Induction of Chromosomal Aneuploids from Brewery Shochu Yeast with Novel Brewery Characteristics
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Selection of Candidate Aneuploids from Shochu Yeast Using Nocodazole
3.2. Metabolome Analysis of the Broth Brewed with the Obtained Aneuploids
3.3. Flavor Analysis of the Broth Brewed with the Obtained Aneuploids
3.4. Sensory Evaluation of Broth Brewed with the Obtained Aneuploids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inui, T. Ryukyu awamori hakko kin chosa hokokusho. J. Chem. Soc. Jpn. 1901, 4, 1421–1430. (In Japanese) [Google Scholar]
- Usami, K. Awamorishu jozo kenkyu hokoku. Tokyo Univ. Bull. 1901, 3, 15–37. (In Japanese) [Google Scholar]
- Sugama, S. Shochu yeast. J. Brew. Soc. Jpn. 1972, 67, 672–677. (In Japanese) [Google Scholar]
- Yoshidome, T.; Maki, N.; Yonetamari, T.; Yoshida, K.; Nakahara, K. Traditional shochu brewing using dry shochu-yeast. J. Brew. Soc. Jpn. 2001, 96, 433–439. (In Japanese) [Google Scholar] [CrossRef][Green Version]
- Gallone, B.; Steensels, J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; Roncoroni, M.; Voordeckers, K.; Miraglia, L.; et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 2016, 166, 1397–1410.e16. [Google Scholar] [CrossRef]
- Peter, J.; Chiara, D.M.; Friedrich, A.; Yue, X.J.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Pavelka, N.; Rancati, G.; Zhu, J.; Bradford, W.D.; Saraf, A.; Florens, L.; Sanderson, B.W.; Hattem, G.L.; Li, R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010, 468, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, N.; Honda, S.; Fujiwara, M.; Yoshimura, Y.; Nakamura, T. Polyploid engineering by increasing mutant gene dosage in yeasts. Microb. Biotechnol. 2021, 14, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, M.; Fujimaru, Y.; Taguchi, S.; Ferdouse, J.; Sawada, K.; Kimura, Y.; Terasawa, Y.; Agrimi, G.; Anai, T.; Noguchi, H.; et al. Chromosomal aneuploidy improves the brewing characteristics of sake yeast. Appl. Environ. Microbiol. 2017, 83, e01620–17. [Google Scholar] [CrossRef]
- Takashita, H.; Fujihara, E.; Furutera, M.; Kajiwara, Y.; Shimoda, M.; Matsuoka, M.; Ogawa, T.; Kawamoto, S.; Ono, K. Competitive advantage and tolerance of selected shochu yeast in barley shochu mash. J. Biosci. Bioeng. 2013, 116, 79–84. [Google Scholar] [CrossRef]
- Kitagaki, H.; Kitamoto, K. Breeding research on sake yeasts in Japan: History, recent technological advances, and future perspectives. Annu. Rev. Food Sci. 2013, 4, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Omori, T.; Umemoto, Y.; Ogawa, K.; Kajiwara, Y.; Shimoda, M.; Wada, H. A novel method for screening high glycerol- and ester-producing brewing yeasts (Saccharomyces cerevisiae) by heat shock treatment. J. Ferment. Bioeng. 1997, 83, 64–69. [Google Scholar] [CrossRef]
- Omori, T.; Ogawa, K.; Shimoda, M. Breeding of high glycerol-producing shochu yeast (Saccharomyces cerevisiae) with acquired salt tolerance. J. Ferment. Bioeng. 1995, 79, 560–565. [Google Scholar] [CrossRef]
- Kanai, M.; Kawata, T.; Yoshida, Y.; Kita, Y.; Ogawa, T.; Mizunuma, M.; Watanabe, D.; Shimoi, H.; Mizuno, A.; Yamada, O.; et al. Sake yeast YHR032W/ERC1 haplotype contributes to high S-adenosylmethionine accumulation in sake yeast strains. J. Biosci. Bioeng. 2017, 123, 8–14. [Google Scholar] [CrossRef]
- Miyashita, K.; Sakamoto, K.; Kitagaki, H.; Iwashita, K.; Ito, K.; Shimoi, H. Cloning and analysis of the AWA1 gene of a nonfoaming mutant of a sake yeast. J. Biosci. Bioeng. 2004, 97, 14–18. [Google Scholar] [CrossRef]
- Mimura, N.; Isogai, A.; Iwashita, K.; Bamba, T.; Fukusaki, E. Gas chromatography/mass spectrometry based component profiling and quality prediction for Japanese sake. J. Biosci. Bioeng. 2014, 118, 406–414. [Google Scholar] [CrossRef]
- Setoguchi, T.; Kamiwatari, T. General components and general flavor compounds in sweet-potato-shochu and brown-sugar-shochu—Analysis of Honkakushochu. J. Brew. Soc. Jpn. 2014, 109, 49–59. (In Japanese) [Google Scholar] [CrossRef]
- De Brabander, M.J.; Van de Velre, R.M.L.; Aerts, F.E.M.; Borgers, M.; Janssen, P.A.J. The effects of methyl [5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl] carbamate, (R 17934; NSC 238159), a new synthetic antitumoral drug interfering with microtubules, on mammalian cells cultured in vitro. Cancer Res. 1976, 36, 905–916. [Google Scholar]
- Zimmermann, F.K.; Mayer, V.W.; Scheel, I. Induction of aneuploidy by oncodazole (nocodazole), an anti-tubulin agent, and acetone. Mutat. Res. Lett. 1984, 141, 15–18. [Google Scholar] [CrossRef]
- Mattson, A.M.; Jensen, C.O.; Dutcher, R.A. Triphenyltetrazolium chloride as a dye for vital tissues. Science 1947, 106, 294–295. [Google Scholar] [CrossRef]
- Furukawa, T.; Akiyama, H. A microbial control of sake brewing. Part IV. On the TTC agar overlay method for grouping of yeast (1). Nippon Nogeikagaku Kaishi 1963, 37, 398–402. (In Japanese) [Google Scholar] [CrossRef]
- Torres, E.M.; Sokolsky, T.; Tucker, C.M.; Chan, L.Y.; Boselli, M.; Dunham, M.J.; Amon, A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 2007, 317, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Dutcher, S.K.; Wood, J.S.; Garvik, B. The fidelity of mitotic chromosome reproduction in S. cerevisiae. Rec. Adv. Yeast Mol. BioI. 1982, 1, 28–38. [Google Scholar]
- Pflügl, S.; Marx, H.; Mattanovich, D.; Sauer, M. 1,3-Propanediol production from glycerol with Lactobacillus diolivorans. Bioresour. Technol. 2012, 119, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; van der Burg, M.; Szuhai, K.; Kops, G.J.; Medema, R.H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011, 333, 1895–1898. [Google Scholar] [CrossRef]
- Makrantoni, V.; Marston, A.L. Cohesin and chromosome segregation. Curr. Biol. 2018, 28, R688–R693. [Google Scholar] [CrossRef]
- Fujimaru, Y.; Kusaba, Y.; Zhang, N.; Dai, H.; Yamamoto, Y.; Takasaki, M.; Kakeshita, T.; Kitagaki, H. Extra copy of the mitochondrial cytochrome-c peroxidase gene confers a pyruvate-underproducing characteristic of sake yeast through respiratory metabolism. J. Biosci. Bioeng. 2021, 131, 640–646. [Google Scholar] [CrossRef]
- Ough, C.S.; Bell, A.A. Effects of nitrogen fertilization of grapevines on amino acid metabolism and higher-alcohol formation during grape juice fermentation. Am. J. Enol. Vitic. 1980, 31, 122–123. [Google Scholar]
- Okumura, J. Maillard reaction and flavor formation. J. Brew. Soc. Jpn. 1993, 88, 178–187. (In Japanese) [Google Scholar] [CrossRef]
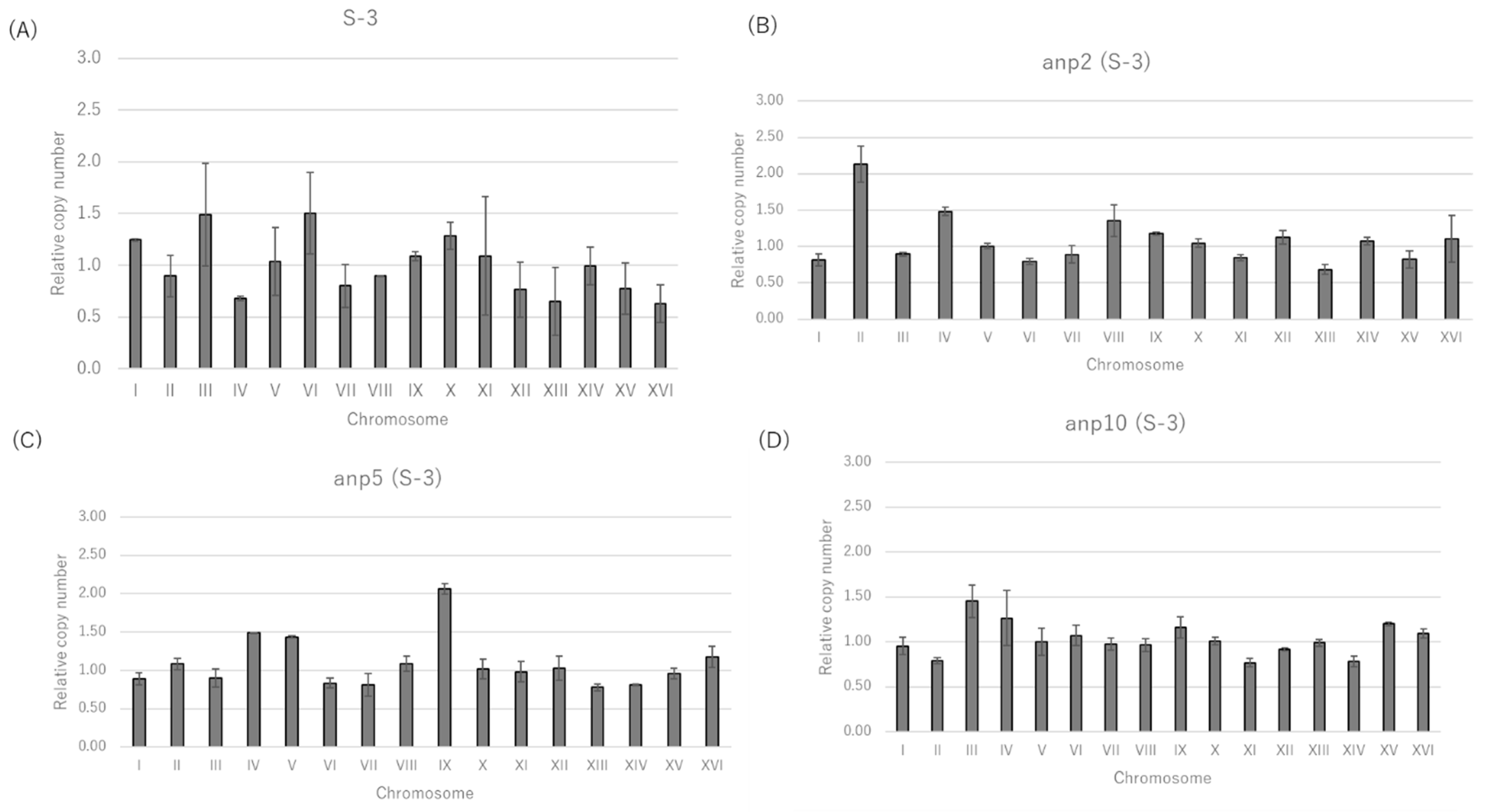
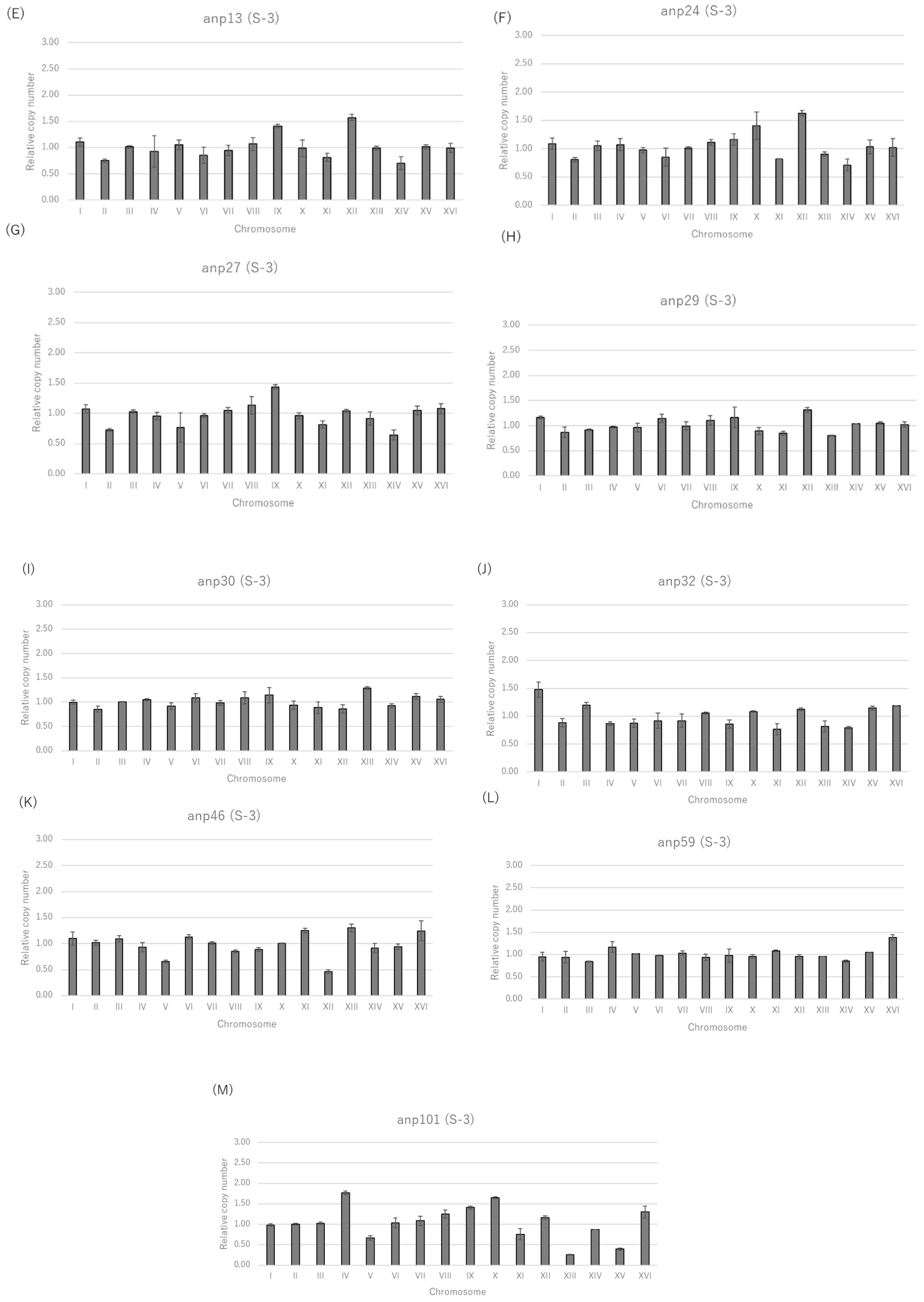

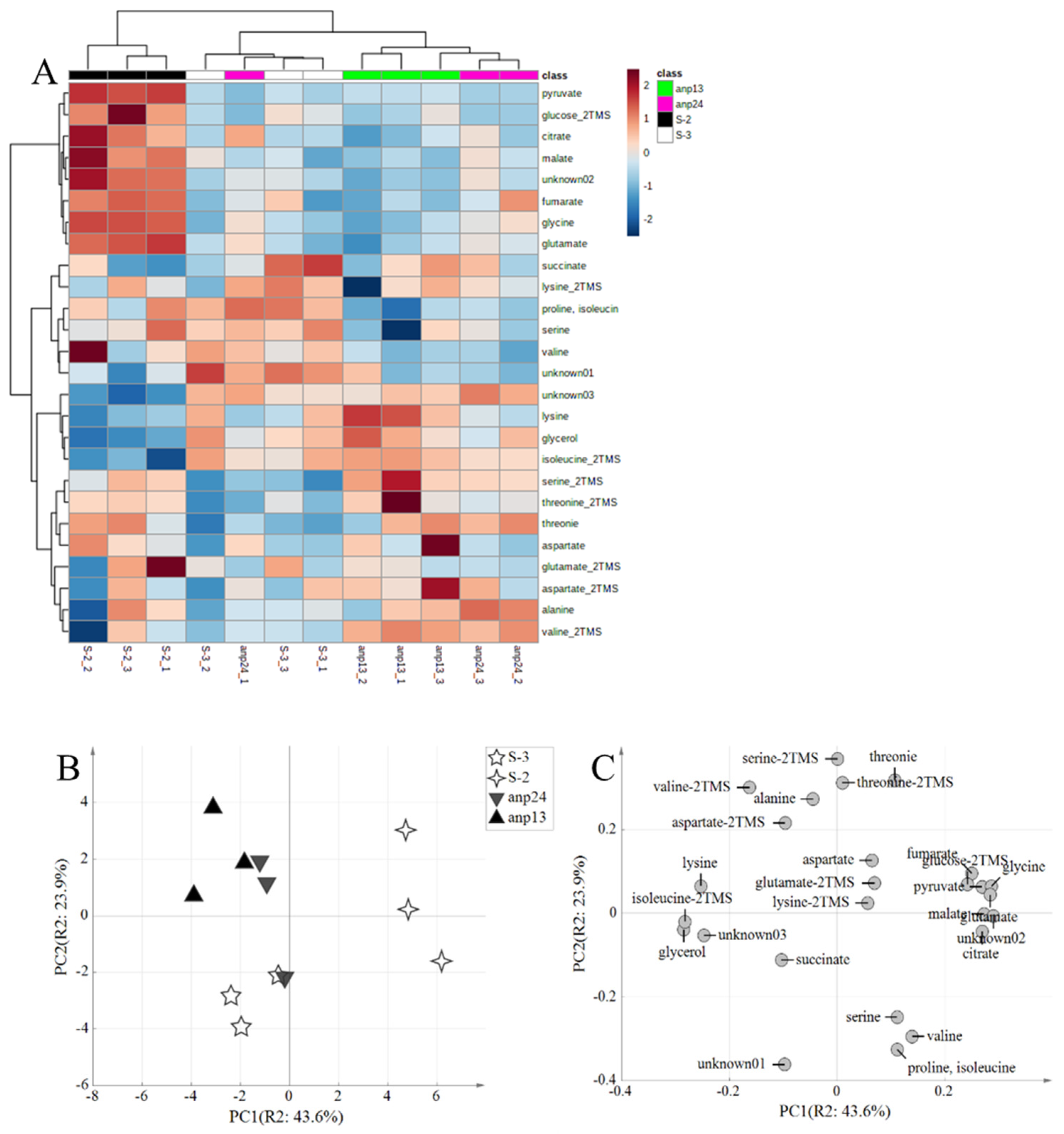

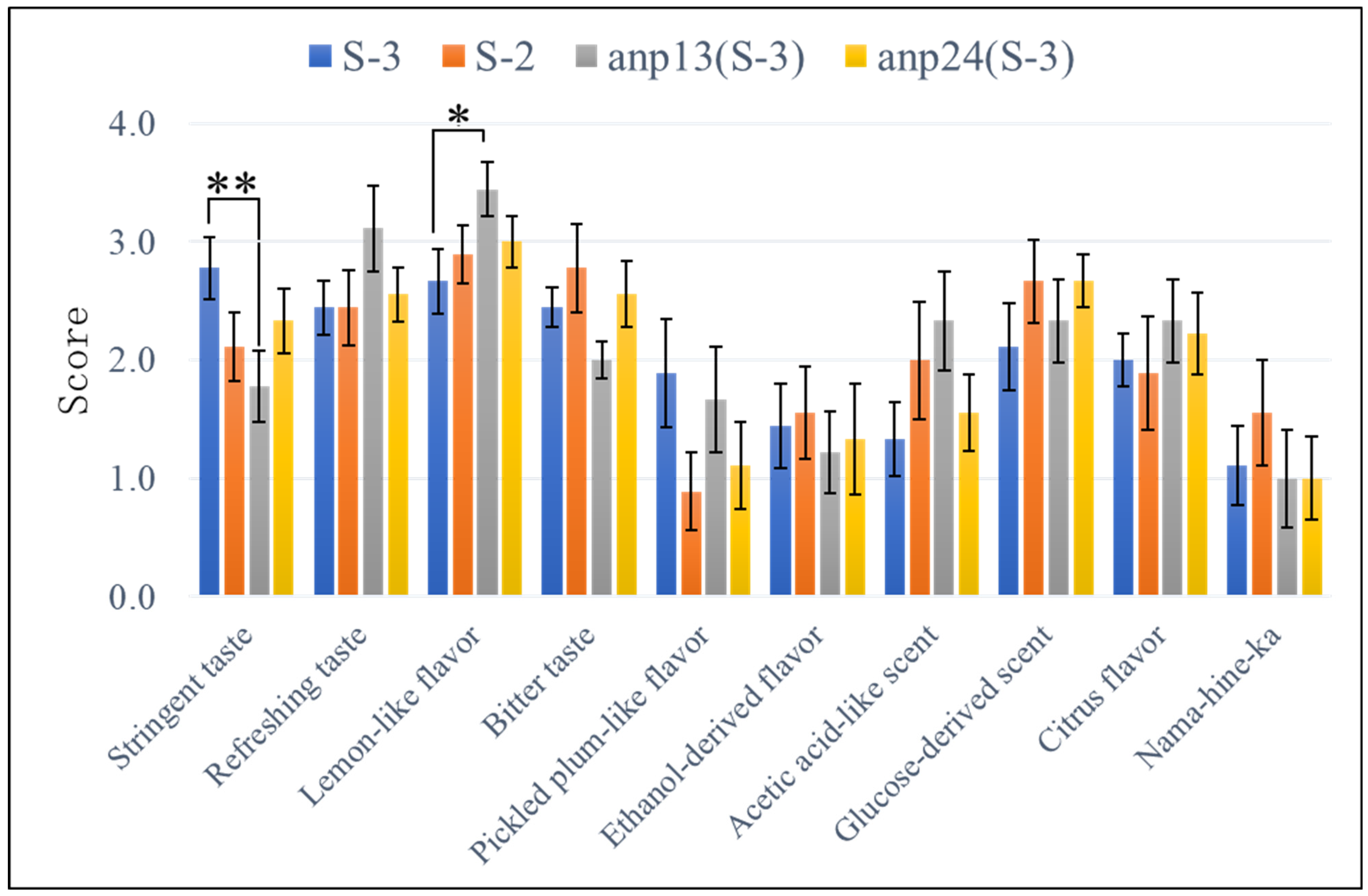
| Nocodazole | ||||||
|---|---|---|---|---|---|---|
| Untreated (Control) | Treated | |||||
| 1st | 2nd | 3rd | 1st | 2nd | 3rd | |
| Colony-forming unit | 5000 | 5000 | 5000 | 3000 | 5000 | 5000 |
| TTC straining | 12 | 2 | 6 | 8 | 36 | 30 |
| Growth rate survey | 1 | 2 | 5 | 4 | 12 | 21 |
| Karyotype analysis | 1 | 0 | 0 | 2 | 7 | 2 |
| Appearance frequency of aneuploids | 2.0 × 10−4 | 0 | 0 | 6.7 × 10−4 | 1.4 × 10−3 | 4.0 × 10−4 |
| Mean (±SE) | 6.7 × 10−5 (±1.6 × 10−5) | 8.2 × 10−4 (±7.0 × 10−5) * | ||||
| Mean ± SEM a | ||||||
|---|---|---|---|---|---|---|
| μ (h−1) | Qs (gg−1h−1) | Qetoh (gg−1h−1) | Qs/x (gg−1) | Qetoh/x (gg−1) | CDM (h−1) | |
| S-3 | 0.16 ± 0.01 | 1.15 ± 0.21 | 1.23 ± 0.02 | 55.32 ± 10.03 | 58.86 ± 1.12 | 0.84 ± 0.04 |
| S-2 | 0.13 ± 0.01 * | 1.42 ± 0.09 | 1.43 ± 0.03 *** | 68.83 ± 4.42 | 68.83 ± 1.24 *** | 0.69 ± 0.02 *** |
| anp13 (S-3) | 0.15 ± 0.00 | 1.47 ± 0.10 | 1.32 ± 0.02 | 70.77 ± 4.69 | 63.20 ± 1.15 | 0.75 ± 0.01 * |
| anp24 (S-3) | 0.16 ± 0.01 | 1.25 ± 0.18 | 1.38 ± 0.05 ** | 60.07 ± 8.76 | 66.34 ± 2.61 ** | 0.78 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusaba, Y.; Otsuka, A.; Dai, H.; Inaba, S.; Kitagaki, H. Induction of Chromosomal Aneuploids from Brewery Shochu Yeast with Novel Brewery Characteristics. Fermentation 2022, 8, 62. https://doi.org/10.3390/fermentation8020062
Kusaba Y, Otsuka A, Dai H, Inaba S, Kitagaki H. Induction of Chromosomal Aneuploids from Brewery Shochu Yeast with Novel Brewery Characteristics. Fermentation. 2022; 8(2):62. https://doi.org/10.3390/fermentation8020062
Chicago/Turabian StyleKusaba, Yuki, Akira Otsuka, Huanghuang Dai, Shigeki Inaba, and Hiroshi Kitagaki. 2022. "Induction of Chromosomal Aneuploids from Brewery Shochu Yeast with Novel Brewery Characteristics" Fermentation 8, no. 2: 62. https://doi.org/10.3390/fermentation8020062
APA StyleKusaba, Y., Otsuka, A., Dai, H., Inaba, S., & Kitagaki, H. (2022). Induction of Chromosomal Aneuploids from Brewery Shochu Yeast with Novel Brewery Characteristics. Fermentation, 8(2), 62. https://doi.org/10.3390/fermentation8020062







