using Rice Bran. Int. J. Adv. Res. Eng. Technol. 2020, 11, 113–126. [Google Scholar]
- Martinez-Martinez, M.; Coscolin, C.; Santiago, G.; Chow, J.; Stogios, P.J.; Bargiela, R.; Gertler, C.; Navarro-Fernández, J.; Bollinger, A.; Thies, S.; et al. Determinants and Prediction of Esterase Substrate Promiscuity Patterns. ACS Chem. Biol. 2018, 13, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, A.; Alias, S.A.; Wong, C.V.; Pang, K.L.; Convey, P. Extracellular hydrolase enzyme production by soil fungi from King George Island, Antarctica. Polar Biol. 2011, 34, 1535–1542. [Google Scholar] [CrossRef]
- Krishnan, A.; Convey, P.; Gonzalez-Rocha, G.; Alias, S.A. Production of extracellular hydrolase enzymes by fungi from King George Island. Polar Biol. 2016, 39, 65–76. [Google Scholar] [CrossRef]
- Gao, B.; Mao, Y.; Zhang, L.; He, L.; Wei, d. A novel saccharifying a-amylase of Antarctic psychrotolerant fungi Geomyces pannorum: Gene cloning, functional expression, and characterization. Starch/Stärke 2016, 68, 20–28. [Google Scholar] [CrossRef]
- Gesheva, V. Distribution of psychrophilic microorganisms in soils of Terra Nova Bay and Edmonson Point, Victoria Land and their biosynthetic capabilities. Polar Biol. 2009, 32, 1287–1291. [Google Scholar] [CrossRef]
- Gesheva, V.; Vasileva-Tonkova, E. Production of enzymes and antimicrobial compounds by halophilic Antarctic Nocardioides sp. grown on different carbon sources. World J. Microbiol. Biotechnol. 2012, 28, 2069–2076. [Google Scholar] [CrossRef] [Green Version]
- Margesin, R.; Neuner, G.; Storey, K.B. Cold-loving microbes, plants, and animals- fundamental and applied aspects. Naturwissenschaften 2007, 94, 77–99. [Google Scholar] [CrossRef]
- Birgisson, H.; Delgado, O.; Garcia Arroyo, L.; Hatti-Kaul, R.; Mattiasson, B. Cold-adapted yeasts as producers of cold-active polygalacturonases. Extremophiles 2003, 7, 185–193. [Google Scholar] [CrossRef]
- Carrasco, M.; Rozas, J.M.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Diversity and extracellular enzymatic activities of yeasts isolated from King George Island, the sub-Antarctic region. BMC Microbiol. 2012, 12, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Zak, D.R.; Kling, G.W. Microbial community composition and function across an Arctic tundra landscape. Ecology 2006, 87, 1659–1670. [Google Scholar] [CrossRef]
- Bancerz, R.; Ginalska, G.; Fiedurek, J.; Gromada, A. Cultivation conditions and properties of extracellular crude lipase from the psychrotrophic fungus Penicillium chrysogenum 9’. J. Ind. Microbiol. Biotechnol. 2005, 32, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Gawas-Sakhalkar, P.; Singh, S.M. Fungal community associated with Arctic moss, Tetraplodon mimoides and its rhizosphere: Bioprospecting for production of industrially useful enzymes. Curr. Sci. 2011, 100, 1701–1705. [Google Scholar]
- Zaferanloo, B.; Bhattacharjee, S.; Ghorbani, M.M.; Mahon, P.J.; Palombo, E.A. Amylase production by Preussia minima, a fungus of endophytic origin: Optimization of fermentation conditions and analysis of fungal secretome by LC-MS. BMC Mcrobiol. 2014, 14, 55–66. [Google Scholar] [CrossRef]
- Punekar, N.S. Exploiting Enzymes: Technology and Applications. In ENZYMES: Catalysis, Kinetics and Mechanisms; Springer: Singapore, 2018; pp. 14–31. [Google Scholar]
- Rousk, J.; Brookes, P.C.; Bååth, E. Contrasting Soil pH Effects on Fungal and Bacterial Growth Suggest Functional Redundancy in Carbon Mineralization. Appl. Environ. Microbiol. 2009, 75, 1589–1596. [Google Scholar] [CrossRef] [Green Version]
- Rajoka, M.I.; Akhtar, M.W.; Hanif, A.; Khalid, A.M. Production and characterization of a highly active cellobiase from Aspergillus niger grown in solid state fermentation. World J. Microbiol. Biotechnol. 2006, 22, 991–998. [Google Scholar] [CrossRef]
- Dojnov, B.; Božić, N.; Nenadović, V.; Ivanović, J.; Vujčić, Z. Purification and properties of midgut α-amylase isolated from Morimus funereus (Coleoptera: Cerambycidae) larvae. Comp. Biochem. Physiol. 2008, 149, 153–160. [Google Scholar] [CrossRef]
- Krishnan, A.; Convey, P.; Gonzalez, M.; Smykla, J.; Alias, S.A. Effects of temperature on extracellular hydrolase enzymes from soil microfungi. Polar Biol. 2018, 41, 537–551. [Google Scholar] [CrossRef]
- Wong, H.J.; Mohamad-Fauzi, N.; Rizman-Idid, M.; Convey, P.; Alias, S.A. Protective mechanisms and responses of micro-fungi towards ultraviolet-induced cellular damage. Polar Sci. 2018, 20, 19–34. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Verma, V.K.; Chaturvedi, V.; Verma, P. GH10 XynF1 and Xyn11A: The predominant xylanase identified in the profiling of extracellular proteome of Aspergillus oryzae LC1. Ann. Microbiol. 2018, 68, 731–742. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Kobayashi, M.; Sasaki, Y.; Kobayashi, S. Purification of amylases and other enzymes by a forced-affinity chromatography method. Biosci. Biotechnol. Biochem. 1997, 61, 813–816. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Storms, R.; Tsang, A. Microplate-based carboxymethylcellulose assay for endoglucanase activity. Anal. Biochem. 2006, 342, 176–178. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Hassan, N.; Rafiq, M.; Hayat, M.; Shah, A.A.; Hasan, F. Psychrophilic and psychrotrophic fungi: A comprehensive review. Rev Environ. Sci. Biotechnol. 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Robinson, C.H. Cold adaptation in Arctic and Antarctic fungi. New Phytol. 2001, 151, 341–353. [Google Scholar] [CrossRef]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.-A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R. Soc. B 2002, 357, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000, 297, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Gault, S.; Higgins, P.M.; Cockell, C.S.; Gillies, K. A meta-analysis of the activity, stability, and mutational characteristics of temperature-adapted enzymes. Biosci. Rep. 2021, 41, 1–10. [Google Scholar] [CrossRef]
- Bisswanger, H. Enzyme assays. Perspect. Sci. 2014, 1, 41–55. [Google Scholar] [CrossRef] [Green Version]
- Cavicchioli, R.; Siddiqui, K.S. Cold-adapted enzymes. In Enzyme Technology; Pandey, A., Webb, C., Soccol, C.R., Larroche, C., Eds.; Springer Science: New York, NY, USA, 2006; pp. 615–638. [Google Scholar]
- Ramli, A.M.; Azhar, M.A.; Shamsir, M.S.; Rabu, A.; Abdul Murad, A.M.; Mahadi, N.M.; Illias, R.M. Sequence and structural investigation of a novel psychrophilic a-amylase from Glaciozyma antarctica PI12 for cold adaptation analysis. J. Mol. Model. 2013, 19, 3369–3383. [Google Scholar] [CrossRef] [PubMed]
- Ramli, A.M.; Mahadi, N.M.; Shamsir, M.S.; Rabu, A.; Joyce-Tan, K.H.; Abdul Murad, A.M.; Illias, R.M. Structural prediction of a novel chitinase from the psychrophilic Glaciozyma antarctica PI12 and an analysis of its structural properties and function. J. Comput. Aided Mol. Des. 2012, 26, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.F.; dos Santos, J.A.; Vianna, M.V.; Vieira, J.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Iefuji, H.; Chino, M.; Kato, M.; Iimura, Y. Raw-starch-digesting and thermostable α-amylase from the yeast Cryptococcus sp. S-2: Purification, characterization, cloning and sequencing. Biochem. J. 1996, 318, 989–996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipton, K.F.; Dixon, H.F. Effects of pH on enzymes. In Methods in Enzymology; Daniel, L.P., Ed.; Academic Press: Cambridge, MA, USA, 1979; Volume 63, pp. 183–234. [Google Scholar]
- King, G.M. Characterization of β-Glucosidase Activity in Intertidal Marine Sediments. Appl. Environ. Microbiol. 1986, 51, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Chróst, R.J. Environmental control of the synthesis and activity of aquatic microbial ectoenzymes. In Microbial Enzymes in Aquatic Environments; Chróst, R.J., Ed.; Springer: New York, NY, USA, 1991; pp. 29–59. [Google Scholar]
- Münster, U. Extracellular enzyme activity in eutrophic and polyhumic lakes. In Microbial Enzymes in Aquatic Environments; Chróst, R.J., Ed.; Springer: New York, NY, USA, 1991; pp. 96–122. [Google Scholar]
- Liu, X.D.; Xu, Y. A novel raw starch digesting α-amylase from a newly isolated Bacillus sp. YX-1: Purification and characterization. Bioresour. Technol. 2008, 99, 4315–4320. [Google Scholar] [CrossRef]
- Pandey, A.; Nigam, P.; Soccol, C.R.; Soccol, V.T.; Singh, D.; Mohan, R. Advances in microbial amylases. Appl. Biochem. Biotechnol. 2000, 31, 135–152. [Google Scholar] [CrossRef]
- Abe, J.I.; Nakajima, K.; Nagano, H.; Hizukuri, S.; Obata, K. Properties of the raw-starch digesting amylase of Aspergillus sp. K-27: A synergistic action of glucoamylase and alpha-amylase. Carbohydr. Res. 1988, 175, 85–92. [Google Scholar] [CrossRef]
- Wang, X.; Kan, G.; Shi, C.; Xie, Q.; Ju, Y.; Wang, R.; Qiao, Y.; Ren, X. Purification and characterization of a novel wild-type α-amylase from Antarctic sea ice bacterium Pseudoalteromonas sp. M175. Protein Expr. Purif. 2019, 164, 105444. [Google Scholar] [CrossRef]
- Vihinen, M.; Mantsala, P. Microbial amylolytic enzymes. Crit. Rev. Biochem. Mol. 1989, 24, 329–418. [Google Scholar] [CrossRef] [PubMed]
- Xian, L.; Wang, F.; Luo, X.; Feng, Y.L.; Feng, J.X. Purification and Characterization of a Highly Efficient Calcium-Independent α-Amylase from Talaromyces pinophilus 1-95. PLoS ONE 2015, 10, e0121531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keskin, S.; Ertunga, S. Purification, immobilization and characterization of thermostable α-amylase from a thermophilic bacterium Geobacillus sp. TF14. Turk. J. Biochem. 2017, 42, 633–642. [Google Scholar] [CrossRef]
- Grootegoed, J.A.; Lauwers, A.M.; Heinen, W. Separation and partial purification of extracellular amylase and protease from Bacillus caldolyticus. Arch. Microbiol. 1973, 90, 223. [Google Scholar] [CrossRef]
- Ratanakhanokchai, K.; Kaneko, J.; Kamio, Y.; Izaki, K. Purification and properties of a maltotetraose and maltotriose producing amylase from Chloroflexus aurantiacus. Appl. Environ. Microbiol. 1992, 58, 2490–2494. [Google Scholar] [CrossRef] [Green Version]
- IPCC. Summary for Policymakers. In Climate Change 2013: The Physical Science Basis; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Marshall, W.A. Aerial Transport of Keratinaceous Substrate and Distribution of the Fungus Geomyces pannorum in Antarctic Soils. Microb. Ecol. 1998, 36, 212–219. [Google Scholar] [CrossRef]
- Bridge, P.L.; Spooner, B.M.; Roberts, P.J. Non-lichenized fungi from the Antarctic region. Mycotaxon 2008, 106, 485–490. [Google Scholar]
- Arenz, B.E.; Blanchette, R.A. Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross Sea Region and McMurdo Dry Valleys. Soil Biol. Biochem. 2011, 43, 308–315. [Google Scholar] [CrossRef]
- Convey, P.; Coulson, S.J.; Worland, M.R.; Sjöblom, A. The importance of understanding annual and shorter term temperature patterns and variation in the upper layers of polar soils for terrestrial biota. Polar Biol. 2018, 41, 1587–1605. [Google Scholar] [CrossRef] [Green Version]
- Misiak, M.; Goodall-Copestake, W.P.; Sparks, T.H.; Worland, M.R.; Boddy, L.; Magan, N.; Convey, P.; Hopkins, D.W.; Newsham, K.K. Inhibitory effects of climate change on the growth and extracellular enzyme activities of a widespread Antarctic soil fungus. Glob. Chang. Biol. 2021, 27, 1111–1125. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- De Mot, R.; Verachtert, H. Purification and characterization of extracellular a-amylase and glucoamylase from the yeast Candida antarctica CBS 6678. Eur. J. Biochem. 1987, 164, 643–654. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Mao, Y.; Zhang, L.; Wang, H.; Alias, S.A.; Gao, B.; Wei, D. Functional expression of a novel α-amylase from Antarctic psychrotolerant fungus for baking industry and its magnetic immobilization. BMC Biotechnol. 2017, 17, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samie, N.; Noghabi, K.A.; Gharegozloo, Z.; Zahiri, H.S.; Ahmadian, G.; Sharafi, H.; Behrozi, R.; Vali, H. Psychrophilic a-amylase from Aeromonas veronii NS07 isolated from farm soils. Process Biochem. 2012, 47, 1381–1387. [Google Scholar] [CrossRef]
- Roohi; Kuddus, M.; Saima. Cold-active detergent-stable extracellular α-amylase from Bacillus cereus GA6: Biochemical characteristics and its perspectives in laundry detergent formulation. J. Biochem. Technol. 2013, 4, 636–644. [Google Scholar]
- Kuddus, M.; Arif, J.M.; Ramteke, P.W. An overview of coldactive microbial a-amylase: Adaptation strategies and biotechnological potentials. Biotechnology 2011, 3, 246–258. [Google Scholar] [CrossRef]
- Chessa, J.P.; Feller, G.; Gerday, C. Purification and characterization of the heat-labile a-amylase secreted by the psychrophilic bacterium TAC 240B. Can. J. Microbiol. 1999, 45, 452–457. [Google Scholar] [CrossRef]
- Smith, M.R.; Zahnley, J.C. Production of amylase by Arthrobacter psychrolactophilus. J. Ind. Microbiol. Biotechnol. 2005, 32, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Rathour, R.; Gupta, J.; Tyagi, B.; Thakur, I.S. Production and characterization of psychrophilic α-amylase from a psychrophilic bacterium, Shewanella sp. ISTPL2. Amylase 2020, 4, 1–10. [Google Scholar] [CrossRef]


| Strain | Sample | Enzyme Activity (µmo/min/mL) | Total Protein Content (mg/mL) | Specific Activity (U/mg) | Purification Fold | Recovery % |
|---|---|---|---|---|---|---|
| Antarctic Pseudogymnoascus sp. | Crude | 40.64 | 15.80 | 2.57 | 1 | 100 |
| Partially purified enzyme | 3.71 | 0.74 | 5.01 | 1.95 | 9.13 | |
| Arctic Pseudogymnoascus sp. | Crude enzyme | 18.67 | 22.67 | 0.82 | 1 | 100 |
| Partially purified enzyme | 3.1 | 0.13 | 23.85 | 29.09 | 16.60 |
| Origin | Growth Condition | Enzyme Characteristics | Specific Activity (U/mg) | MW (kDa) | Recombinant/Wild Type Enzyme | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Temperature | pH | Optimum Temperature | Optimum pH | |||||
| Fungi | ||||||||
| Pseudogymnoascus sp. | 15 | 6 | 15 | 5–6 | 5.01 & 23.85 | 68 | Wild type | Present study |
| Moesziomyces antarcticus | 29 | NR | 62 | 4.2 | NR | 50 | Wild type | [82] |
| Geomyces pannorum R1-2 (currently known as Pseudogymnoascus pannorum) | 20 | NR | 70 | 6.0 | 9.78 × 103 | NR | Recombinant | [29] |
| Geomyces pannorum R1-2 (currently known as Pseudogymnoascus pannorum) | NR | NR | 40 | 5 | 12.8 × 103 | 52 | Recombinant | [83] |
| Bacteria | ||||||||
| Aeromonas veronii (Bacteria) | 10 | NR | 10 | 4 | 430 | 63 | Wild type | [84] |
| Bacillus cereus | NR | NR | 20 | 10 | 175.92 | 55 | Wild type | [85] |
| Microbacterium foliorum GA2 and Bacillus cereus GA6 | 20 | NR | 20 | 9 & 10 | NR | NR | Wild type | [86] |
| strain TAC 240B | NR | NR | 7.5 | 71 | 50 | Wild type | [87] | |
| Arthrobacter psychrolactophilus | 22 | 7 | 20 | NR | 105 & 26 | Wild type | [88] | |
| Shewanella sp. ISTPL2 (bacteria) | 10 | 6.9 | 4 | 8 | 36,690.47 | 45 | Wild type | [89] |
| Pseudoalteromonas sp. M175 | 15 | 8 | 30 | 7.5 | 289.79 | 61 | Wild type | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnan, A.; Alias, Z.; Convey, P.; González-Aravena, M.; Smykla, J.; Rizman-Idid, M.; Alias, S.A. Temperature and pH Profiling of Extracellular Amylase from Antarctic and Arctic Soil Microfungi. Fermentation 2022, 8, 601. https://doi.org/10.3390/fermentation8110601
Krishnan A, Alias Z, Convey P, González-Aravena M, Smykla J, Rizman-Idid M, Alias SA. Temperature and pH Profiling of Extracellular Amylase from Antarctic and Arctic Soil Microfungi. Fermentation. 2022; 8(11):601. https://doi.org/10.3390/fermentation8110601
Chicago/Turabian StyleKrishnan, Abiramy, Zazali Alias, Peter Convey, Marcelo González-Aravena, Jerzy Smykla, Mohammed Rizman-Idid, and Siti Aisyah Alias. 2022. "Temperature and pH Profiling of Extracellular Amylase from Antarctic and Arctic Soil Microfungi" Fermentation 8, no. 11: 601. https://doi.org/10.3390/fermentation8110601






