Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Information
2.2. Bacterial Growth and DNA Extraction
2.3. Genome Sequencing, Assembly, and Bioinformatics Analysis
2.4. Cell Survival and L-Malic Acid Consumption in Synthetic Wine
2.4.1. Cell Culture and Acclimation
2.4.2. Vinification Assays at Laboratory Scale
2.5. Reproducibility of Results
3. Results
3.1. General Features of the Genome
3.2. Genes Encoding Enzymes Related to the Winemaking Process
3.3. Genes Encoding Proteins Related to Stress Responses
3.4. Genes Encoding Proteins That Deteriorate Wine
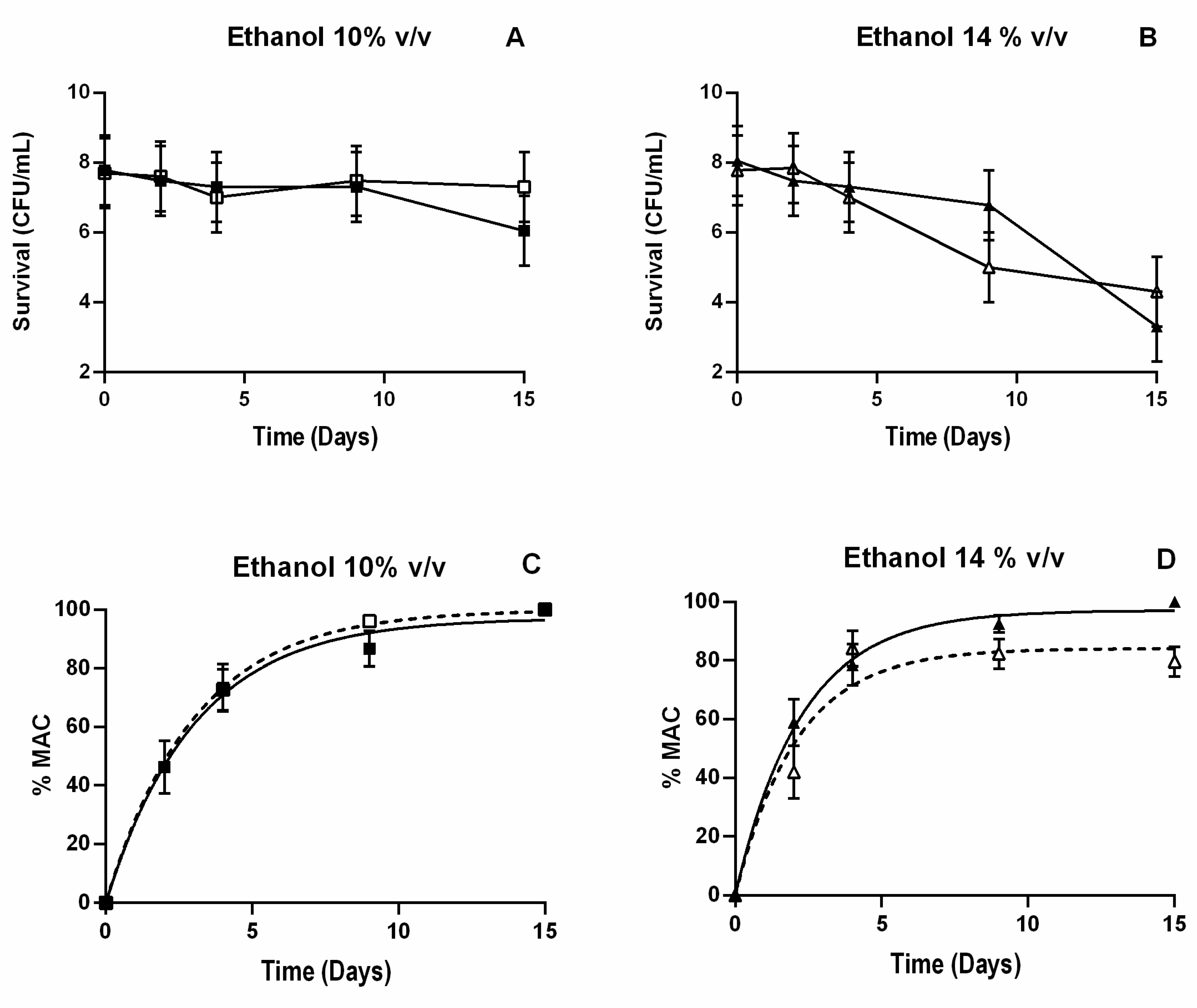
3.5. Cell Viability and L-Malic Acid Consumption
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumby, K.M.; Bartle, L.; Grbin, P.R.; Jiranek, V. Measures to improve wine malolactic fermentation. Appl. Microbiol. Biotechnol. 2015, 103, 2033–2051. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Franquès, J.; Araque, I.; Palahí, E.; del Carmen Portillo, M.; Reguant, C.; Bordons, A. Presence of Oenococcus oeni and other lactic acid bacteria in grapes and wines from Priorat (Catalonia, Spain). LWT 2017, 81, 326–334. [Google Scholar] [CrossRef]
- Cappello, M.S.; Zapparoli, G.; Logrieco, A.; Bartowsky, E.J. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int. J. Food Microbiol. 2017, 243, 16–27. [Google Scholar] [CrossRef]
- Lorentzen, M.P.; Lucas, P.M. Distribution of Oenococcus oeni populations in natural habitats. Appl. Microbiol. Biotechnol. 2019, 103, 2937–2945. [Google Scholar] [CrossRef]
- Lerm, E.; Engelbrecht, L.; Du Toit, M. Selection and characterisation of Oenococcus oeni and Lactobacillus plantarum South African wine isolates for use as malolactic fermentation starter cultures. S. Afr. J. Enol. Vitic. 2011, 32, 280–295. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Chiang, S.S.; Pan, T.M. Beneficial effects of Lactobacillus paracasei subsp. paracasei NTU 101 and its fermented products. Appl. Microbiol. Biotechnol. 2011, 93, 903–916. [Google Scholar] [CrossRef]
- Valdés La Hens, D.; Bravo-Ferrada, B.M.; Delfederico, L.; Caballero, A.C.; Semorile, L.C. Prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation in Patagonian red wines revealed by polymerase chain reaction-denaturing gradient gel electrophoresis with two targeted gene. Aust. J. Grape Wine Res. 2015, 21, 49–56. [Google Scholar] [CrossRef]
- du Plessis, H.W.; Dicks, L.M.T.; Pretorius, I.S.; Lambrechts, M.G.; Du Toit, M. Identification of lactic acid bacteria isolated from South African brandy base wines. Int. J. Food Microbiol. 2004, 91, 19–29. [Google Scholar] [CrossRef]
- Kačániová, M.; Kunová, S.; Sabo, J.; Ivanišová, E.; Žiarovská, J.; Felšöciová, S.; Fatrcová-Šramková, K.; Terentjeva, M. Isolation and Identification of Lactic Acid Bacteria in Wine Production by MALDI-TOF MS Biotyper. Acta Hortic. Regiotect. 2020, 23, 21–24. [Google Scholar] [CrossRef]
- López-Seijas, J.; García-Fraga, B.; da Silva, A.F.; Zas-García, X.; Lois, L.C.; Gago-Martínez, A.; Sieiro, C. Evaluation of malolactic bacteria associated with wines from Albariño variety as potential Starters: Screening for quality and safety. Foods 2020, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.; Tymczyszyn, E.E.; Semorile, L.C.; Valdes La Hens, D.; Delfederico, L.; Hollmann, A.; Bravo-Ferrada, B. Lactobacillus plantarum as a malolactic starter culture in winemaking: A new (old) player? Electron. J. Biotechnol. 2019, 38, 10–18. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; Valdés La Hens, D.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Brizuela, N.S.; La Hens, D.V.; Tymczyszyn, E.E.; Semorile, L. Growth and consumption of l-malic acid in wine-like medium by acclimated and non-acclimated cultures of Patagonian Oenococcus oeni strains. Folia Microbiol. 2016, 61, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; La Hens, D.V.; Hollmann, A.; Delfederico, L.; Caballero, A.; Tymczyszyn, E.E.; Semorile, L. Comparative vinification assays with selected Patagonian strains of Oenococcus oeni and Lactobacillus plantarum. LWT 2017, 77, 348–355. [Google Scholar] [CrossRef]
- Manera, C.; Olguin, N.T.; Bravo-Ferrada, B.M.; Tymczyszyn, E.E.; Delfederico, L.; Bibiloni, H.; Caballero, A.C.; Semorile, L.; La Hens, D.V. Survival and implantation of indigenous psychrotrophic Oenococcus oeni strains during malolactic fermentation in a Patagonian Pinot noir wine. LWT 2019, 108, 353–360. [Google Scholar] [CrossRef]
- Brizuela, N.S.; Bravo-Ferrada, B.M.; Curilén, Y.; Delfederico, L.; Caballero, A.; Semorile, L.; Tymczyszyn, E.E. Advantages of using blend cultures of native L. plantarum and O. oeni strains to induce malolactic fermentation of Patagonian Malbec wine. Front. Microbiol. 2018, 9, 2109. [Google Scholar] [CrossRef]
- Mendoza, S.N.; Canon, P.M.; Contreras, A.; Ribbeck, M.; Agosin, E. Genome-scale reconstruction of the metabolic network in Oenococcus oeni to assess wine malolactic fermentation. Front. Microbiol. 2017, 8, 534. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; El Khoury, M.; Favier, M.; Romano, A.; Biasioli, F.; Spano, G.; Sherman, D.J.; Bouchez, O.; Coton, E.; Coton, M.; et al. Phylogenomic Analysis of Oenococcus oeni Reveals Specific Domestication of Strains to Cider and Wines. Genome Biol. Evol. 2015, 7, 1506–1518. [Google Scholar] [CrossRef]
- Romero, J.; Ilabaca, C.; Ruiz, M.; Jara, C. Oenococcus oeni in Chilean Red Wines: Technological and Genomic Characterization. Front. Microbiol. 2018, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.G.; Brizuela, N.S.; Tymczyszyn, E.E.; Hollmann, A.; Valdés La Hens, D.; Semorile, L.; Bravo-Ferrada, B.M. Complete Genome Sequencing of Lactobacillus plantarum UNQLp 11 Isolated from a Patagonian Pinot Noir Wine. S. Afr. J. Enol. Vitic. 2020, 41, 197–209. [Google Scholar] [CrossRef]
- Delfederico, L.; Hollmann, A.; Martínez, M.; Iglesias, N.G.; De Antoni, G.; Semorile, L. Molecular identification and typing of lactobacilli isolated from kefir grains. J. Dairy Res. 2006, 73, 20. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; van Hylckama Vlieg, J.E.T. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 2011, 10, S3. [Google Scholar] [CrossRef]
- Altermann, E.; Russell, W.M.; Azcarate-Peril, M.A.; Barrangou, R.; Buck, B.L.; McAuliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M.; et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A Medium for the Cultivation of Lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Tymczyszyn, E.E.; Gómez-Zavaglia, A.; Semorile, L. Effect of acclimation medium on cell viability, membrane integrity and ability to consume malic acid in synthetic wine by oenological Lactobacillus plantarum strains. J. Appl. Microbiol. 2014, 116, 360–367. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; Van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Stott, C.M.; Bobay, L.M. Impact of homologous recombination on core genome phylogenies. BMC Genom. 2020, 21, 829. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, J.; Yang, P.; Wu, Z.; Zhang, J.; Du, G. Enhanced acid-stress tolerance in Lactococcus lactis NZ9000 by overexpression of ABC transporters. Microb. Cell Factories 2019, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Nauta, A.; Francke, C.; Siezen, R.J. Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria. Appl. Environ. Microbiol. 2008, 74, 4590–4600. [Google Scholar] [CrossRef] [PubMed]
- Chambellon, E.; Rijnen, L.; Lorquet, F.; Gitton, C.; Van Hylckama Vlieg, J.E.; Wouters, J.A.; Yvon, M. The D-2-hydroxyacid dehydrogenase incorrectly annotated PanE is the sole reduction system for branched-chain 2-keto acids in Lactococcus lactis. J. Bacteriol. 2009, 191, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J. Appl. Microbiol. 2005, 99, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Styger, G.; Prior, B.; Bauer, F.F. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 2011, 38, 1145. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.Z.; Zhou, T.; Xu, J.Q.; Wang, Y.X.; Sun, M.M.; He, Y.J.; Pan, S.W.; Xiong, W.; Peng, Z.K.; Gao, X.H.; et al. Structure, kinetic properties and biological function of mechanosensitive Piezo channels. Cell Biosci. 2021, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Biggin, P.C.; Sansom, M.S.P. Mechanosensitive Channels: Stress Relief. Curr. Biol. 2003, 13, R183–R185. [Google Scholar] [CrossRef]
- Stefanovic, E.; McAuliffe, O. Comparative genomic and metabolic analysis of three Lactobacillus paracasei cheese isolates reveals considerable genomic differences in strains from the same niche. BMC Genom. 2018, 19, 205. [Google Scholar] [CrossRef]
- Todorov, S.D.; Dicks, L.M.T. Screening for bacteriocin-producing lactic acid bacteria from boza, a traditional cereal beverage from Bulgaria: Comparison of the bacteriocins. Process Biochem. 2006, 41, 11–19. [Google Scholar] [CrossRef]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. 16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry. BMC Microbiol. 2016, 16, 105. [Google Scholar] [CrossRef]
- Martin, F.P.J.; Sprenger, N.; Montoliu, I.; Rezzi, S.; Kochhar, S.; Nicholson, J.K. Dietary modulation of gut functional ecology studied by fecal metabonomics. J. Proteome Res. 2010, 9, 5284–5295. [Google Scholar] [CrossRef] [PubMed]
- Frolova, M.; Yudin, S.; Makarov, V.; Glazunova, O.; Alikina, O.; Markelova, N.; Kolzhetsov, N.; Dzhelyadin, T.; Shcherbakova, V.; Trubitsyn, V.; et al. Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity. Life 2021, 11, 1246. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Torres, L.; Landete, J.M.; Huedo, P.; Peirotén, Á.; Langa, S.; Rodríguez-Minguez, E.; Medina, M.; Arahal, D.R.; Arqués, J.L. Complete genome sequences of Lacticaseibacillus paracasei INIA P272 (CECT 8315) and Lacticaseibacillus rhamnosus INIA P344 (CECT 8316) isolated from breast-fed infants reveal probiotic determinants. Gene 2022, 840, 146743. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial genome analysis: The COG approach. Brief. Bioinform. 2019, 20, 1063–1070. [Google Scholar] [CrossRef]
- Spano, G.; Massa, S. Environmental Stress Response in Wine Lactic Acid Bacteria: Beyond Bacillus subtilis. Crit. Rev. Microbiol. 2006, 32, 77–86. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Beneduce, L.; Weidmann, S.; Grieco, F.; Guzzo, J.; Spano, G. Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett. Appl. Microbiol. 2010, 50, 327–334. [Google Scholar] [CrossRef]
- Hedberg, M.; Hasslöf, P.; Sjöström, I.; Twetman, S.; Stecksén-Blicks, C. Sugar fermentation in probiotic bacteria—An in vitro study. Oral Microbiol. Immunol. 2008, 23, 482–485. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Monedero, V.; Zúñiga, M. Malic enzyme and malolactic enzyme pathways are functionally linked but independently regulated in Lactobacillus casei BL23. Appl. Environ. Microbiol. 2013, 79, 5509–5518. [Google Scholar] [CrossRef]
- Spano, G.; Rinaldi, A.; Ugliano, M.; Moio, L.; Beneduce, L.; Massa, S. A beta-glucosidase gene isolated from wine Lactobacillus plantarum is regulated by abiotic stresses. J. Appl. Microbiol. 2005, 98, 855–861. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Henschke, P.A. The ‘buttery’ attribute of wine—Diacetyl—Desirability, spoilage and beyond. Int. J. Food Microbiol. 2004, 96, 235–252. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Arribas, M.V.; Carmen Polo, M. Occurrence of lactic acid bacteria and biogenic amines in biologically aged wines. Food Microbiol. 2008, 25, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, D.; Milli, A.; Rinalducci, S.; Zolla, L.; Zapparoli, G. Proteomicanalysis of Oenococcus oeni freeze-dried culture to assesstheimportance of cellacclimation to conduct malolactic fermentation in wine. Electrophoresis 2009, 30, 2988–2995. [Google Scholar] [CrossRef]
- Solieri, L.; Genova, F.; De Paola, M.; Giudici, P. Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: A framework for selection of new starter cultures. J. Appl. Microbiol. 2010, 108, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Lerm, E.; Engelbrecht, L.; Du Toit, M. Malolacticfermentation: TheABC’s of MLF. S. Afr. J. Enol. Vitic. 2010, 31, 186–212. [Google Scholar]
- Costantini, A.; Rantsiou, K.; Majumder, A.; Jacobsen, S.; Pessione, E.; Svensson, B.; Garcia-Moruno, E.; Cocolin, L. Complementing DIGE proteomics and DNA subarrayanalyses to shed light on Oenococcus oeni adaptation to ethanol in wine-simulated conditions. J. Proteom. 2015, 123, 114–127. [Google Scholar] [CrossRef]
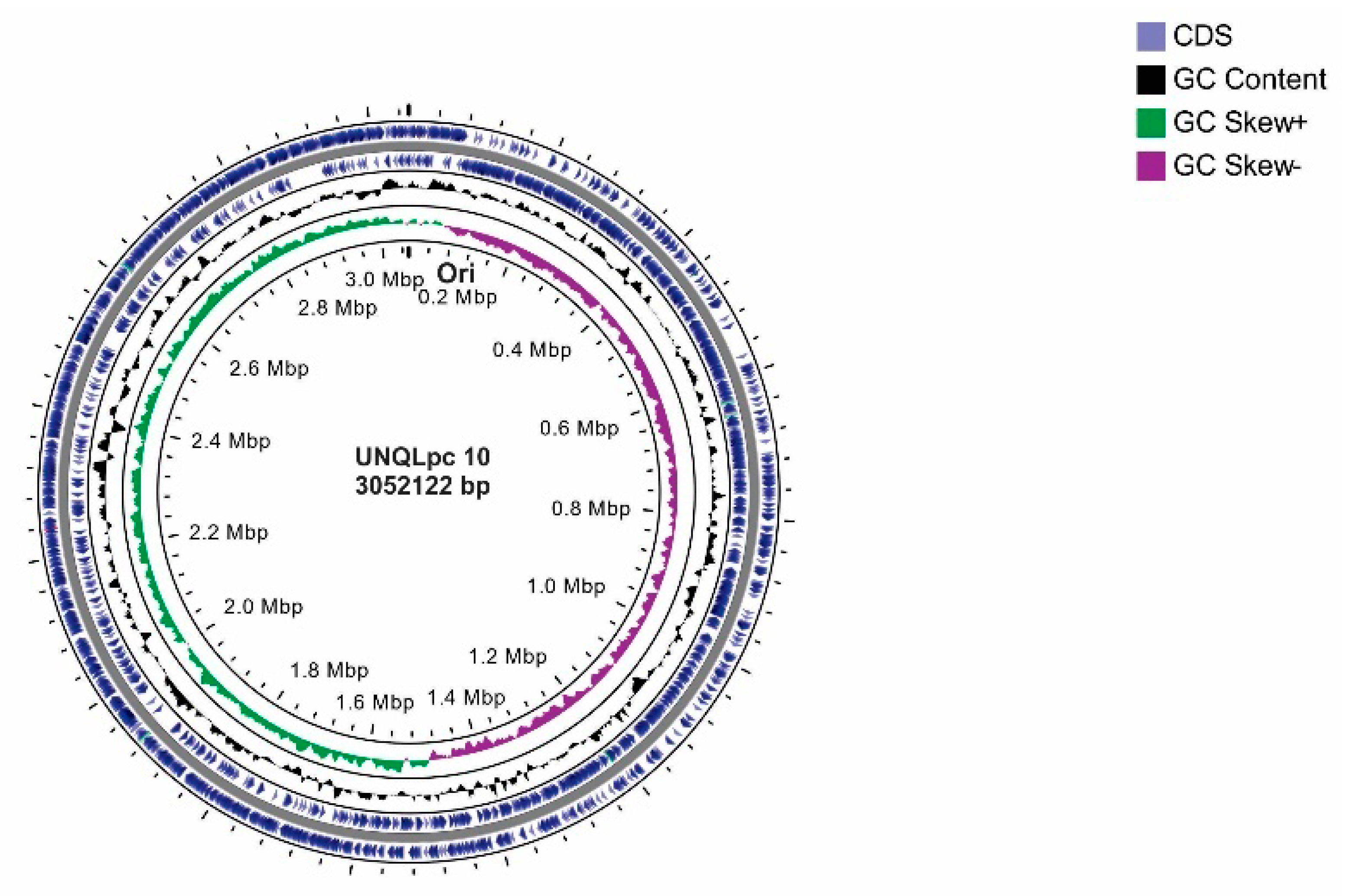
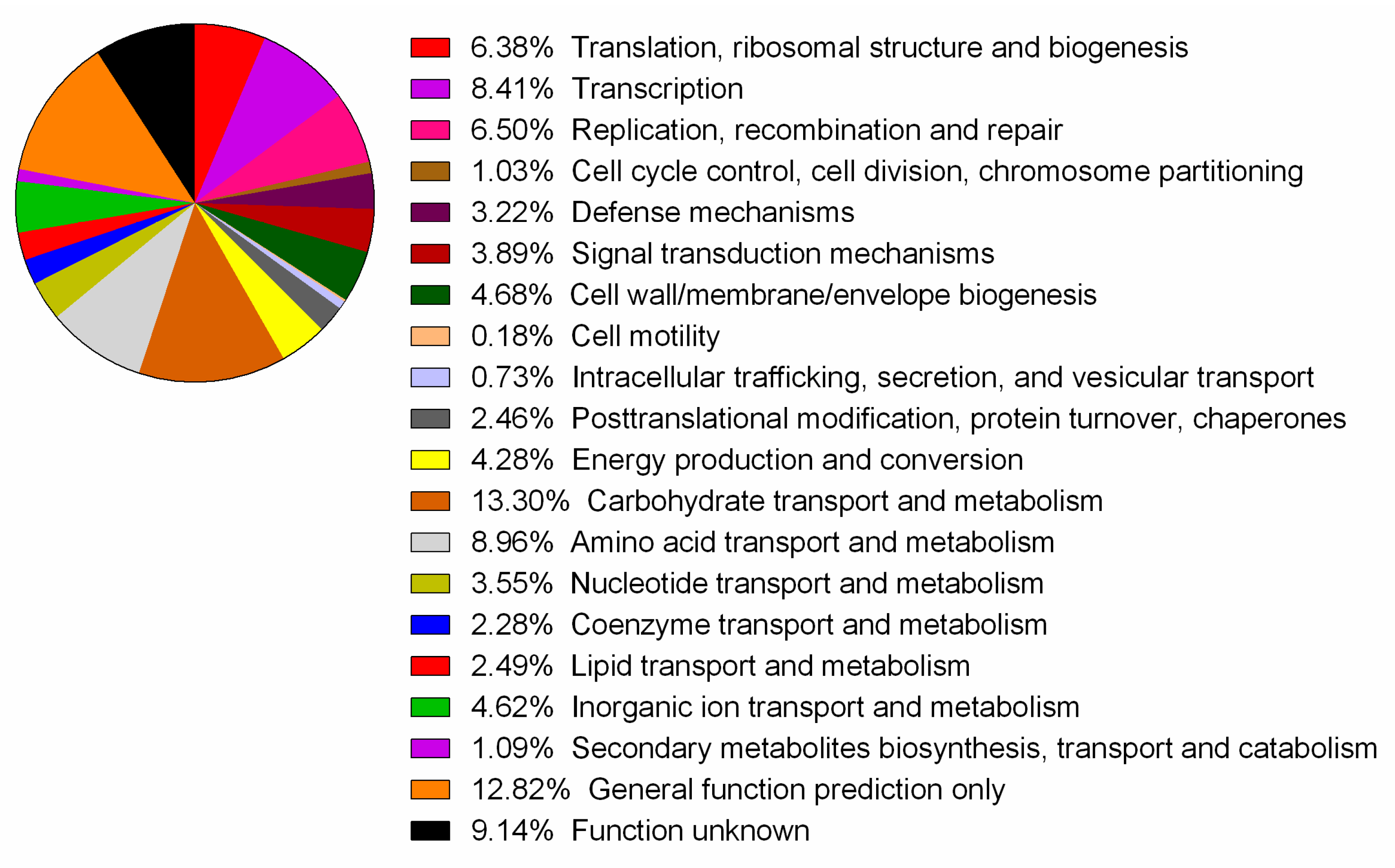
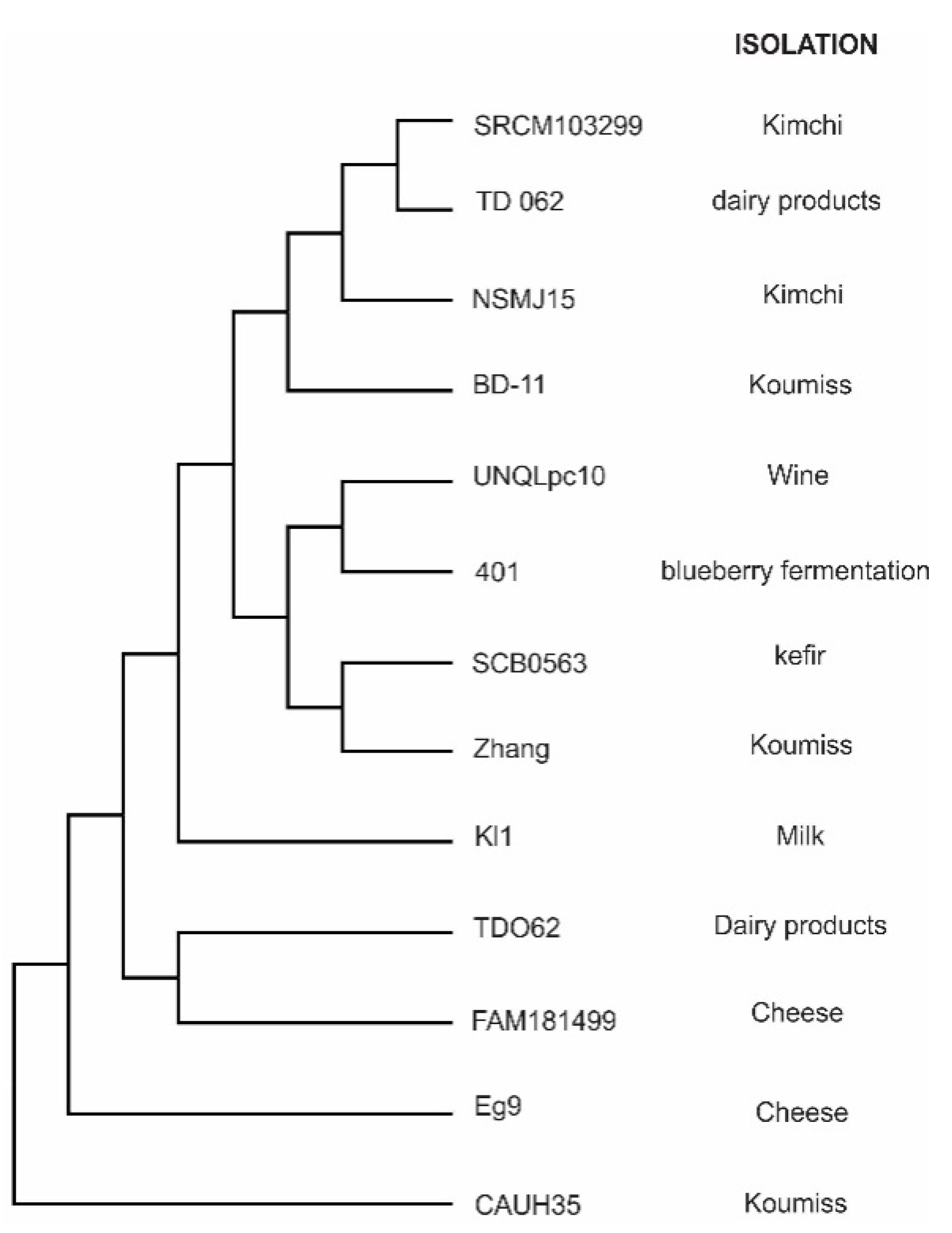
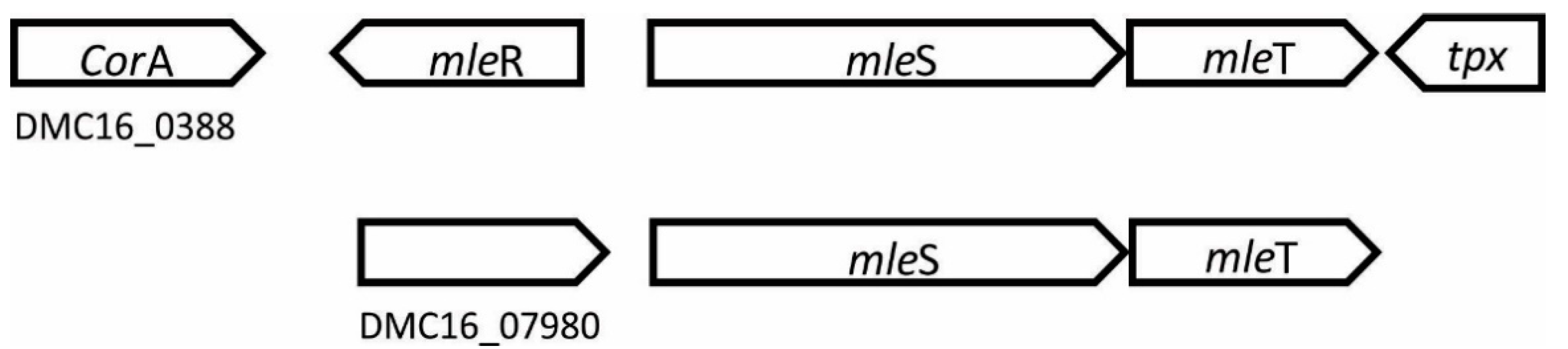
| Name | Function | Genes in UNQLpc10 |
|---|---|---|
| Sugar Metabolism | ||
| Phosphoenolpyruvate | Carbohydrate transport and metabolism | 28 |
| Sugar-ABC transporters | Mainly mediate the transport of nutrients and other molecules into cells or the pumping of toxins and lipids across membranes [31] | 4 |
| L-lactate dehydrogenase | Participates in anaerobic energy metabolism, reducing pyruvate (from glycolysis) to regenerate NAD+ | 3 |
| Related Enzymes of Flavor Development | ||
| Branched-chain aminotransferase | Conversion of valine, leucine, and isoleucine into keto acid components. The keto acids are then further converted into aldehydes, alcohols, and esters, which are important aroma compounds [32] | 1 |
| Aromatic aminotransferase | Conversion of tyrosine, tryptophan, and phenylalanine into Keto acid components [32] | 1 |
| Aspartate transaminase | Conversion of aspartate into keto acid components. | 1 |
| Glutamate dehydrogenase | Catalyzes the deamination of glutamate to oxoglutaric acid related to amino acid (branched-chain amino acids, aromatic amino acids, and methionine) degradation Pathway [32] | 1 |
| D-hydroxyacid dehydrogenase | Catalyzes the reduction of two keto branched-chain acids to hydro acids of interest in flavor formation [33] | 1 |
| Esterase | Catalyze the biosynthesis of esters derived from short-chain fatty acids [32] | 1 |
| Homoserine dehydrogenase | Has homoserine trans-acetylase activity and involved in the Biosynthesis of methionine [32] | 1 |
| Homoserine kinase | Involved in the onset of methionine biosynthesis [32] | 1 |
| 6-phospho-beta-glucosidase | Hydrolytic activity in glycosylated compounds, acts on the glucosidic bonds β (1–4) [34] | 3 |
| Alpha-glucosidase | Hydrolyticactivityonterminal, non-reducing(1→4)- linked alpha-D-glucose residues with release of D-glucose [34] | 5 |
| Malolactic enzyme | Involved in the malolactic fermentation of wine, which results in a natural decrease in acidity and favorable changes in wine flavors | 2 |
| Other Enzymes of Oenological Interest | ||
| Membrane intrinsic proteins | Regulate a large set of developmental and physiological processes and stress responses within cells. | 5 |
| Heat-shock genes | Environmental stress response | 8 |
| Ethyl stress response | Environmental stress response | 2 |
| Wine | Ethanol 10% | Ethanol 14% | ||
|---|---|---|---|---|
| Acclimation | No | Yes | No | Yes |
| %MACf | 97.18 ± 3.58 | 100 ± 1.00 | 84.10 ± 7.11 * | 97.12 ± 2.16 |
| K | 0.328 ± 0.037 | 0.330 ± 0.013 | 0.475 ± 0.150 | 0.440 ± 0.035 |
| R2 | 0.99 | 0.99 | 0.94 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iglesias, N.G.; Navarro, M.E.; Brizuela, N.S.; Valdés La Hens, D.; Semorile, L.C.; Tymczyszyn, E.E.; Bravo Ferrada, B.M. Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter. Fermentation 2022, 8, 726. https://doi.org/10.3390/fermentation8120726
Iglesias NG, Navarro ME, Brizuela NS, Valdés La Hens D, Semorile LC, Tymczyszyn EE, Bravo Ferrada BM. Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter. Fermentation. 2022; 8(12):726. https://doi.org/10.3390/fermentation8120726
Chicago/Turabian StyleIglesias, Nestor Gabriel, Marina Edith Navarro, Natalia Soledad Brizuela, Danay Valdés La Hens, Liliana Carmen Semorile, Emma Elizabeth Tymczyszyn, and Bárbara Mercedes Bravo Ferrada. 2022. "Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter" Fermentation 8, no. 12: 726. https://doi.org/10.3390/fermentation8120726
APA StyleIglesias, N. G., Navarro, M. E., Brizuela, N. S., Valdés La Hens, D., Semorile, L. C., Tymczyszyn, E. E., & Bravo Ferrada, B. M. (2022). Analysis of the Genome Architecture of Lacticaseibacillus paracasei UNQLpc 10, a Strain with Oenological Potential as a Malolactic Starter. Fermentation, 8(12), 726. https://doi.org/10.3390/fermentation8120726







