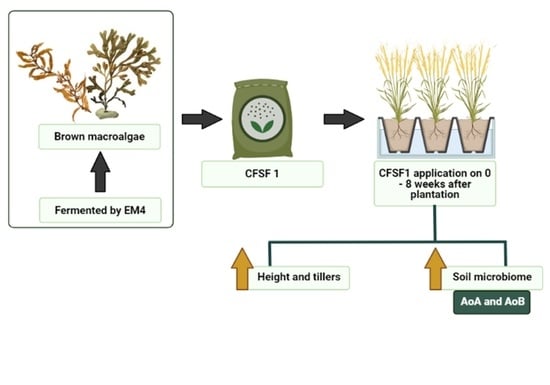
Seaweed Fertilizer Prepared by EM-Fermentation Increases Abundance of Beneficial Soil Microbiome in Paddy (Oryzasativa L.) during Vegetative Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Seaweed Fertilizer Raw Material Preparation
2.2. Seaweed Fertilizer Fermentation Process
2.3. Pot Experimental Design
2.4. Determination of Soil and Paddy Leaves Chemical Properties
2.5. Soil Sampling, DNA Isolation and 16S rRNA Gene Amplification
2.6. Sequencing and Data Processing
2.7. Statistical Analyses
3. Results and Discussion
3.1. Fermented Fertilizer Production
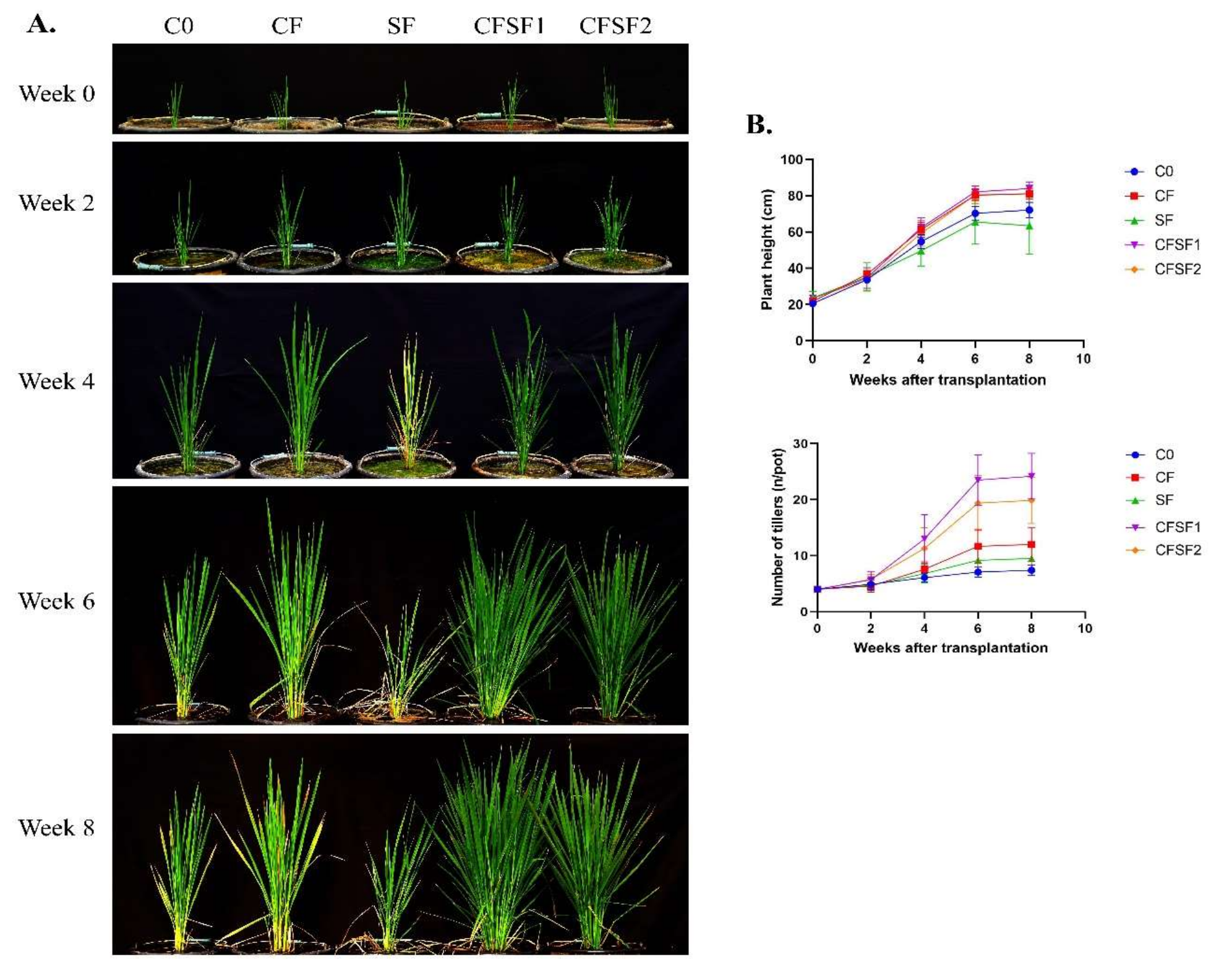
3.2. Effects of SF on Vegetative Growth of Paddy Plants
3.3. Effects of SF on N Uptake of Paddy in Vegetative Stage
3.4. Sequencing Quality Check
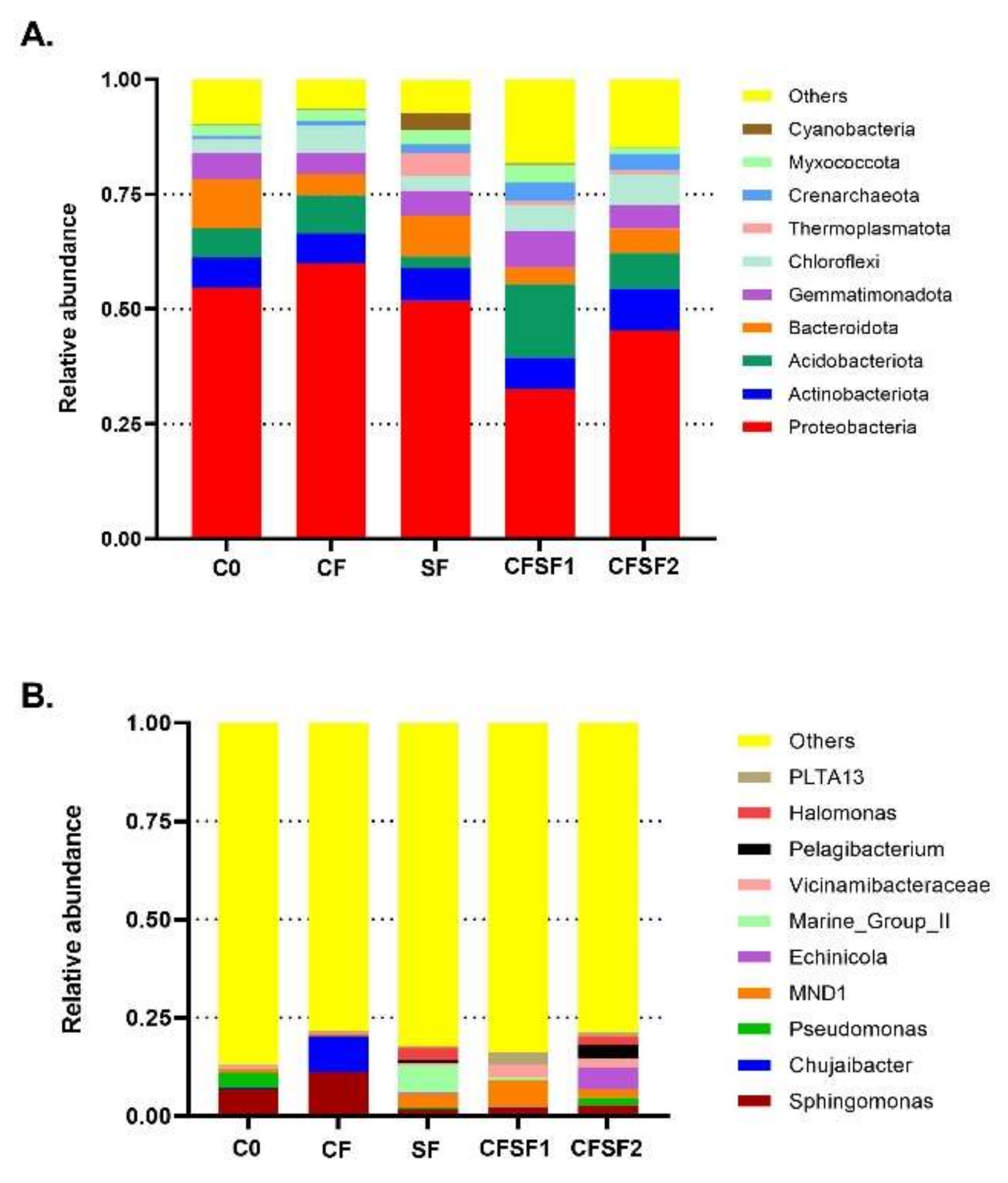
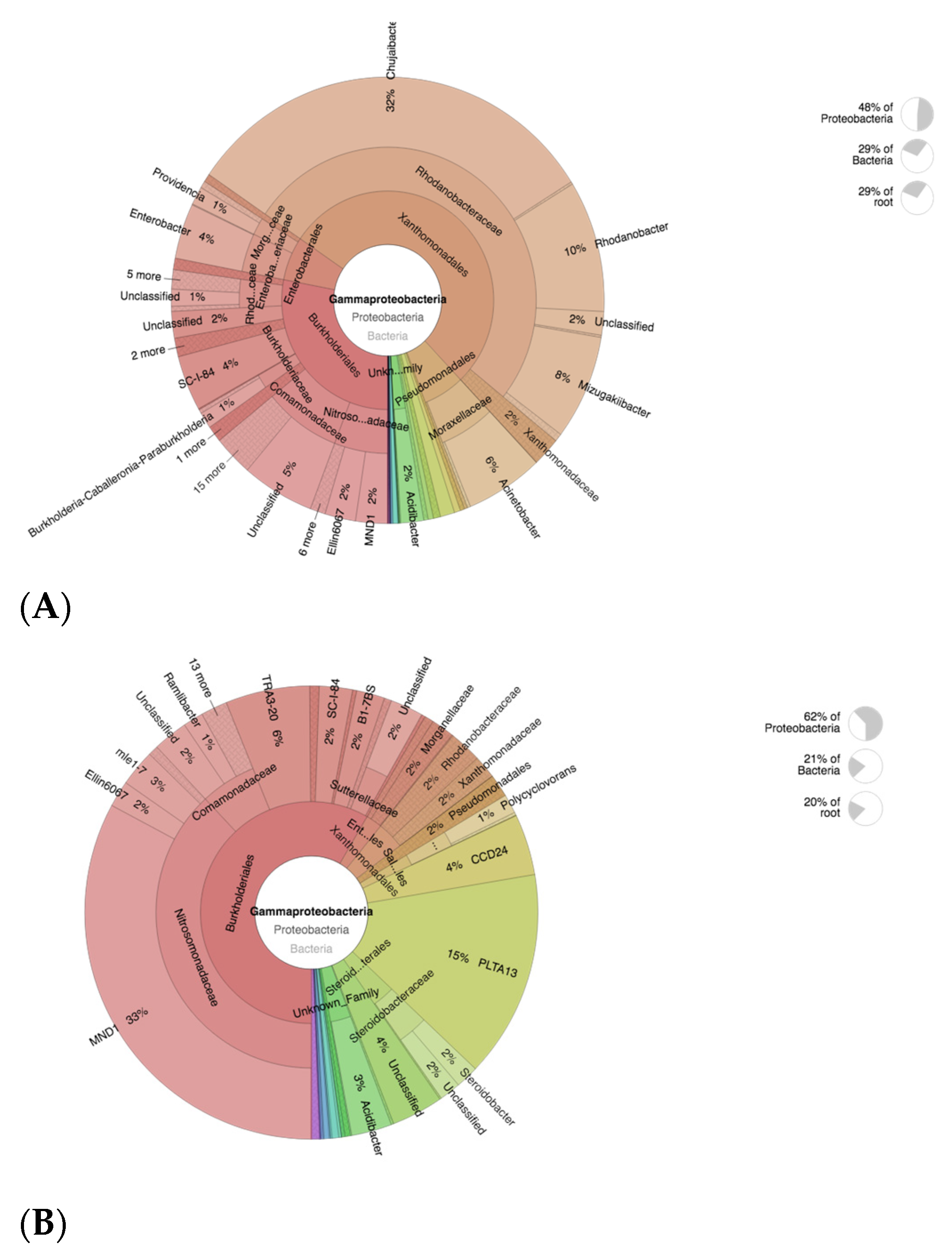
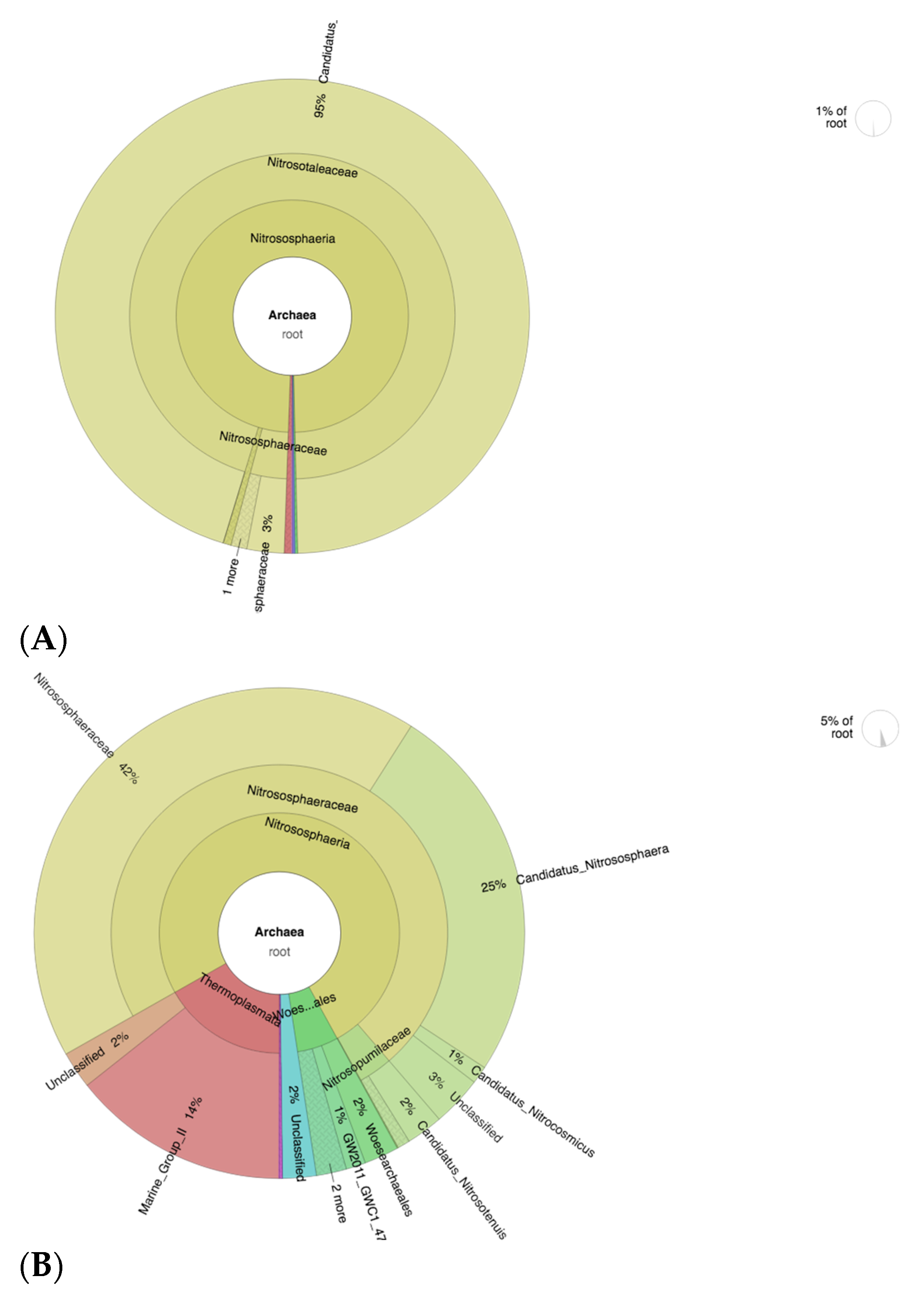
3.5. OTU Identification and Taxonomic Annotation
3.6. Alpha Diversity Analyses
3.7. Beta Diversity Analyses
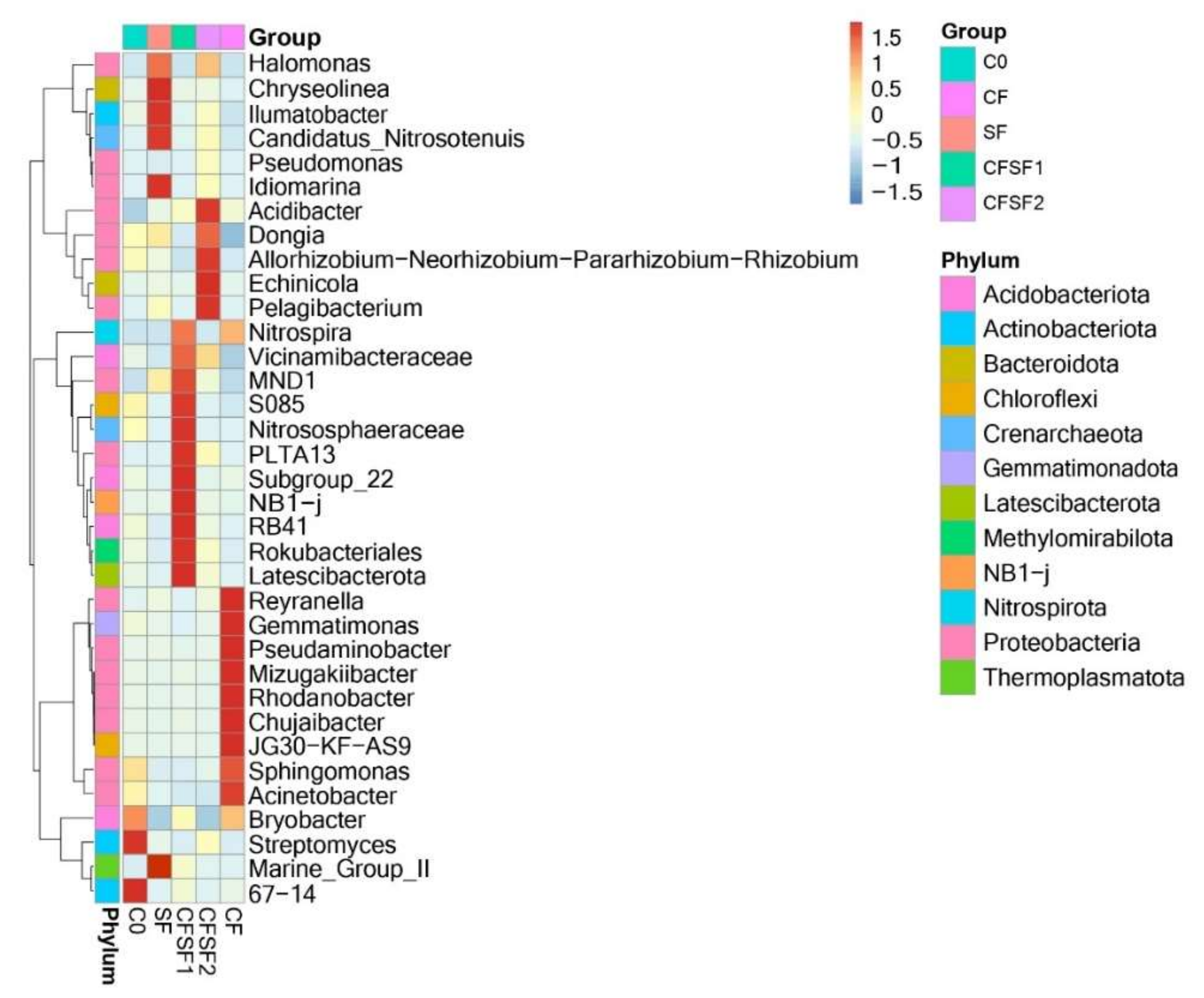
3.8. Taxonomic Abundance Cluster Heatmap
3.9. Abundance of Nitrifying Bacteria and Archaea in SF-Treated Soil of Paddy Plants
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hakim, R.; Haryanto, T.; Sari, D.W. Technical Efficiency among Agricultural Households and Determinants of Food Security in East Java, Indonesia. Sci. Rep. 2021, 11, 4141. [Google Scholar] [CrossRef]
- Rahman, M.M.; Nahar, K.; Ali, M.M.; Sultana, N.; Karim, M.M.; Adhikari, U.K.; Rauf, M.; Azad, M.A.K. Effect of Long-Term Pesticides and Chemical Fertilizers Application on the Microbial Community Specifically Anammox and Denitrifying Bacteria in Rice Field Soil of Jhenaidah and Kushtia District, Bangladesh. Bull. Environ. Contam. Toxicol. 2020, 104, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Ugulu, I.; Ahmad, K.; Khan, Z.I.; Munir, M.; Wajid, K.; Bashir, H. Effects of Organic and Chemical Fertilizers on the Growth, Heavy Metal/Metalloid Accumulation, and Human Health Risk of Wheat (Triticum aestivum L.). Environ. Sci. Pollut. Res. Int. 2021, 28, 12533–12545. [Google Scholar] [CrossRef] [PubMed]
- Rimmer, M.A.; Larson, S.; Lapong, I.; Purnomo, A.H.; Pong-Masak, P.R.; Swanepoel, L.; Paul, N.A. Seaweed Aquaculture in Indonesia Contributes to Social and Economic Aspects of Livelihoods and Community Wellbeing. Sustainability 2021, 13, 10946. [Google Scholar] [CrossRef]
- Craigie, J.S. Seaweed Extract Stimuli in Plant Science and Agriculture. J. Appl. Phycol. 2011, 23, 371–393. [Google Scholar] [CrossRef]
- Crouch, I.J.; Smith, M.T.; van Staden, J.; Lewis, M.J.; Hoad, G.V. Identification of Auxins in a Commercial Seaweed Concentrate. J. Plant Physiol. 1992, 139, 590–594. [Google Scholar] [CrossRef]
- Ateweberhan, M.; Bruggemann, J.H.; Breeman, A.A.M. Seasonal Module Dynamics in Sargassum subrepandum (Fucales, Phaeophyta). J. Phycol. 2008, 44, 269–283. [Google Scholar] [CrossRef]
- Sunarpi, H.; Kurnianingsih, R.; Ghazali, M.; Fanani, R.A.; Sunarwidhi, A.L.; Widyastuti, S.; Nikmatullah, A.; Prasedya, E.S. Evidence for the Presence of Growth-Promoting Factors in Lombok Turbinaria murayana Extract Stimulating Growth and Yield of Tomato Plants (Lycopersicum esculentum Mill.). J. Plant Nutr. 2020, 43, 1813–1823. [Google Scholar] [CrossRef]
- Sunarpi, H.; Pebriani, S.A.; Ambana, Y.; Putri, F.E.; Nikmatullah, A.; Ghazali, M.; Kurnianingsih, R.; Prasedya, E.S. Effect of Inorganic Fertilizer and Brown Alga Solid Ectract on Growth and Yield of Rice Plants. AIP Conf. Proc. 2019, 2199, 070006. [Google Scholar] [CrossRef]
- Rebours, C.; Marinho-Soriano, E.; Zertuche-González, J.A.; Hayashi, L.; Vásquez, J.A.; Kradolfer, P.; Soriano, G.; Ugarte, R.; Abreu, M.H.; Bay-Larsen, I.; et al. Seaweeds: An Opportunity for Wealth and Sustainable Livelihood for Coastal Communities. J. Appl. Phycol. 2014, 26, 1939–1951. [Google Scholar] [CrossRef] [Green Version]
- Ali, O.; Ramsubhag, A.; Jayaraman, J. Biostimulant Properties of Seaweed Extracts in Plants: Implications towards Sustainable Crop Production. Plants 2021, 10, 531. [Google Scholar] [CrossRef]
- Chontal, M.A.H.; Collado, C.J.L.; Orozco, N.R.; Velasco, J.V.; Gabriel, A.L.; Romero, G.L. Nutrient Content of Fermented Fertilizers and Its Efficacy in Combination with Hydrogel in Zea mays L. Int. J. Recycl. Org. Waste Agric. 2019, 8, 309–315. [Google Scholar] [CrossRef]
- Monteiro, P.; Lomartire, S.; Cotas, J.; Pacheco, D.; Marques, J.C.; Pereira, L.; Gonçalves, A.M.M. Seaweeds as a Fermentation Substrate: A Challenge for the Food Processing Industry. Processes 2021, 9, 1953. [Google Scholar] [CrossRef]
- Hirsch, P.R.; Mauchline, T.H. The Importance of the Microbial N Cycle in Soil for Crop Plant Nutrition. Adv. Appl. Microbiol. 2015, 93, 45–71. [Google Scholar] [CrossRef]
- Arioli, T.; Mattner, S.W.; Winberg, P.C. Applications of Seaweed Extracts in Australian Agriculture: Past, Present and Future. J. Appl. Phycol. 2015, 27, 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higa, T. Effective microorganisms: A biotechnology for mankind. In Proceedings of the 1st International Conference on Kyusei Nature Farming; Parr, J.F., Hornick, S.B., Whitman, S.E., Eds.; USDA: Washington, DC, USA, 1991; pp. 8–14. [Google Scholar]
- Dobrinčić, A.; Balbino, S.; Zorić, Z.; Pedisić, S.; Kovačević, D.B.; Garofulić, I.E.; Dragović-Uzelac, V. Advanced Technologies for the Extraction of Marine Brown Algal Polysaccharides. Mar. Drugs 2020, 18, 168. [Google Scholar] [CrossRef] [Green Version]
- Sunarpi, H.; Nikmatullah, A.; Sunarwidhi, A.L.; Sapitri, I.; Ilhami, B.T.K.; Widyastuti, S.; Prasedya, E.S. Growth and Yield of Rice Plants (Oryza sativa) Grown in Soil Media Containing Several Doses of Inorganic Fertilizers and Sprayed with Lombok Brown Algae Extracts. IOP Conf. Ser. Earth Environ. Sci. 2020, 594, 012032. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1983; pp. 539–579. ISBN 978-0-89118-977-0. [Google Scholar]
- Jiang, Y.; Huang, X.; Zhang, X.; Zhang, X.; Zhang, Y.; Zheng, C.; Deng, A.; Zhang, J.; Wu, L.; Hu, S.; et al. Optimizing Rice Plant Photosynthate Allocation Reduces N2O Emissions from Paddy Fields. Sci. Rep. 2016, 6, 39333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, G.; Todorov, T.I. Distribution of Nutrient and Toxic Elements in Brown and Polished Rice. Food Chem. 2019, 289, 299–307. [Google Scholar] [CrossRef]
- Pavlovska, M.; Prekrasna, I.; Parnikoza, I.; Dykyi, E. Soil Sample Preservation Strategy Affects the Microbial Community Structure. Microbes Environ. 2021, 36, ME20134. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Van de Poel, B.; Smet, D.; Van Der Straeten, D. Ethylene and Hormonal Cross Talk in Vegetative Growth and Development. Plant Physiol. 2015, 169, 61–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Y.; Ling, N.; Xue, C.; Chen, H.; Ruan, Y.; Guo, J.; Zhu, C.; Wang, M.; Shen, Q.; Guo, S. Long-Term Fertilization Regimes Change Soil Nitrification Potential by Impacting Active Autotrophic Ammonia Oxidizers and Nitrite Oxidizers as Assessed by DNA Stable Isotope Probing. Environ. Microbiol. 2019, 21, 1224–1240. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Ling, N.; Chen, H.; Duan, Y.; Wang, S.; Shen, Q.; Vandenkoornhuyse, P. Long-Term Chemical-Only Fertilization Induces a Diversity Decline and Deep Selection on the Soil Bacteria. mSystems 2020, 5, e00337-20. [Google Scholar] [CrossRef]
- De Oliveira e Silva, A.M.; Vidal-Novoa, A.; Batista-González, A.E.; Pinto, J.R.; Portari Mancini, D.A.; Reina-Urquijo, W.; Mancini-Filho, J. In Vivo and in Vitro Antioxidant Activity and Hepatoprotective Properties of Polyphenols from Halimeda opuntia (Linnaeus) Lamouroux. Redox Rep. 2012, 17, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Meng, X.; Li, T.; Raza, W.; Liu, D.; Shen, Q. The Growth Promotion of Peppers (Capsicum annuum L.) by Trichoderma Guizhouense NJAU4742-Based Biological Organic Fertilizer: Possible Role of Increasing Nutrient Availabilities. Microorganisms 2020, 8, E1296. [Google Scholar] [CrossRef] [PubMed]
- Dahunsi, S.O.; Oranusi, S.; Efeovbokhan, V.E.; Adesulu-Dahunsi, A.T.; Ogunwole, J.O. Crop Performance and Soil Fertility Improvement Using Organic Fertilizer Produced from Valorization of Carica papaya Fruit Peel. Sci. Rep. 2021, 11, 4696. [Google Scholar] [CrossRef] [PubMed]
- Benítez García, I.; Dueñas Ledezma, A.K.; Martínez Montaño, E.; Salazar Leyva, J.A.; Carrera, E.; Osuna Ruiz, I. Identification and Quantification of Plant Growth Regulators and Antioxidant Compounds in Aqueous Extracts of Padina durvillaei and Ulva lactuca. Agronomy 2020, 10, 866. [Google Scholar] [CrossRef]
- Metay, A.; Magnier, J.; Guilpart, N.; Christophe, A. Nitrogen Supply Controls Vegetative Growth, Biomass and Nitrogen Allocation for Grapevine (Cv. Shiraz) Grown in Pots. Funct. Plant Biol. 2014, 42, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Mbandi, S.K.; Hesse, U.; Rees, D.J.G.; Christoffels, A. A Glance at Quality Score: Implication for de Novo Transcriptome Reconstruction of Illumina Reads. Front. Genet. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielak, A.M.; Cipriano, M.A.P.; Kuramae, E.E. Acidobacteria Strains from Subdivision 1 Act as Plant Growth-Promoting Bacteria. Arch. Microbiol. 2016, 198, 987–993. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, J.; Huang, Z.; Su, G.; Li, X.; Sun, Z.; Qin, Y. Impact of Short-Term Application of Seaweed Fertilizer on Bacterial Diversity and Community Structure, Soil Nitrogen Contents, and Plant Growth in Maize Rhizosphere Soil. Folia Microbiol. 2020, 65, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Ghaderiardakani, F.; Collas, E.; Damiano, D.K.; Tagg, K.; Graham, N.S.; Coates, J.C. Effects of Green Seaweed Extract on Arabidopsis Early Development Suggest Roles for Hormone Signalling in Plant Responses to Algal Fertilisers. Sci. Rep. 2019, 9, 1983. [Google Scholar] [CrossRef] [PubMed]
- Renaut, S.; Masse, J.; Norrie, J.P.; Blal, B.; Hijri, M. A Commercial Seaweed Extract Structured Microbial Communities Associated with Tomato and Pepper Roots and Significantly Increased Crop Yield. Microb. Biotechnol. 2019, 12, 1346–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juhmani, A.-S.; Vezzi, A.; Wahsha, M.; Buosi, A.; Pascale, F.D.; Schiavon, R.; Sfriso, A. Diversity and Dynamics of Seaweed Associated Microbial Communities Inhabiting the Lagoon of Venice. Microorganisms 2020, 8, 1657. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A Precious Bio-Resource in Agriculture, Ecosystem, and Environmental Sustainability. Front. Microbiol. 2016, 7, 529. [Google Scholar] [CrossRef] [Green Version]
- Burson, A.; Stomp, M.; Greenwell, E.; Grosse, J.; Huisman, J. Competition for Nutrients and Light: Testing Advances in Resource Competition with a Natural Phytoplankton Community. Ecology 2018, 99, 1108–1118. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Madamwar, D.; Incharoensakdi, A. Bloom Dynamics of Cyanobacteria and Their Toxins: Environmental Health Impacts and Mitigation Strategies. Front. Microbiol. 2015, 6, 1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orellana, L.H.; Ben Francis, T.; Krüger, K.; Teeling, H.; Müller, M.-C.; Fuchs, B.M.; Konstantinidis, K.T.; Amann, R.I. Niche Differentiation among Annually Recurrent Coastal Marine Group II Euryarchaeota. ISME J. 2019, 13, 3024–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, L.; Goubet, A.; Madigou, C.; Pernelle, J.-J.; Palmier, K.; Labadie, K.; Lemainque, A.; Michot, O.; Astoul, L.; Barbier, P.; et al. Colonization Kinetics and Implantation Follow-up of the Sewage Microbiome in an Urban Wastewater Treatment Plant. Sci. Rep. 2020, 10, 11634. [Google Scholar] [CrossRef]
- Köberl, M.; Dita, M.; Martinuz, A.; Staver, C.; Berg, G. Members of Gammaproteobacteria as Indicator Species of Healthy Banana Plants on Fusarium Wilt-Infested Fields in Central America. Sci. Rep. 2017, 7, 45318. [Google Scholar] [CrossRef]
- Sinong, G.F.; Yasuda, M.; Nara, Y.; Lee, C.G.; Dastogeer, K.M.G.; Tabuchi, H.; Nakai, H.; Djedidi, S.; Okazaki, S. Distinct Root Microbial Communities in Nature Farming Rice Harbor Bacterial Strains With Plant Growth-Promoting Traits. Front. Sustain. Food Syst. 2021, 4, 314. [Google Scholar] [CrossRef]
- Michas, A.; Pastore, G.; Chiba, A.; Grafe, M.; Clausing, S.; Polle, A.; Schloter, M.; Spohn, M.; Schulz, S. Phosphorus Availability Alters the Effect of Tree Girdling on the Diversity of Phosphorus Solubilizing Soil Bacterial Communities in Temperate Beech Forests. Front. For. Glob. Chang. 2021, 4, 79. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Qi, G.; Ma, G.; Zhao, X. Biochar Amendment Controlled Bacterial Wilt through Changing Soil Chemical Properties and Microbial Community. Microbiol. Res. 2020, 231, 126373. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Nupur; Wu, N.; Madsen, A.M.; Chen, X.; Gardiner, A.T.; Koblížek, M. Gemmatimonas Groenlandica sp. Nov. Is an Aerobic Anoxygenic Phototroph in the Phylum Gemmatimonadetes. Front. Microbiol. 2021, 11, 3395. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. Cra20 Increases Plant Growth Rate and Alters Rhizosphere Microbial Community Structure of Arabidopsis thaliana Under Drought Stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kini, K.; Agnimonhan, R.; Dossa, R.; Soglonou, B.; Gbogbo, V.; Ouedraogo, I.; Kpemoua, K.; Traoré, M.; Silue, D. First Report of Sphingomonas sp. Causing Bacterial Leaf Blight of Rice in Benin, Burkina Faso, The Gambia, Ivory Coast, Mali, Nigeria, Tanzania and Togo. New Dis. Rep. 2017, 35, 32. [Google Scholar] [CrossRef] [Green Version]
- Sparks, A.H.; Forbes, G.A.; Hijmans, R.J.; Garrett, K.A. Climate Change May Have Limited Effect on Global Risk of Potato Late Blight. Glob. Chang. Biol. 2014, 20, 3621–3631. [Google Scholar] [CrossRef] [PubMed]
- Podlesnaya, G.V.; Krasnopeev, A.Y.; Potapov, S.A.; Tikhonova, I.V.; Shtykova, Y.R.; Suslova, M.Y.; Timoshkin, O.A.; Belykh, O.I. Diversity of Nitrifying Bacteria in Microbial Communities from Water and Epilithic Biofilms of the Lake Baikal Littoral Zone. Limnol. Freshw. Biol. 2020, 4, 1008–1010. [Google Scholar] [CrossRef]
- Chi, Z.; Hou, L.; Li, H. Effects of Pollution Load and Salinity Shock on Nitrogen Removal and Bacterial Community in Two-Stage Vertical Flow Constructed Wetlands. Bioresour. Technol. 2021, 342, 126031. [Google Scholar] [CrossRef]
- Kostka, J.E.; Green, S.J.; Rishishwar, L.; Prakash, O.; Katz, L.S.; Mariño-Ramírez, L.; Jordan, I.K.; Munk, C.; Ivanova, N.; Mikhailova, N.; et al. Genome Sequences for Six Rhodanobacter Strains, Isolated from Soils and the Terrestrial Subsurface, with Variable Denitrification Capabilities. J. Bacteriol. 2012, 194, 4461–4462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Sun, L.; Wang, Y.; Fan, K.; Xu, Q.; Li, Y.; Ma, Q.; Wang, J.; Ren, W.; Ding, Z. Cow Manure Application Effectively Regulates the Soil Bacterial Community in Tea Plantation. BMC Microbiol. 2020, 20, 190. [Google Scholar] [CrossRef] [PubMed]
- Barot, S.; Allard, V.; Cantarel, A.; Enjalbert, J.; Gauffreteau, A.; Goldringer, I.; Lata, J.-C.; Le Roux, X.; Niboyet, A.; Porcher, E. Designing Mixtures of Varieties for Multifunctional Agriculture with the Help of Ecology. A Review. Agron. Sustain. Dev. 2017, 37, 13. [Google Scholar] [CrossRef] [Green Version]
- Niu, H.; Pang, Z.; Fallah, N.; Zhou, Y.; Zhang, C.; Hu, C.; Lin, W.; Yuan, Z. Diversity of Microbial Communities and Soil Nutrients in Sugarcane Rhizosphere Soil under Water Soluble Fertilizer. PLoS ONE 2021, 16, e0245626. [Google Scholar] [CrossRef]
- Bollmann, A.; Bullerjahn, G.S.; McKay, R.M. Abundance and Diversity of Ammonia-Oxidizing Archaea and Bacteria in Sediments of Trophic End Members of the Laurentian Great Lakes, Erie and Superior. PLoS ONE 2014, 9, e97068. [Google Scholar] [CrossRef] [Green Version]
- Prosser, J.I.; Head, I.M.; Stein, L.Y. The Family Nitrosomonadaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 901–918. ISBN 978-3-642-30197-1. [Google Scholar]
- Abbas, T.; Zhou, H.; Zhang, Q.; Li, Y.; Liang, Y.; Di, H.; Zhao, Y. Anammox Co-Fungi Accompanying Denitrifying Bacteria Are the Thieves of the Nitrogen Cycle in Paddy-Wheat Crop Rotated Soils. Environ. Int. 2019, 130, 104913. [Google Scholar] [CrossRef] [PubMed]
- Mannaa, M.; Han, G.; Jeon, H.W.; Kim, J.; Kim, N.; Park, A.R.; Kim, J.-C.; Seo, Y.-S. Influence of Resistance-Inducing Chemical Elicitors against Pine Wilt Disease on the Rhizosphere Microbiome. Microorganisms 2020, 8, 884. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Dörr de Quadros, P.; Camargo, F.A.O.; Triplett, E. Drivers of Archaeal Ammonia-Oxidizing Communities in Soil. Front. Microbiol. 2012, 3, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| pH | OM (mg/g) | N (%) | P (mg/kg) | K (mg/kg) | Soil Texture |
|---|---|---|---|---|---|
| 7.61 | 11.42 | 3.19 | 481.76 | 82.54 | Clay |
| Chemical Parameters | Before Fermentation | After Fermentation |
|---|---|---|
| pH | 7.57 a | 7.42 a |
| Water content (%) | 46.63 a | 31.49 b |
| Organic C (%) | 33.58 a | 43.14 b |
| N (%) | 1.06 a | 1.88 b |
| C:N (%) | 23.54 a | 22.99 a |
| P (%) | 2.23 a | 2.34 a |
| K (%) | 2.25 a | 3.16 b |
| Ca (mg/kg) | 315.23 a | 421.56 b |
| Mg (mg/kg) | 335.76 a | 432.67 b |
| B (mg/kg) | 9.89 a | 19.32 b |
| Na (mg/kg) | ND | ND |
| Cl (mg/kg) | ND | ND |
| Treatment | Raw Tags | Clean Tags | Effective Tags | Base (nt) | AvgLen (nt) | Q20 | GC% | Effective% |
|---|---|---|---|---|---|---|---|---|
| C0 | 127,434 | 125,464 | 71,995 | 29,902,893 | 415 | 97.67 | 55.92 | 45.54 |
| CF | 157 | 127,442 | 66,042 | 27,523,135 | 417 | 97.67 | 55.47 | 38.96 |
| SF | 149,362 | 147,557 | 90,751 | 37,439,023 | 413 | 98.31 | 55.68 | 57.72 |
| CFSF1 | 151,979 | 149,896 | 112,710 | 47,254,458 | 419 | 98.26 | 57.27 | 70.75 |
| CFSF2 | 151,420 | 149,896 | 95,880 | 39,851,907 | 416 | 98.33 | 55.86 | 60.85 |
| Treatment | Observed Species | Shannon | Simpson | Chao1 | ACE | Goods Coverage | PD Whole Tree |
|---|---|---|---|---|---|---|---|
| C0 | 2283 | 8.823 | 0.992 | 2505.289 | 2542.541 | 0.996 | 228.506 |
| CF | 2578 | 8.991 | 0.992 | 2752.444 | 2779.912 | 0.993 | 247.965 |
| SF | 2937 | 8.412 | 0.980 | 3215.766 | 3414.616 | 0.994 | 257.892 |
| CFSF1 | 3697 | 9.838 | 0.996 | 3940.166 | 4049.955 | 0.998 | 476.768 |
| CFSF2 | 3621 | 9.776 | 0.996 | 4211.104 | 4249.748 | 0.995 | 446.746 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasedya, E.S.; Kurniawan, N.S.H.; Kirana, I.A.P.; Ardiana, N.; Abidin, A.S.; Ilhami, B.T.K.; Jupri, A.; Widyastuti, S.; Sunarpi, H.; Nikmatullah, A. Seaweed Fertilizer Prepared by EM-Fermentation Increases Abundance of Beneficial Soil Microbiome in Paddy (Oryzasativa L.) during Vegetative Stage. Fermentation 2022, 8, 46. https://doi.org/10.3390/fermentation8020046
Prasedya ES, Kurniawan NSH, Kirana IAP, Ardiana N, Abidin AS, Ilhami BTK, Jupri A, Widyastuti S, Sunarpi H, Nikmatullah A. Seaweed Fertilizer Prepared by EM-Fermentation Increases Abundance of Beneficial Soil Microbiome in Paddy (Oryzasativa L.) during Vegetative Stage. Fermentation. 2022; 8(2):46. https://doi.org/10.3390/fermentation8020046
Chicago/Turabian StylePrasedya, Eka Sunarwidhi, Nanda Sofian Hadi Kurniawan, Indah Alibiah Putri Kirana, Nur Ardiana, Angga Susmana Abidin, Bq Tri Khairina Ilhami, Ahmad Jupri, Sri Widyastuti, Haji Sunarpi, and Aluh Nikmatullah. 2022. "Seaweed Fertilizer Prepared by EM-Fermentation Increases Abundance of Beneficial Soil Microbiome in Paddy (Oryzasativa L.) during Vegetative Stage" Fermentation 8, no. 2: 46. https://doi.org/10.3390/fermentation8020046
APA StylePrasedya, E. S., Kurniawan, N. S. H., Kirana, I. A. P., Ardiana, N., Abidin, A. S., Ilhami, B. T. K., Jupri, A., Widyastuti, S., Sunarpi, H., & Nikmatullah, A. (2022). Seaweed Fertilizer Prepared by EM-Fermentation Increases Abundance of Beneficial Soil Microbiome in Paddy (Oryzasativa L.) during Vegetative Stage. Fermentation, 8(2), 46. https://doi.org/10.3390/fermentation8020046