Fermentation in Minimal Media and Fungal Elicitation Enhance Violacein and Deoxyviolacein Production in Two Janthinobacterium Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Media for Bacterial Cultivation
2.2. Bacterial Strains
2.3. Cryo-Culture Preparation
2.4. Pre-Culture Preparation
2.5. Bacterial Growth Measurement
2.6. Large-Scale Fermentation
2.7. Extraction, Fractionation, and Isolation of Violacein and Deoxyviolacein
2.8. Mass Spectrometry Analysis
2.9. Antibacterial Assay
2.10. Statistical Analysis
3. Results
3.1. Bioinformatic Analysis of the Strains and Isolation of Violacein and Deoxyviolacein
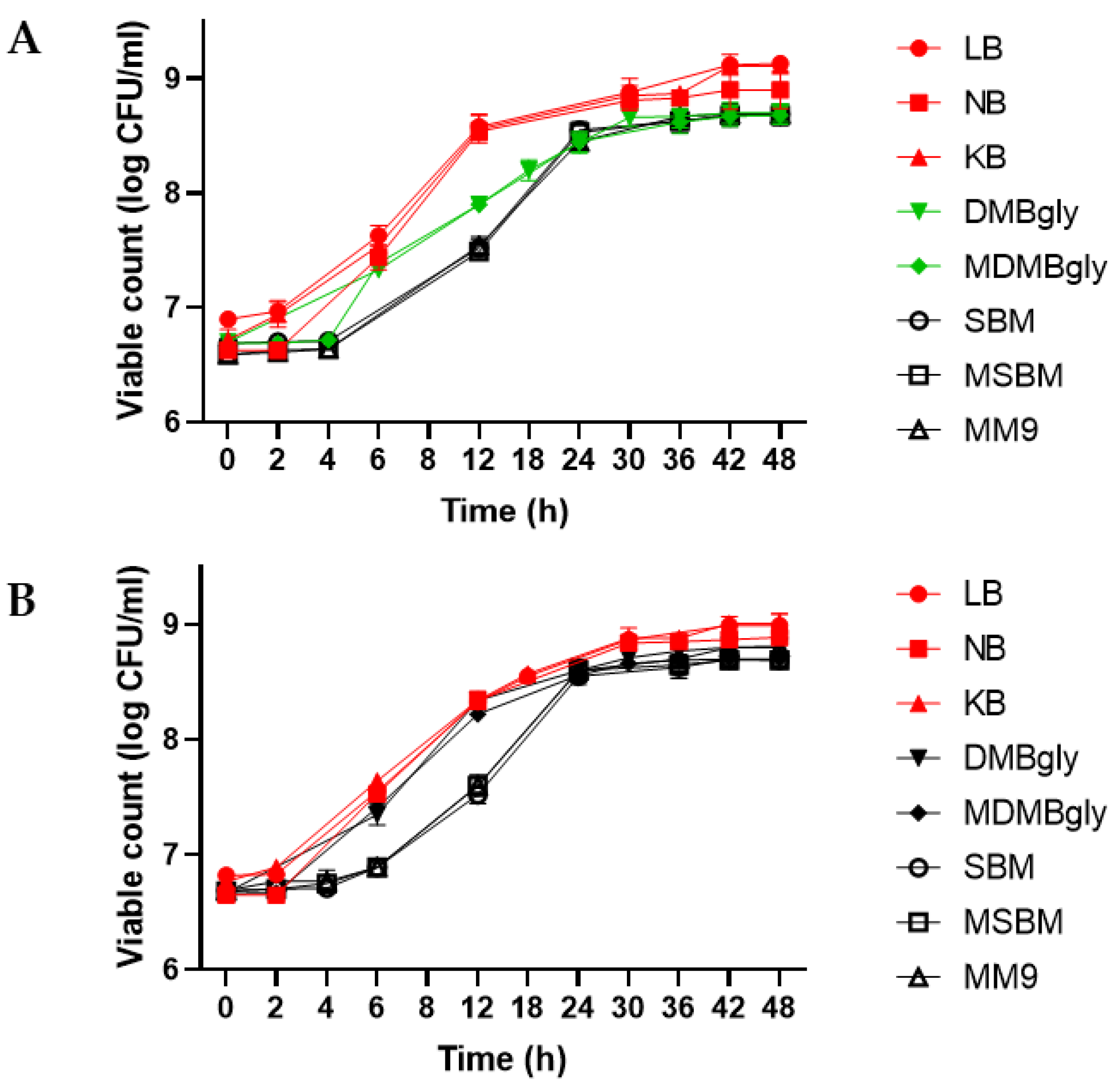
3.2. The Growth Rate of Janthinobacterium agaricidamnosum DSM 9628 and Janthinobacterium lividum DSM 1552 in Different Culture Media
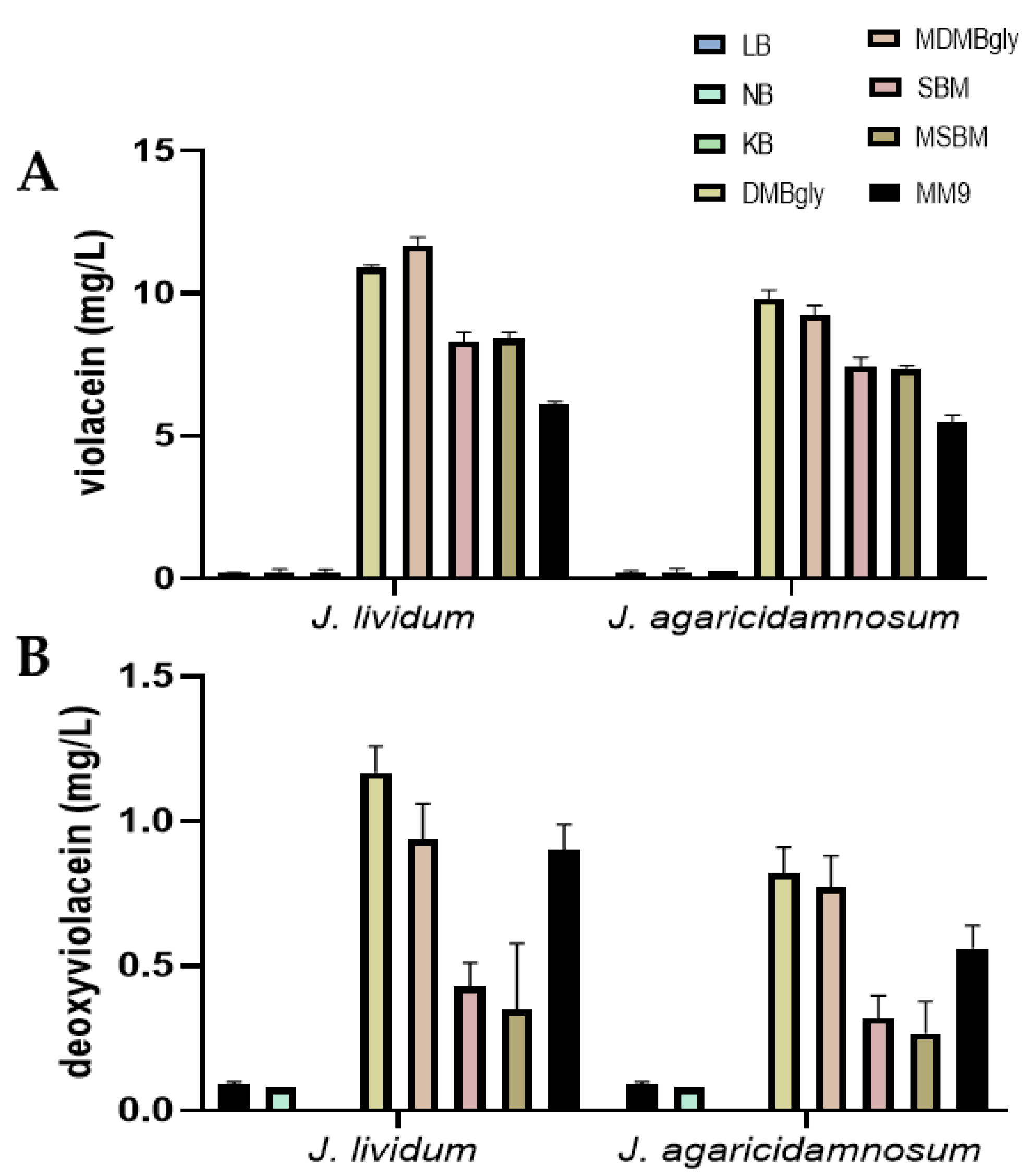
3.3. The Production Rate of Violacein and Deoxyviolacein in Different Culture Media
3.4. Agitation during Culture and Fungal Elicitation Could Not Increase Violacein Production
3.5. Antibacterial Activities of Violacein and Deoxyviolacein Isolated from J. agaricidamnosum DSM 9628 and J. lividum DSM 1552
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sen, T.; Barrow, C.J.; Deshmukh, S.K. Microbial pigments in the food industry—Challenges and the way forward. Front. Nutr. 2019, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Tuli, H.S.; Chaudhary, P.; Beniwal, V.; Sharma, A.K. Microbial pigments as natural color sources: Current trends and future perspectives. J. Food Sci. 2015, 52, 4669–4678. [Google Scholar] [CrossRef]
- Kanelli, M.; Mandic, M.; Kalakona, M.; Vasilakos, S.; Kekos, D.; Nikodinovic-Runic, J.; Topakas, E. Microbial production of violacein and process optimization for dyeing polyamide fabrics with acquired antimicrobial properties. Front. Microbiol. 2018, 9, 1495. [Google Scholar] [CrossRef] [PubMed]
- Füller, J.J.; Röpke, R.; Krausze, J.; Rennhack, K.E.; Daniel, N.P.; Blankenfeldt, W.; Schulz, S.; Jahn, D.; Moser, J. Biosynthesis of violacein, structure and function of l-Tryptophan oxidase VioA from Chromobacterium violaceum. J. Biol. Chem. 2016, 291, 20068–20084. [Google Scholar] [CrossRef] [PubMed]
- Wille, G.; Steglich, W. A short synthesis of the bacterial pigments violacein and deoxyviolacein. Synthesis 2001, 2001, 759–762. [Google Scholar] [CrossRef]
- Hashimi, S.M.; Xu, T.; Wei, M.Q. Violacein anticancer activity is enhanced under hypoxia. Oncol. Rep. 2015, 33, 1731–1736. [Google Scholar] [CrossRef]
- Rodrigues, A.L.; Trachtmann, N.; Becker, J.; Lohanatha, A.F.; Blotenberg, J.; Bolten, C.J.; Korneli, C.; de Souza Lima, A.O.; Porto, L.M.; Sprenger, G.A. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 2013, 20, 29–41. [Google Scholar] [CrossRef]
- Sasidharan, A.; Sasidharan, N.K.; Amma, D.B.N.S.; Vasu, R.K.; Nataraja, A.V.; Bhaskaran, K. Antifungal activity of violacein purified from a novel strain of Chromobacterium sp. NIIST (MTCC 5522). J. Microbiol. 2015, 53, 694–701. [Google Scholar]
- Wang, H.; Wang, F.; Zhu, X.; Yan, Y.; Yu, X.; Jiang, P.; Xing, X.-H. Biosynthesis and characterization of violacein, deoxyviolacein and oxyviolacein in heterologous host, and their antimicrobial activities. Biochem. Eng. J. 2012, 67, 148–155. [Google Scholar] [CrossRef]
- Andrighetti-Fröhner, C.; Antonio, R.; Creczynski-Pasa, T.; Barardi, C.; Simões, C. Cytotoxicity and potential antiviral evaluation of violacein produced by Chromobacterium violaceum. Memórias Inst. Oswaldo Cruz 2003, 98, 843–848. [Google Scholar] [CrossRef]
- Tsukimoto, A.; Saito, R.; Hashimoto, K.; Takiguchi, Y.; Miyano-Kurosaki, N. Violacein induces various types of cell death in highly differentiated human hepatocellular carcinoma and hepatitis C virus replicon cell lines. Int. J. Anal. Bio-Sci. 2021, 9, 46–56. [Google Scholar]
- Konzen, M.; De Marco, D.; Cordova, C.A.; Vieira, T.O.; Antonio, R.V.; Creczynski-Pasa, T.B. Antioxidant properties of violacein: Possible relation on its biological function. Bioorganic Med. Chem. 2006, 14, 8307–8313. [Google Scholar] [CrossRef] [PubMed]
- Vishnu, T.; Palaniswamy, M. Systematic Approach on Evaluating the in vitro Antioxidant Activity of Violacein; Novel Isolate Chromobacterium Vaccinii CV5. Biomed. Pharmacol. J. 2018, 11, 703–709. [Google Scholar]
- Choi, S.Y.; Kim, S.; Lyuck, S.; Kim, S.B.; Mitchell, R.J. High-level production of violacein by the newly isolated Duganella violaceinigra str. NI28 and its impact on Staphylococcus aureus. Sci. Rep. 2015, 5, 15598. [Google Scholar]
- Subramaniam, S.; Ravi, V.; Sivasubramanian, A. Synergistic antimicrobial profiling of violacein with commercial antibiotics against pathogenic micro-organisms. Pharm. Biol. 2014, 52, 86–90. [Google Scholar] [CrossRef]
- Cauz, A.C.; Carretero, G.P.; Saraiva, G.K.; Park, P.; Mortara, L.; Cuccovia, I.M.; Brocchi, M.; Gueiros-Filho, F. Violacein targets the cytoplasmic membrane of bacteria. ACS Infect. Dis. 2019, 5, 539–549. [Google Scholar] [CrossRef]
- Choi, S.Y.; Yoon, K.-H.; Lee, J.I.; Mitchell, R.J. Violacein: Properties and production of a versatile bacterial pigment. BioMed Res. Int. 2015, 2015, 465056. [Google Scholar] [CrossRef]
- Pemberton, J.M.; Vincent, K.M.; Penfold, R.J. Cloning and heterologous expression of the violacein biosynthesis gene cluster from Chromobacterium violaceum. Curr. Microbiol. 1991, 22, 355–358. [Google Scholar] [CrossRef]
- Zhang, X.; Enomoto, K. Characterization of a gene cluster and its putative promoter region for violacein biosynthesis in Pseudoalteromonas sp. 520P1. Appl. Microbiol. Biotechnol. 2011, 90, 1963–1971. [Google Scholar] [CrossRef]
- Park, H.; Park, S.; Yang, Y.-H.; Choi, K.-Y. Microbial synthesis of violacein pigment and its potential applications. Crit. Rev. Biotechnol. 2021, 41, 879–901. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, P.; Lu, Y.; Ruan, Z.; Jiang, R.; Xing, X.-H.; Lou, K.; Wei, D. Optimization of culture conditions for violacein production by a new strain of Duganella sp. B2. Biochem. Eng. J. 2009, 44, 119–124. [Google Scholar] [CrossRef]
- Hoshino, T. Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: Biosynthetic mechanism and pathway for construction of violacein core. Appl. Microbiol. Biotechnol. 2011, 91, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Hakvåg, S.; Fjærvik, E.; Klinkenberg, G.; Borgos, S.E.F.; Josefsen, K.D.; Ellingsen, T.E.; Zotchev, S.B. Violacein-producing Collimonas sp. from the sea surface microlayer of costal waters in Trøndelag, Norway. Mar. Drugs 2009, 7, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Miess, H.; Frediansyah, A.; Göker, M.; Gross, H. Draft genome sequences of six type strains of the genus Massilia. Microbiol. Resour. Announc. 2020, 9, e00226-20. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xiong, H.; Lee, O.O.; Qi, S.H.; Qian, P.Y. Effect of agitation on violacein production in Pseudoalteromonas luteoviolacea isolated from a marine sponge. Lett. Appl. Microbiol. 2007, 44, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Pantanella, F.; Berlutti, F.; Passariello, C.; Sarli, S.; Morea, C.; Schippa, S. Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 2007, 102, 992–999. [Google Scholar] [CrossRef]
- Mendes, A.S.; de Carvalho, J.E.; Duarte, M.C.; Durán, N.; Bruns, R.E. Factorial design and response surface optimization of crude violacein for Chromobacterium violaceum production. Biotechnol. Lett. 2001, 23, 1963–1969. [Google Scholar] [CrossRef]
- Gohil, N.; Bhattacharjee, G.; Gayke, M.; Narode, H.; Alzahrani, K.J.; Singh, V. Enhanced production of violacein by Chromobacterium violaceum using agro-industrial waste soybean meal. J. Appl. Microbiol. 2022, 132, 1121–1133. [Google Scholar] [CrossRef]
- Alem, D.; Marizcurrena, J.J.; Saravia, V.; Davyt, D.; Martinez-Lopez, W.; Castro-Sowinski, S. Production and antiproliferative effect of violacein, a purple pigment produced by an Antarctic bacterial isolate. World J. Microbiol. Biotechnol. 2020, 36, 120. [Google Scholar] [CrossRef]
- Frediansyah, A.; Straetener, J.; Brötz-Oesterhelt, H.; Gross, H. Massiliamide, a cyclic tetrapeptide with potent tyrosinase inhibitory properties from the Gram-negative bacterium Massilia albidiflava DSM 17472T. J. Antibiot. 2021, 74, 269–272. [Google Scholar] [CrossRef]
- Frediansyah, A.; Romadhoni, F.; Nurhayati, R.; Wibowo, A.T. Fermentation of Jamaican cherries juice using Lactobacillus plantarum elevates antioxidant potential and inhibitory activity against Type II diabetes-related enzymes. Molecules 2021, 26, 2868. [Google Scholar] [CrossRef]
- Jiao, J.; Du, J.; Frediansyah, A.; Jahanshah, G.; Gross, H. Structure elucidation and biosynthetic locus of trinickiabactin from the plant pathogenic bacterium Trinickia caryophylli. J. Antibiot. 2020, 73, 28–34. [Google Scholar] [CrossRef]
- Lincoln, S.P.; Fermor, T.R.; Tindall, B. Janthinobacterium agaricidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int. J. Syst. Evol. Microbiol. 1999, 49, 1577–1589. [Google Scholar] [CrossRef]
- Stein, J.; Schlosser, N.; Bardl, B.; Peschel, G.; Meyer, F.; Kloss, F.; Rosenbaum, M.A.; Regestein, L. Scalable Downstream Method for the Cyclic Lipopetide Jagaricin; Wiley Online Library: New York, NY, USA, 2021. [Google Scholar]
- Graupner, K.; Lackner, G.; Hertweck, C. Genome sequence of mushroom soft-rot pathogen Janthinobacterium agaricidamnosum. Genome Announc. 2015, 3, e00277-15. [Google Scholar] [CrossRef]
- Cazoto, L.L.; Martins, D.; Ribeiro, M.G.; Durán, N.; Nakazato, G. Antibacterial activity of violacein against Staphylococcus aureus isolated from bovine mastitis. J. Antibiot. 2011, 64, 395–397. [Google Scholar] [CrossRef]
- Kim, J.; Kim, K.H. Effects of minimal media vs. complex media on the metabolite profiles of Escherichia coli and Saccharomyces cerevisiae. Process Biochem. 2017, 57, 64–71. [Google Scholar]
- Antônio, R.V.; Creczynski-Pasa, T.B.J.G.M.R. Genetic analysis of violacein biosynthesis by Chromobacterium violaceum. Genet. Mol. Res. 2004, 3, 85–91. [Google Scholar] [PubMed]
- Silva-Santisteban, B.O.Y.; Maugeri Filho, F. Agitation, aeration, and shear stress as key factors in inulinase production by Kluyveromyces marxianus. Enzyme Microb. Technol. 2005, 36, 717–724. [Google Scholar] [CrossRef]
- Heinemann, B.; Howard, A.J.; Palocz, H. Influence of dissolved oxygen levels on production of L-asparaginase and prodigiosin by Serratia marcescens. Appl. Microbiol. 1970, 19, 800–804. [Google Scholar] [CrossRef]



| Cultivation Media | Constituent per L |
|---|---|
| Luria–Bertani (LB) | 10 g tryptone, 5 g yeast extract, 10 g NaCl; final pH 7.5 at 25 °C |
| Nutrient Broth (NB) | 5 g peptone, 2 g yeast extract, 5 g NaCl; final pH 7.4 at 25 °C |
| King’s B Medium (KB) | 20 g proteose peptone, 1.5 g K2HPO4, 1.5 g MgSO4, 15 g glycerol; final pH 7.5 at 25 °C |
| Davis Minimal Broth with Glycerol (DMBgly) | 10.6 g Davis minimal broth without dextrose, 1.83 mL of 100% glycerol; pH adjustment not necessary |
| Modified DMBgly (MDMBgly) | 5.3 g Davis minimal broth without dextrose, 0.56 g HEPES buffer, 5.1 g sucrose, 1 mg methionine, 1 mL trace metal mix A5 with Co; final pH 6.6 at 25 °C |
| Shipworm Basal Medium (SBM) without sea salt | 15.3 mg KH2PO4, 10 mg Na2CO3, 2.5 mg Na2MoO4.2H2O, 0.5 mg EDTA, 3 mg C6H8FeNO7, 5.2 g HEPES buffer, 1 mL trace metal mix A5 with Co; pH adjustment not necessary |
| Modified SBM (MSBM) without sea salt | 15.3 mg KH2PO4, 10 mg Na2CO3, 2.5 mg Na2MoO4.2H2O, 0.5 mg EDTA, 5.2 g HEPES buffer, 1 mL trace metal mix A5 with Co, 0.267 g NH4Cl, 5 g sucrose, 5 mg L-methionine; final pH 7.2 at 25 °C |
| MM9: | 900 ml solution C, 20 ml solution A, 20 ml solution B, 16.7 ml 100mM L-leucine, 5 ml 60mM L-histidine, 10 ml 100 mM L-lysine, 10 ml 40 mM L-tryptophan, 10 ml 40 mM L-methionine, 20 ml 50% (w/v) glucose, 1 mL trace metal mix A5 with Co; pH adjustment not necessary |
| Bacterial Strain | Submitter | Sources | NCBI Reference Sequence | Genome Size (Gb) | GC Content (%) |
|---|---|---|---|---|---|
| J. agaricidamnosum DSM 9628 | NITE Biological Resource Center, Japan | mushroom Agaricus bisporus | NZ_BCTH00000000.1 | 5.88 | 61.1 |
| J. lividum DSM 1552 | Georg-August-University Goettingen, Germany | from the soil, Michigan, USA | NZ_LRHW00000000.1 | 6.71 | 62.4 |
| Antibacterial Activities | MIC (µg/mL) | ||||
|---|---|---|---|---|---|
| Violacein | Deoxyviolacein | Tetracycline (+) | |||
| Jl | Ja | Jl | Ja | ||
| Pseudomonas aeruginosa DSM 50071 | 19.81 | 19.5 | 24.79 | 23.4 | 1.3 |
| Yersinia tuberculosis YPIII | >64 | >64 | >64 | >64 | 1.4 |
| Paenibacillus sp. AD64 | 14.49 | 14.2 | 15.35 | 16 | 1.1 |
| Bacillus subtilis ATCC 6051 | 16 | 16 | 16 | 16 | 1.1 |
| Staphylococcus aureus USA300 LAC | 7.89 | 8 | 8.02 | 8 | 4.2 |
| Mycobacterium phlei 01 | >64 | >64 | >64 | >64 | 5.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frediansyah, A.; Manuhara, Y.S.W.; Kristanti, A.N.; Luqman, A.; Wibowo, A.T. Fermentation in Minimal Media and Fungal Elicitation Enhance Violacein and Deoxyviolacein Production in Two Janthinobacterium Strains. Fermentation 2022, 8, 714. https://doi.org/10.3390/fermentation8120714
Frediansyah A, Manuhara YSW, Kristanti AN, Luqman A, Wibowo AT. Fermentation in Minimal Media and Fungal Elicitation Enhance Violacein and Deoxyviolacein Production in Two Janthinobacterium Strains. Fermentation. 2022; 8(12):714. https://doi.org/10.3390/fermentation8120714
Chicago/Turabian StyleFrediansyah, Andri, Yosephine Sri Wulan Manuhara, Alfinda Novi Kristanti, Arif Luqman, and Anjar Tri Wibowo. 2022. "Fermentation in Minimal Media and Fungal Elicitation Enhance Violacein and Deoxyviolacein Production in Two Janthinobacterium Strains" Fermentation 8, no. 12: 714. https://doi.org/10.3390/fermentation8120714
APA StyleFrediansyah, A., Manuhara, Y. S. W., Kristanti, A. N., Luqman, A., & Wibowo, A. T. (2022). Fermentation in Minimal Media and Fungal Elicitation Enhance Violacein and Deoxyviolacein Production in Two Janthinobacterium Strains. Fermentation, 8(12), 714. https://doi.org/10.3390/fermentation8120714







