Abstract
This study identified the genetic diversity of amylase-producing lactic acid bacteria from brown rice (Oryza nivara) Wakawondu cultivar based on the 16S rRNA gene. The ten lactic acid bacteria strains were isolated from the fermented Wakawondu rice washing water; two isolates, SBM3D and SBM4A, displayed strong amylase activity. The two isolates had the same characteristics according to both morphological and biochemical examination. The effect of fermentation time on SBM3D bacterial isolates revealed that bacterial growth at 12 h with OD values and enzyme activity, respectively, of 0.856 and 175 mU/mL, was nearly identical to the growth at 27 h with OD values of 0.886 and 176 mU/mL consecutively. Meanwhile, the bacterial isolate of SBM4A showed a significant increase in growth at 15 h with an OD value of 0.552 and enzyme activity of 99 U/mL. The maximum growth was seen at 18 h with an OD value of 0.657 and enzyme activity of 126 mU/mL. Cladogram of an SBM3D isolate with Pediococcus pentosaceus strains SL001 CP039378.1 and SRCM102740 CP028269.1 forming a sister group clad. Pediococcus pentosaceus strains SRCM102739 CP028266.1 and SRCM102738 CP028264.1 form a sister group in the cladogram of the SBM4A isolate. SBM3D and SBM4A, which are amylase-producing Pediococcus pentosaceus, can be used in food, chemical, health, and other industries.
1. Introduction
Wakawondu is one of the local upland brown rice cultivars originating from North Buton Regency, Southeast Sulawesi. Starch-rich rice washing water from Wakawondu can be utilized as a medium for fermentation. Lactic acid bacteria (LAB), which may use starch as an energy source, overgrow in the medium during the spontaneous fermentation process. LAB was isolated from the cloudy liquid in the middle of the fermented rice washing water. This process which acts as an intermediary for the formation of hormones auxin and gibberellins, contains abundant nutrients, including carbohydrates in the form of 85% starch, protein, cellulose, phosphorus, and vitamins [1]. According to Asngad et al. [2], waste is used to make syrup through fermentation with the addition of rosella plants as natural dyes. Presently, rice washing water has been used as a source of isolates to obtain lactic acid bacteria (LAB) and bioethanol. Therefore, this process produces more useful products [3].
Lactic acid bacteria are a category of gram-positive, acid-tolerant, mostly nonsporulating, rod- or cocci-shaped bacteria that can ferment carbohydrates like starch and simple sugars to produce lactic acid as the primary metabolic end-product [4]. These bacteria significantly contribute to the food world, with LAB widely used as a starter for fermented beverages, meat, and vegetables. LAB significantly changes fermented product texture, aroma, color, digestibility, and nutritional quality [5]. LAB can utilize starch as a substrate, known as amylolytic LAB, in changing the product characteristics to produce lactic acid, specific enzymes, and aromatic compounds [6]. According to preliminary studies, LAB can produce extracellular amylase and ferment starch directly to lactic acid. This fact is because the amylolytic LAB ferments the enzyme hydrolysis of carbohydrate substrates (starch) to ferment sugar into lactic acid [7,8]. In the food, chemical, pharmaceutical, and health industries, the application of amylase is relatively high and still shows dominance among the enzymes on the market. Amylase enzymes are widely used in the production of glucose syrup, crystalline glucose, dissolving and forming starch sugar for alcoholic fermentation, as well as inhibiting spoilage in the bakery industry. In the chemical industry, amylase enzymes are used as additives to remove impurities, reduce paper starch viscosity, and prevent textile fiber swelling. In the pharmaceutical and health fields, amylase enzymes are used to increase the digestibility of fiber which aids digestion [9]. Ardhi et al. [10] isolated the amylase enzyme from thermophilic bacteria, which is useful in several applications requiring high temperatures, for example, in gelatinization, liquefaction, and saccharification.
One of the methods widely used to analyze bacterial detection is the PCR technique. Aris [11] used this strategy to examine the DNA profile of the 16S rRNA gene, which is a molecular marker identical in all bacteria. Each bacterial species share the specific properties of this distinctive 16S rRNA because it is used as a universal, representative, and practical molecular systematic parameter to construct phylogenetic relationships at the species level [12]. A phylogenetic tree is used to delimit the taxa of each connected group. Kasi et al. [13] identify lactic acid bacteria by determining the phylogenetic tree. According to Barus et al. [14], 31 BAL isolates extracted from cassava tapai showed high identity compared to the 16S rDNA sequences in the Genebank database. In the food industry, LAB isolates from rice washing water can be utilized, for instance, to enhance flour quality [15]. To use the LAB from fermented Wakawondu rice washing water in the food sector and other associated industries, it must first be described to identify the LAB.
2. Materials and Methods
2.1. Materials
The materials used in this research were Merck’s DeMan Rogosa Sharpe Agar (MRS-A), DeMan Rogosa Sharpe Broth (MRS-B), NB, Merck (Darmstadt, Germany) distilled water, 96% alcohol, 70% alcohol, 0.9% sodium chloride, 0.3% calcium carbonate, sodium tartrate, DNA Extraction Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, D6005) (Zymo Research Corporation, Orange, CA, USA), MyTaq HS Red Mix PCR master mix, 2× (Bioline, BIO-25048 Meridian River Hills, Cincinnati, OH, USA), and Agarose Gel 1% Merck (Darmstadt, Germany)
2.2. Methods
2.2.1. Preparation of Samples of Wakawondu Rice Washing Water
To wash the rice uniformly, 100 g of Wakawondu rice was poured into a beaker glass with a 1000 mL capacity, 200 mL of distilled water was added, and the liquid was slowly spun for 5 min. Next, 100 mL of the washed water was added to a beaker that had been rubber-banded shut and sealed with a filter or porous cloth. Following this preparation, samples of the water used to wash rice were allowed to ferment without the addition of yeast for three days at room temperature under cover of darkness. The cloudy liquid in the middle of fermented rice washing water served as a sample [16,17].
2.2.2. Bacterial Isolation
Lactic acid bacteria (LAB) were isolated from 0.1 mL of samples using the pour plate method. It was placed into 0.9 mL of sterile distilled water aseptically and then stirred using a vortex until the solution was homogeneous. The obtained suspension (dilution 10−1) was diluted by the stratified dilution method up to 10−4 with 0.2 mL of each dilution series and put into a sterile petri dish. Furthermore, ±5 ml of MRSA medium was added with 0.3% calcium carbonate and shaken to ensure the calcium carbonate was evenly distributed in the medium. After obtaining a hardened MRSA medium, the Petri dishes were placed in an incubator at 37 °C and incubated for 48 h. Colonies that formed clear zones on MRSA medium, presumed to be LAB, were purified using a needle. It was further inoculated on a new agar medium by the streak plate method and incubated at 37 °C for 48 h. Amylase activity-high isolates were chosen for additional analysis. The purification step was conducted to obtain pure isolates (single colonies), with the addition of culture maintenance on all isolates obtained. Pure cultures were stored on slanted MRSA media at a refrigerator temperature of 40 °C [18].
2.3. Morphological and Biochemical Characteristics of Selected LAB Isolates
The morphological characters of selected LAB isolates observed include color, shape, edge, elevation, size of the colony, and cell shape. Then, the isolate was analyzed biochemically, which included a starch hydrolysis test using MRS agar media enriched with starch as a carbohydrate source, a Gram reaction test using 3% potassium hydroxide solution, a motility test using semi-solid media which was incubated for 48 h at 35 °C, and catalase test using 3% hydrogen peroxide. The ability of LAB to ferment glucose, sucrose, and lactose contained in the medium was tested using the Triple Sugar Iron Agar (TSIA) test, and Methyl Red (MR) test was used to determine the presence of mixed acid fermentation from glucose through a pathway of lactic, acetic, formic, and succinic acid. The tests were carried out following the standard protocols [18,19,20].
2.4. Amylase Enzyme Quantitative Test
2.4.1. Amylase Enzyme Extraction
The amylase enzyme extraction process was carried out by putting 1 mL of bacteria into 25 mL of liquid media. Bacterial cultures were incubated on a shaker incubator for 18 h at 37 °C. Cultures were centrifuged at 5000 rpm for 10 min to obtain a filtrate containing a crude amylase extract [19].
2.4.2. Amylase Activity Test
The amylase activity was conducted quantitatively by putting 0.1 mL of crude amylase extract into a test tube. Then 1 mL of soluble starch medium was added to the dissolved phosphate buffer. Incubated at 37 °C for 30 min. Furthermore, 1 mL DNS was added and then incubated at boiling water for 15 min until the solution turned brownish red. The test tube was filled to 10 mL with sterile water after 1 mL of 40% KNa-Tartrate was added. The absorbance of the sample solution was measured using a NanoDrop 2000/2000c Spectrofotometer (Thermo Scientific, Waltham, MA, USA) at a wavelength of 540 nm [20].
2.5. Bacterial DNA Isolation
The samples overgrown with colonies were then isolated for their DNA using Genomic DNA extraction with the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, D6005, Zymo Research, Orange, CA, USA). The quantitative method was used to test the DNA samples to determine their purity. Approximately 5 μL of the isolated DNA was dripped onto the NanoDrop 2000/2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA).
The Agilent Surecycler 8800 thermal spectrophotometer (Thermo Scientific, Waltham, MA, USA) cycler was used to determine the DNA amplification. Primers used were 27F 5′ forward primer–AGAGTTTGATCMTGGCTCAG–3′ primary reverse 1492R 5′–GGTTACCTTGTTACGACTT–3′ (Genetics Lab., 2021), PCR was set under predenaturation, denaturation, annealing, extension, and post-extension conditions at temperatures of 95 °C, 95 °C, 52 °C, 72 °C, and 72 °C it completed 35 cycles [21]. An amplification product sample in the amount of 1 mL was added together with 2.5 mL of loading dye. Then it was inserted into the 1% agarose gel wells. Running times for the electrophoresis were 135 V, 1 A, and 25 min. The DNA bands generated after electrophoresis were observed using the UV transilluminator Dolphin-Doc Plus Gel Image System UV 320 nm.
2.6. Sequencing and Analysis of Sequencing Results
DNA sequencing was performed, trimmed, and assembled with the BioEdit program 7.2.5 (Denton, TX, USA) and then converted into FASTA form. Furthermore, the result was analyzed using the Basic Local Alignment Search Tool (BLAST) program on the NCBI website (https://www.ncbi.nlm.nih.gov/) to determine the identity of the reference sequences contained in GeneBank.2.7 Kinship Analysis.
Bacterial relationships were determined using phylogenetic analysis, which was performed by aligning the analyzed DNA sequences with homologous samples downloaded from GeneBank. The phylogenetic tree was then constructed based on the 16S rRNA gene sequence using the Molecular Evolutionary Genetics Analysis (MEGA 11.0.13) software (New York, NY, USA).
3. Results
3.1. LAB Isolates from Wakawondu Rice Washing Water Fermentation Process
A total of 10 LAB strains were isolated from the fermented Wakawondu rice washing water using MRSA containing 0.3% calcium carbonate, as shown by the development of a clear zone surrounding the bacterial colonies. Additionally, these 10 LAB were examined for their capacity to hydrolyze starch, yielding two isolates with the greatest amylase activity, SBM3D and SBM4A. The results showed that the SBM3D and SBM4A isolates had the same colony morphological characteristics. The colonies were milky white in color, round shape, entire edges, convex elevation, and moderate colony size.
3.2. Physiological and Biochemical Characteristics of LAB Isolates
The results of the test showed that both LABs (SBM3D and SBM4A) isolates had the same physiological and biochemical characteristics. These two strains were Gram-positive, cocci in shape, non-motile, and catalase negative. The KOH 3% test of LAB isolates showed that the isolates reacted negatively to the KOH test. This finding was marked by the absence of mucus formation in bacterial colonies, indicating that LAB isolates were a group of gram-positive bacteria. The ability of the LAB to ferment glucose, sucrose, and lactose contained in the medium. Determination of bacterial kinship using the current genotypic identification method using DNA produced a genetic material resistant to evolution. However, the use of DNA was hampered by the inadequate number of samples tested using the Triple Sugar Iron Agar (TSIA) test. The two isolates were able to ferment the glucose present in the TSIA media. For the two BAL strains isolated from the fermented Wakawondu rice washing water, the MR test was positive, and the VP test was negative (Table 1).

Table 1.
Physiological and biochemical properties of the LAB isolated from fermented Wakawondu rice washing water.
3.3. Starch Hydrolysis
The ability of the SBM3D and SBM4A strains to hydrolyze starch revealed that all isolates responded favorably, as evidenced by forming a clear zone around the bacterial growth area. The diameter of the clear zone ranged from 3.6 to 3.7 mm produced by strains SB4A and SBM3D (Table 2).

Table 2.
Starch hydrolysis Activity of the LAB isolated from Wakawondu.
3.4. Bacterial Isolate Amylase Enzyme Activity from Wakawondu
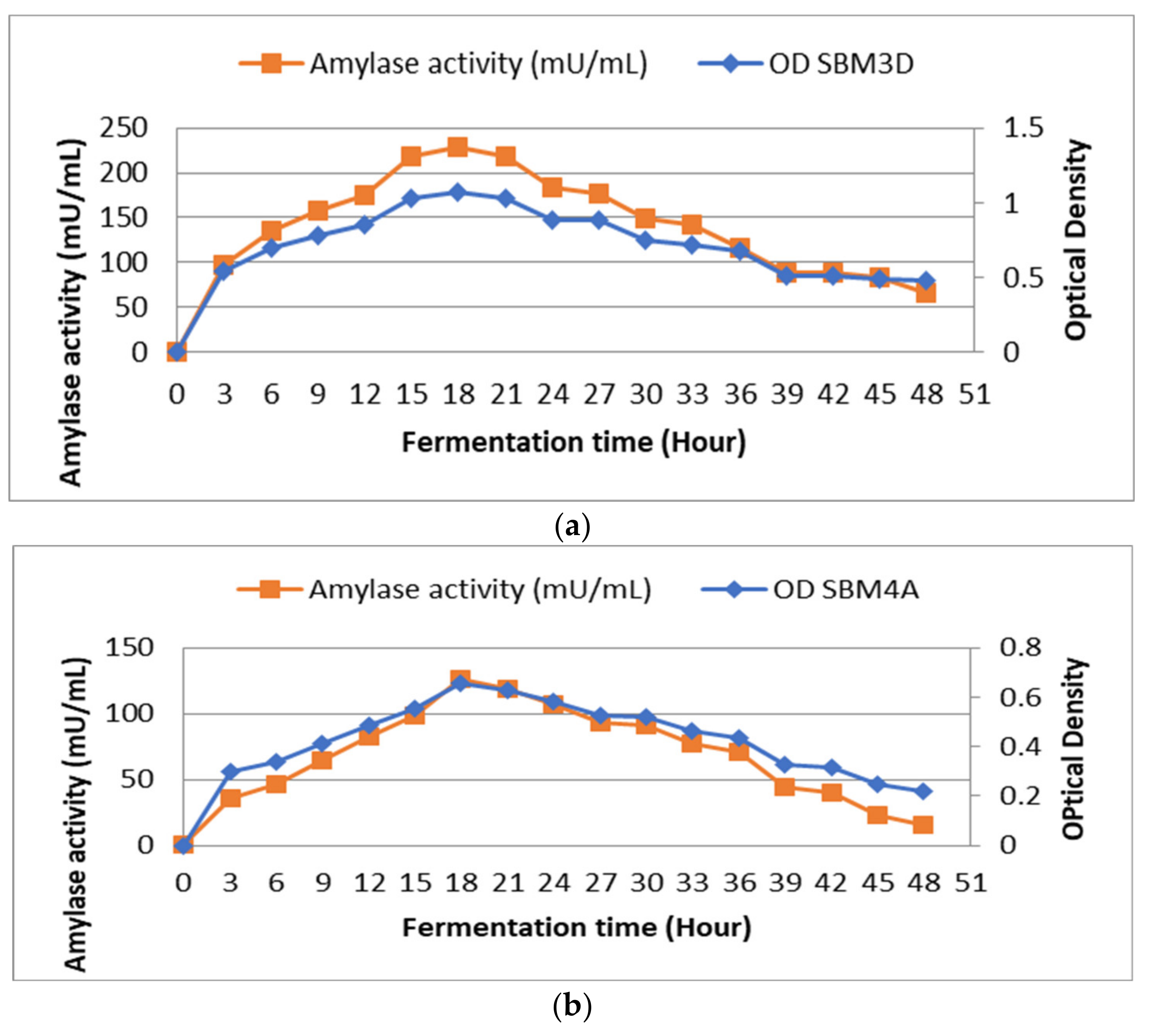
Observations on the curve show the effect of fermentation time on bacterial growth and the production of amylase enzymes. When fermentation duration was compared to bacterial growth, it was found that growth at 12 h with OD values and enzyme activity, respectively, of 0.856 and 175 mU/mL and growth at 27 h with OD values of 0.886 and 176 mU/mL were almost equal (Figure 1a). The effect of fermentation time on the SBM4A bacterial isolate showed that bacterial growth increased significantly at 15 h with an OD value of 0.552 and enzyme activity of 99 mU/mL. Furthermore, the enzyme activity and maximum growth were observed at the 18th h with an OD value of 0.657 and an enzyme activity of 126 mU/mL (Figure 1b).
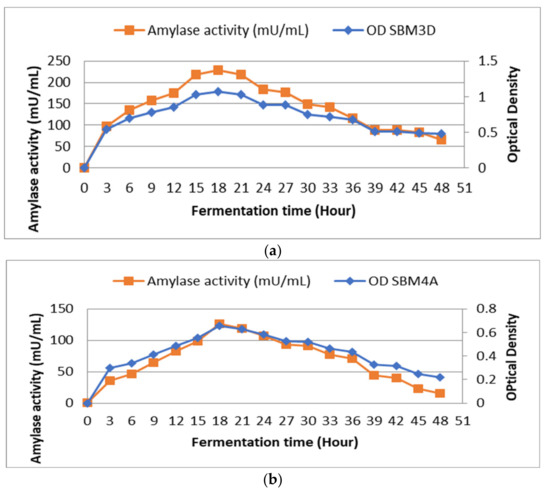
Figure 1.
Effect of fermentation time on the growth and activity amylase enzyme of the BAL strains isolated from Wakawondu (a) SBM3D; and (b) SBM4A.
3.5. Molecular Characteristics and Visualization of PCR Products
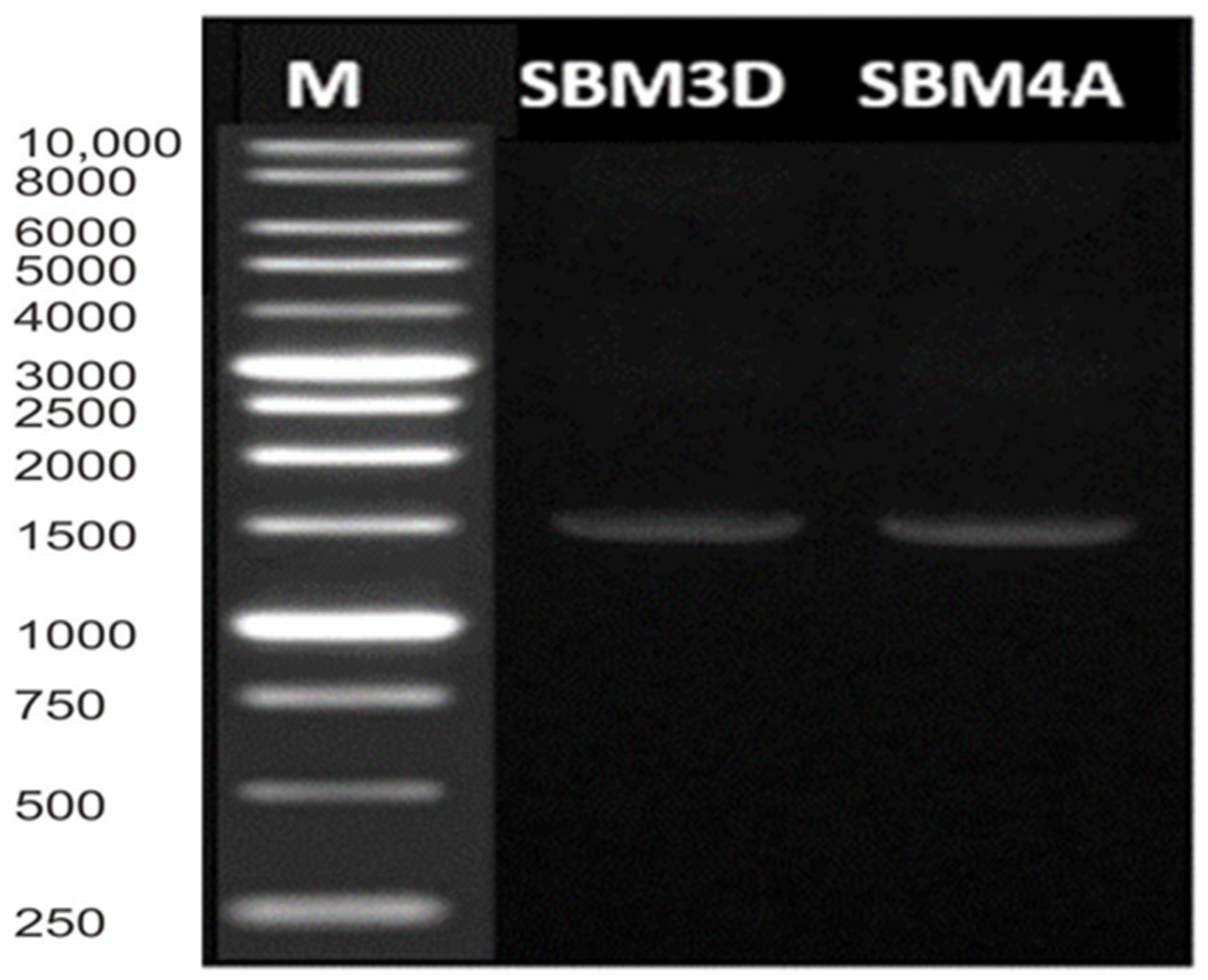
The results of multiplex PCR showed only a single band with a size of 1451 bp for SBM3D and 1452 bp for SBM4A strains, which indicates that their 16S rRNA gene has been well amplified (Figure 2).
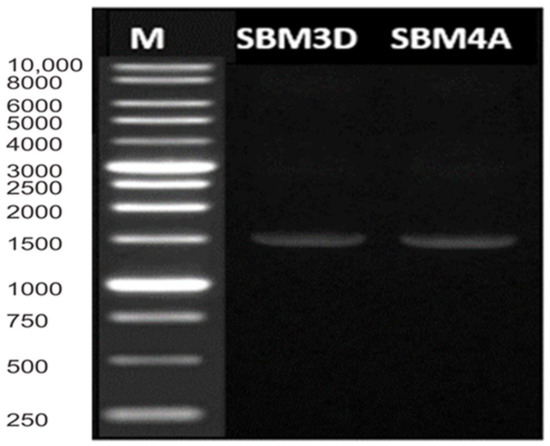
Figure 2.
Multiplex-PCR amplification patterns. Lane M = DL 10,000 DNA Marker.
DNA can be visualized by UV because the agarose gel contains ethidium bromide, whose molecule fluoresces when illuminated with ultraviolet (UV) light in the visible UV range. Ethidium binds to the hydrophobic location of the DNA molecule by inserting it between the base bonds in the double-stranded DNA through Van der Walls interactions.
3.6. Determination of the DNA Sequence
The 16S rRNA gene sequencing of the SBM3D and SBM4A strains in the form of an electropherogram was then translated using the BIOEDIT software to produce nucleotide sequence data for the 16S rRNA gene isolate (Figure 3 and Figure 4).

Figure 3.
Results of the 16S rRNA gene sequencing of SBM3D isolate.

Figure 4.
The 16S rRNA gene sequencing of the SBM4A isolate.
3.7. Relationship Level of LAB Isolates from Wakawondu
The LAB gene sequencing obtained was made into a phylogenetic tree to determine the level of bacterial kinship. The phylogenetic tree construction aims to visualize the relationship between sample organisms based on their evolutionary relationships with comparison organism sequences from NCBI sites [22].
The cladogram shows that the SBM3D isolate exists between Pediococcus pentosaceus strain SRCM102740 CP028269.1 and Pediococcus pentosaceus strain SL001 CP039378.1 to form a sister group clad with a bootstrap value of 99, and form a monophyletic group Pediococcus pentosaceus strain 8362 16S MT538852, Pediococcus pentosaceus 6340 16S MT463749.1, Pediococcus pentosaceus strain GDIAS001 CP046938 (Figure 5).

Figure 5.
Evolutionary relationships of isolate SBM3D regarding bacterial strains in Genebank NCBI by using the maximum parsimony method with a bootstrap value of 100×.
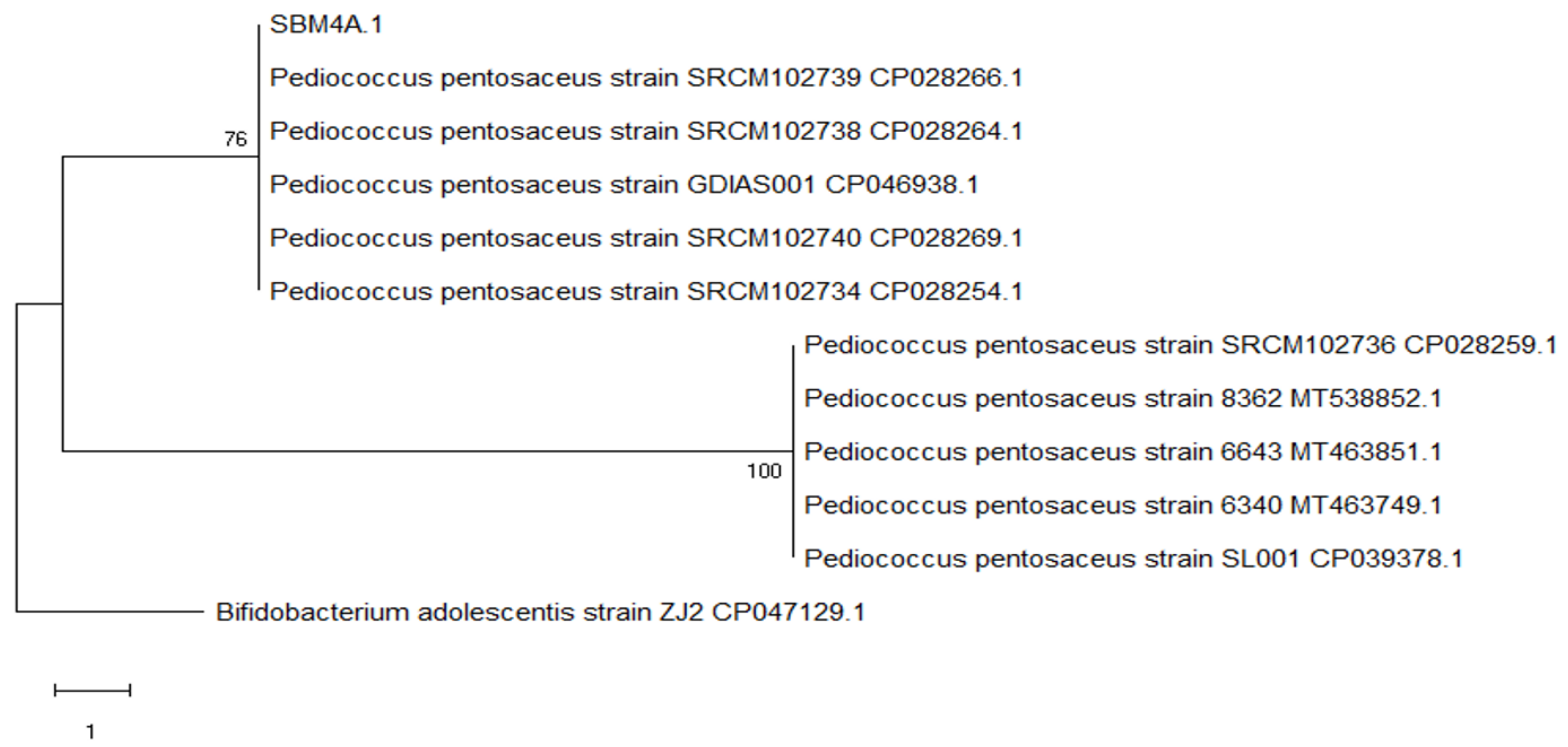
The cladogram shows that SBM4A is between the Pediococcus pentosaceus strain SRCM102739 CP028266.1 and Pediococcus pentosaceus strain SRCM102738 CP028264.1 forming a sister clade group with a bootstrap value of 76, and forming the monophyletic group Pediococcus pentosaceus strain SRCM102736 CP028259.1, Pediococcus pentosaceus strain 8362 16S MT538852.1 and Pediococcus pentosaceus strain 6643 16S MT463851.1 (Figure 6).

Figure 6.
Evolutionary relationships of isolate SBM4A regarding bacterial strains in Genebank NCBI by using the maximum parsimony method with a bootstrap value of 100×.
4. Discussion
From fermented Wakawondu rice washing water, a total of 10 bacterial isolates were collected. The macroscopic observations of bacterial colony morphology on MRSA media are circular with milky white on the edges. The acquired bacterial isolates were then cultured on an MRS agar medium with 0.3% calcium carbonate. Two LAB isolates, SBM3D and SBM4A, had the highest amylase activity when measured by their capacity to hydrolyze starch. These two strains were Gram-positive, cocci in shape, non-motile, and catalase negative. Several studies have been carried out on LAB isolation. For instance, Susilawati [16], Aguswinarto [23], and Kusumaningrum [24] reported that from the white rice washing water, Wikau Maombo, cabbage vegetables, and sago starch fermentation.
Strains with amylolytic ability will hydrolyze starch in the media around their growth sites with the non-formation of dark blue color in the degradation zone for detecting and selecting amylolytic strains. According to Putri et al. [25], the clear zone will appear when iodine solution is added after a few times. The starch in the LAB medium will provide a clear zone around the place of growth caused by exoenzymes and organisms that hydrolyze starch in an agar medium. Therefore, the amylase enzyme in glucose has hydrolyzed the starch grown by bacteria. The starch hydrolysis reaction in bacteria was characterized by the appearance of a clear area around the growth of the inoculated bacteria. Similar results were also obtained by Kusumaningrum et al. [24], who isolated LAB from sago starch and reported that 10 isolates produced clear zones on MRS agar medium.
LAB grows by utilizing the starch present in the medium to provide a clear zone around the place. Putri et al. [25] also reported the presence of 13 LAB isolates from growol fermentation, which produced a clear zone on the MRS agar medium. Meanwhile, Elvira et al. [26] also noted that the hydrolysis test of starch extracted from the Wikau Maombo fermentation obtained 8 LAB isolates, producing a clear zone around LAB colonies. This fact was due to secondary metabolites or other antimicrobial active compounds produced by LAB isolates in a near-death phase.
The bacterial growth curve is related to incubation time, which enables the lactic acid bacteria to undergo several phases as the incubation time progresses. The growth curve of lactic acid bacteria has four stages: lag, log, stationary, and death. The lag phase is the adaptation of the bacteria to the environment, while the log is characterized by rapid growth. The stationary phase is when the bacterial growth rate equals the death rate. Incubation time and bacterial growth phase also play an important role in producing maximum amylase enzyme activity [27].
The SBM3D and SBM4A growth curves started with a lag phase (adaptation) at 0 to 3 h, whereby the bacteria adjusted to the new environment by adding a significant absorbance value. The exponential or lag phase is when the microorganism cells are stable and can divide multiple times. The logarithmic phase occurred from the 3rd to the 18th h for SBM3D and SBM4A bacteria. Research conducted by Ramadhan and Wikandari [27] illustrated that L. mesentroides SU-LS 67 bacteria experienced a log phase of bacterial growth at the incubation time of 18 to 24 h. At the 24-h incubation time, it experienced the maximum amylase enzyme activity of 2.50 U/ml. In this phase, the growth of bacteria takes place very quickly, and it multiplies rapidly, doubling the previous population. In this phase, energy requirements also increase compared to the adaptation phase.
Furthermore, the stationary phase occurs from the 27th to the 48th hour, where the growth curve is relatively constant. In the stationary phase, the number of growing cells equals the number of dead ones with a decrease in size [28]. In the stationary phase, the highest number of bacterial populations is still experiencing a rise in bacterial populations, where lactic acid experience a bacterial growth rate equal to the death rate. Therefore, lactic acid bacteria can still produce maximum enzyme activity at certain incubation times in the log and stationary phases [27]. There are differences in the incubation time of SBM3D and SBM4A bacteria in producing maximum amylase enzyme activity. Varying types of bacteria can cause these differences because each bacterium has a specific ability to adapt to its optimum conditions.
Pediococcus pentosaceus is usually used as a starter for fermented beverages, meat, and vegetables. The nutritional value, texture, flavor, aroma, color, and digestibility of fermented goods are all influenced by Pediococcus pentosaceus. Giving Pediococcus pentosaceus at a dose of 3 mL (3.81 × 107 CFU/g) improved egg quality by increasing the Haugh Units of eggs while not affecting the thickness of the eggshell or the color of the yolk in Pitalah ducks [5]. In comparison to strain TC50, Pediococcus pentosaceus TC48 exhibited a better capacity to ferment silage powder by increasing the content of lactic acid silage. The extracellular supernatant from the two strains, TC48 and TC50, displayed strong antibacterial and probiotic activities and enhanced the best silage quality [29].
In molecular analysis, DNA is one of the main target materials in the cell’s nucleus. Bacterial chromosomes are not surrounded by a bran-bound nucleus but are located in the cytoplasm of the bacterial cell. Bacteria have only one circular chromosome that is not packaged in a true nucleus, with all the genes arranged along the circle.
The selection of DNA isolation methods from microbial groups needs to consider the diversity of creatures, such as the susceptibility of their cell walls to the breakdown method, the content of microbial polysaccharides, and the bonding of DNA with proteins in cells. Gram-negative bacteria have a thinner cell wall than Gram-positive bacteria, which have a polysaccharide layer. Therefore, obtaining the 16S rRNA gene in gram-negative bacteria starts with destroying the cell wall and membrane and separating DNA from other chromosomes, such as proteins.
The 16S rRNA gene was used to determine kinship size because (1) it is a universal machine for protein synthesis, (2) the base sequences are conservative, (3) abundant in the cell, (4) it meets the size for statistical calculations, and (5) the availability of sufficient information. Many positions in the 16S rRNA and 23S-rRNA molecules that evolved independently provided data to predict the phylogenetic relationship of a group of microbes [30].
The 16S rRNA gene sequences of SBM3D and SBM4A isolates were analyzed to determine the identity with the data in GenBank using the BLAST-N (Basic Local Alignment Search Tool-Nucleotide) program. The BLAST parameters checked are query coverage and maximum identity. The results of the query coverage stated that the percentage of the total nucleotide sequence length of SBM3D and SBM4A was good enough to be aligned with the nucleotide sequences owned by GenBank. Meanwhile, the maximum identity parameter was used to determine the percentage identity between the aligned nucleotide sequences of both isolates.
Based on the analysis of the BLAST-N program, it was found that SBM3D isolates 100% identity with the Pediococcus pentosaceus strain 8362 16S, Pediococcus pentosaceus strain 6340 16S, and Pediococcus pentosaceus strain SRCM102734. Meanwhile, SBM4A isolates had a 99.93% identity with Pediococcus pentosaceus strain 8362 16S, Pediococcus pentosaceus strain 6643 16S, and Pediococcus pentosaceus strain 6340 16S. Isolates with a 16S rRNA sequence homology level of more than 97% can represent the same species level. In comparison, those between 93–97% denote the identity of bacteria at the genus at the varying species level. If the homology level is below 93%, it is a new species whose nitrogen base sequence has not been included in the GenBank database.
Pediococcus pentosaceus is a lactic acid bacterium that is tolerant to acid. The development of this bacterium is very important due to its ability as a starter culture to ferment various foods such as meat, vegetables, cheese [31], and dadih [32]. Rosyidah et al. [33] and Pato et al. [34] stated that P. pentosaceus produced high lactic acid and bacteriocin, with a high OD value in the logarithmic phase. The number of similar sequences unites the phylogenetic analysis on lactic acid bacteria. Therefore, the phylogenetic tree SBM3D had a sister clade group Pediococcus pentosaceus strain SRCM102740 CP028269.1 and Pediococcus pentosaceus strain SL001 CP039378.1 forming a sister clade with a bootstrap value of 99, and forming a monophyletic group Pediococcus pentosaceus strain 8362 16S MT538852, Pediococcus pentosace363743 MT53643. 1 and Pediococcus pentosaceus strain GDIAS001 CP046938. While the bacterial isolate SBM4A has a sister group clade with Pediococcus pentosaceus strain SRCM102739 CP028266.1 and Pediococcus pentosaceus strain SRCM102738 CP028264.1 forming a clade sister group with a bootstrap value of 76 and forming a monophyletic group Pediococcus pentosaceus strain SRCM102736, Pediococcus pentosaceus strain CP02825. 16S MT538852.1 and Pediococcus pentosaceus strain 6643 16S MT463851.1
With a bootstrap value of 76, Dharmayanti [35] reported that the identity value is used to test the quality of the model data set, to determine if the identity value is low, and the ability of the sequence to obtain a phylogenetic tree. Muzzazinah [36] stated that a tall and good phylogenetic tree has a bootstrap value above 70. Therefore, SBM3D and SBM4A isolates can be classified into the Pediococcus pentosaceus species.
Preliminary studies reported that the bacterial isolates produced from brown rice washing water are included in the LAB group. The findings of the previous study [37] obtained 10 LAB isolates that had Gram-positive characteristics, cocci-shaped cells, non-motile, catalase-negative, and facultative anaerobes. These characteristics have similarities with the LAB obtained in this study. Sulistiani and Hidayat [38] stated that there were six species of lactic acid bacteria in the tape samples: Pediococcus pentosaceus, Lactobacillus vini, L. plantarum, L. kunkeei, L. fermentum, W. paramesenteroides, and Leuconostoc mesenteroides.
5. Conclusions
SBM3D and SBM4A, amylase-producing LAB from fermented brown rice washing water, were discovered in the current investigation. The cladogram of the SBM3D is between Pediococcus pentosaceus strain SRCM102740 CP028269.1 and Pediococcus pentosaceus strain SL001 CP039378.1 forming a sister group cladogram. SBM4A cladogram between Pediococcus pentosaceus strain SRCM102739 CP028266.1 and Pediococcus pentosaceus strain SRCM102738 CP028264.1 forming a sister group clad. Finally, by sequencing and aligning their 16s RNA, we found two strains of Pediococcus pentosaceus that have potential for use in the food industry and other related industries.
Author Contributions
Conceptualization, S.W.; methodology, S.W., P.E.S. and A.K.; validation, A. and S.; formal analysis, A.K. and W.O.G.P.; investigation, S.W. and P.E.S.; data curation, S.; writing—original draft preparation, W.O.G.P.; writing—review and editing, U.P., S.W. and W.O.G.P.; visualization, W.O.G.P. and S.; supervision, S.W., P.E.S. and A.K.; project administration, S.W. All authors have read and agreed to the published version of the manuscript.
Funding
Ministry of Education, Culture, Research, and Technology of the Republic of Indonesia with Contract Number: 019/E5/PG.02.00.PT/2022.
Data Availability Statement
No applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yulianingsih, R. Effect of rice washing water on growth and yield of purple eggplant (Solanum melongena L.). Piper 2017, 24, 61–68. [Google Scholar]
- Asngad, A.; Astuti, P.; Rahmawati, I.N. Utilization of rice washings water waste of IR-36 and IR-64 (Leri water) for producing syrup by fermentation process with the addition of rosella flowers as natural dyes. Semin. Nas. X Pendidik Biol. FKIP UNS 2013, 64–72. [Google Scholar]
- Chethana, S.H.; Pratap, B.; Roy, S.; Jaiswal, A.; Shruthi, S.D.; Vedamurthy, A.B. Bioethanol Production from Rice Water Waste: A Low-Cost Motor Fuel. Pharmacologyonline 2011, 3, 125–134. [Google Scholar]
- Emmawati, A.; Sri, B.; Nuraida, L.; Syah, D. Characterization of lactic acid bacteria isolates from mandai function as probiotics. Agritech 2015, 35, 146–155. [Google Scholar] [CrossRef]
- Yunenshi, F. The effect of giving Pediococcus pentosaceus probiotics from hybrid cocoa fermentation to reduce cholesterol in pitalah duck eggs. Program Pascasarjana, Universitas Andalas, Padang, Indonesia, 2011.
- Camargo, C.; Colona, P.; Buleon, A.; Molard, D. Functional properties of sour cassava (Manihot utilissima) starch: Polvilho Azedo. J. Sci. Food Agric. 1988, 3, 429–435. [Google Scholar] [CrossRef]
- Reddy, G.; Altaf, M.; Naveena, B.J.; Venkateshwar, M.; Kumar, E.V. Amylolytic bacterial lactic acid fermentation—A Review. Biotechnol. Adv. 2008, 26, 22–34. [Google Scholar] [CrossRef]
- Petrov, K.; Urshev, Z.; Petrova, P. L(+)-Lactic acid production from starch by a novel amylolytic Lactococcus lactis subsp. lactis B84. Food Microbiol. 2008, 25, 550–557. [Google Scholar] [CrossRef]
- Souza, P.M.D.; Magalhães, P.D.O. Application of microbial α-amylase in industry—A Review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef]
- Ardhi, A.; Sidauruk, A.N.; Suraya, N.; Pratiwi, N.W.; Pato, U. Molecular identification of amylase-producing thermophilic bacteria isolated from Bukit Gadang Hot Spring, West Sumatra, Indonesia. Biodiversitas 2020, 21, 994–1000. [Google Scholar] [CrossRef]
- Aris, M. Identification, the Pathogenicity of Bacteria and the Use of Gene 16S rRNA for Ice-Ice Detection on Seaweed Aquaculture (Kappaphycus alvarezii); Institut Pertanian Bogor: Bogor, Indonesia, 2011. [Google Scholar]
- Madigan, M.T.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. Diversity of Bacteria. Brock Biology of Microorganisms; Prentice Hall Inc.: Upper Saddle River, NJ, USA, 2016. [Google Scholar]
- Kasi, P.D.; Ariandi Tenriawaru, E.P. Identification of lactic acid bacteria from sago wastewater with 16S rRNA gene. Maj. Ilm. Biol. Biosf. 2019, 36, 35–40. [Google Scholar]
- Barus, T.; Chalista, S.; Lay, B.W. Identification and genetic diversity of lactic acid bacteria from cassava Tapai based on 16S rRNA gene. Biota 2017, 2, 46–52. [Google Scholar]
- Kenneth, T. Lactic Acid Bacteria. In Today’s Online Textbook of Bacteriology; University of Wisconsin: Madison, WI, USA, 2004; p. 530. [Google Scholar]
- Susilawati, S. Isolation and Characterization of Lactic acid Bacteria (LAB) from Fermented Rice Rinsed Water; Universitas Syarif Hidayatullah: Jakarta, Indonesia, 2016. [Google Scholar]
- Ikeda, H.; Delargy, A.H.; Yokogawa, T.; Urban, J.M.; Burgess, H.A.; Ono, F. Intrinsic Properties of Larval Zebrafish Neurons in Ethanol. PLoS ONE 2013, 8, 1–7. [Google Scholar] [CrossRef]
- Yusmarini Indrati, R.; Utami, T.; Marsono, Y. Isolation and identification of proteolytic lactic acid bacteria from spontaneously fermented soy milk. J. Natur. Indones. 2009, 12, 28–33. [Google Scholar]
- Jennifer, V.; Thiruneelakandan, G. Enzymatic activity of marine Lactobacillus species from the southeast coast of India. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 542–546. [Google Scholar]
- Apriani, K.; Haryani, Y.; Kartika, G.F. Production and test of cellulase activity from cellulolytic bacteria isolates from Indragiri river. JOM Fmipa 2014, 1, 261–267. [Google Scholar]
- Barus, T.; Wijaya, N. Dominant microbiota and its role in the taste of cassava tape. J. Biota 2011, 16, 354–361. [Google Scholar] [CrossRef]
- Pangestika, Y.; Budiharjo, A.; Kusumaningrum, H.P. Phylogenetic analysis of Curcuma zedoaria based on internal transcribed spacer (ITS) gene. J. Biol. 2015, 4, 8–13. [Google Scholar]
- Aguswinarto Wahyuni, S.; Khaeruni, A. The effect of adding lactic acid bacteria (LAB) from Wikau Maombo and fermentation time on the organoleptic characteristics of palm sugar probiotic drinks. J. Sains Dan Teknol. Pangan. 2016, 1, 184–192. [Google Scholar]
- Kusumaningrum, Y.; Ali, A. Isolation and identification of amylolytic lactic acid bacteria from processing industry sago starch. JOM Faperta 2015, 2, 55–60. [Google Scholar]
- Putri, W.D.R.; Haryadi Marseno, D.W.; Cahyanto, M.N. Isolation and characterization of amylolytic lactic acid bacteria during growol fermentation, an Indonesian traditional food. J. Teknol. Pertan. 2012, 13, 52–60. [Google Scholar]
- Elvira, I.; Wahyuni, S.; Asyik, N. Characterization of biochemical properties of lactic acid bacteria isolates produced from the Wikau Maombo fermentation process. J. Sains. Dan Teknol. Pangan. 2016, 1, 121–124. [Google Scholar]
- Ramadhan, B.; Wikandari, P.R. Review Article: Activities enzyme amylase from lactic acid bacteria: Characterization and application. Unesa J. Chem. 2021, 10, 109–120. [Google Scholar] [CrossRef]
- Kumar, M.; Tiwari, S.; Srivastava, S. Purification and characterization of enterocin LR/6, a bacteriocin from Enterococcus faecium LR/6. Appl. Biochem. Biotechnol. 2010, 160, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Soundharrajan, I.; Kim, D.; Kuppusamy, P.; Muthusamy, K.; Lee, H.J.; Choi, K.C. Probiotic and Triticale Silage Fermentation Potential of Pediococcus pentosaceus and Lactobacillus brevis and their Impacts on pathogenic bacteria. Microorganisms 2019, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Bollet, C.; Carlioz, A.; Martelin, R.; Gayral, J.P.; Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000, 38, 3623–3630. [Google Scholar] [CrossRef]
- Yeni, M.A.; Sunarti, T.C. The use of tofu whey for biomass production by Pediococcus pentosaceus E.1222. J. Teknol. Ind. Pertan. 2016, 26, 284–293. [Google Scholar]
- Pato, U.; Riftyan, E.; Jonnaidi, N.N.; Wahyuni, M.S.; Feruni, J.A.; Abdel-Wahhab, M.A. Isolation, characterization, and antimicrobial evaluation of bacteriocin produced by lactic acid bacteria against Erwinia carotovora. Food Sci. Technol. 2022, 42, 1–7. [Google Scholar] [CrossRef]
- Rosyidah, E.; Meryandini, A.; Sunarti, T.C. Isolation of lactic acid and cellulolytic bacteria and their application to improve the quality of corn flour. In Mathematics and Natural Science; IPB University: Bogor, Indonesia, 2013. [Google Scholar]
- Pato, U.; Riftyan, E.; Ayu, D.F.; Jonnaidi, N.N.; Wahyuni, M.S.; Feruni, J.A.; Abdel-Wahhab, M.A. Antibacterial efficacy of lactic acid bacteria and bacteriocin isolated from Dadih against Staphylococcus aureus. Food Sci. Technol. 2022, 42, 1–6. [Google Scholar] [CrossRef]
- Dharmayanti, I. Molecular phylogenetic: Organism taxonomy method based on evolutionary history. Wartazoa 2011, 21, 1–10. [Google Scholar]
- Muzzazinah. The Phylogenetic Method in Indigofera. Pros Semin. Nas Pendidik Biol. Dan Biol. 2017, 25–40. [Google Scholar]
- Adnan, N.S.; Wahyuni, S.; Khaeruni, A. Testing properties of amylolytic and proteolytic of lactic acid bacteria (LAB) isolates from fermented brown rice washing water cultivars of Waka Wondu. J. Sains Dan Teknol. Pangan. 2017, 2, 759–769. [Google Scholar]
- Sulistiani, H.I. Molecular identification of lactic acid bacteria from Tempe and Tape based on 16S rRNA gene sequences. Maj. Ilm Biol. Biosf. A Sci. J. 2020, 37, 69–77. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).