Optimization of Alpha-Amylase Production by a Local Bacillus paramycoides Isolate and Immobilization on Chitosan-Loaded Barium Ferrite Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Soil Samples and Isolation of Alpha-Amylase Producers
2.2. Dinitrosalicylic Acid Assay (DNS) to Detect Alpha-Amylase Activity
2.3. Identification of the Most Potent Alpha-Amylase Producer MS009
2.4. Optimization of Alpha-Amylase Production
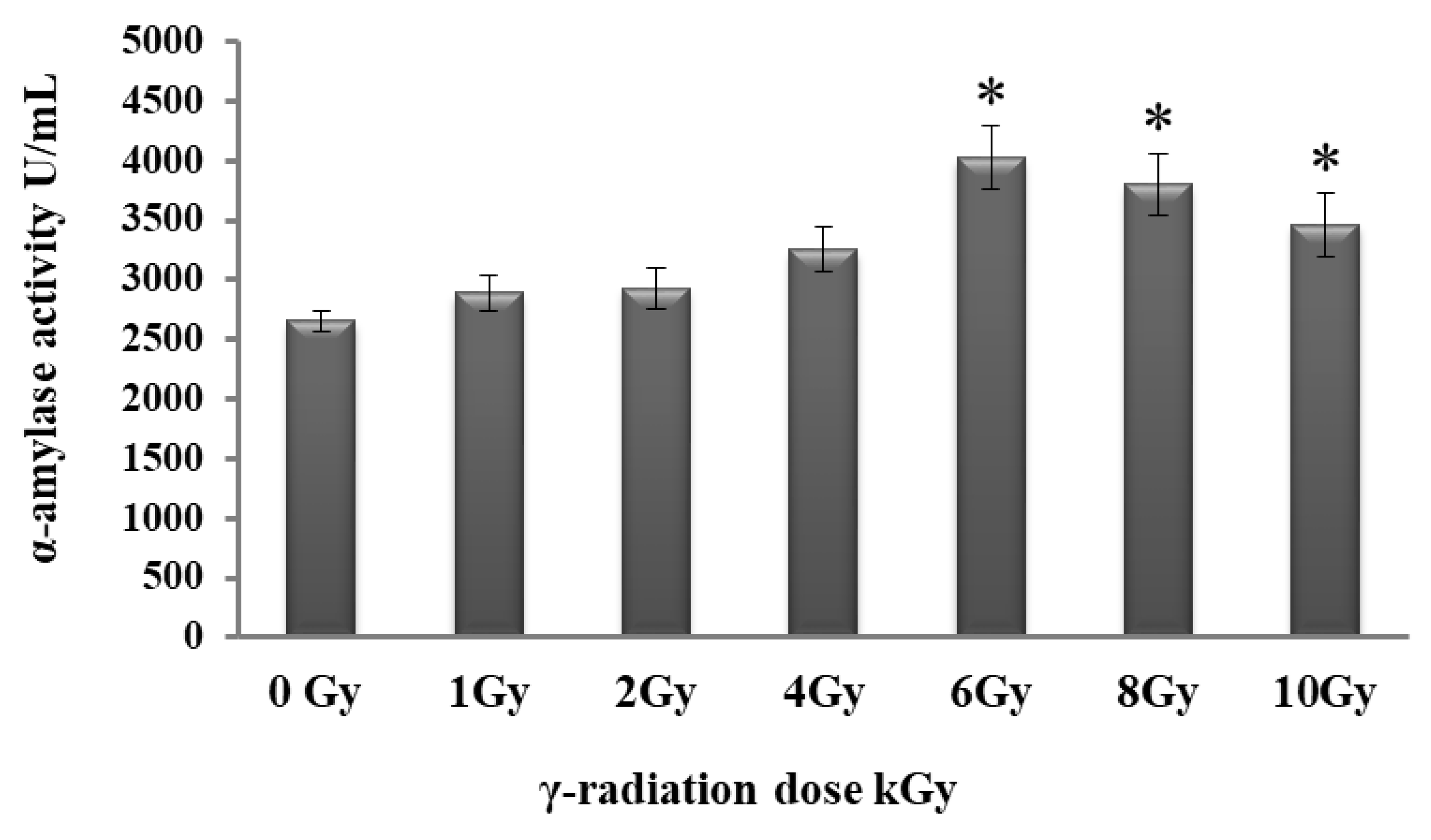
2.5. Effect of Gamma Irradiation on Alpha-Amylase Production
2.6. Partial Purification of Alpha-Amylase
2.7. Immobilization of the Partially Purified Alpha-Amylase on Chitosan-Loaded Barium Ferrite Nanoparticles (CLBFNPs)
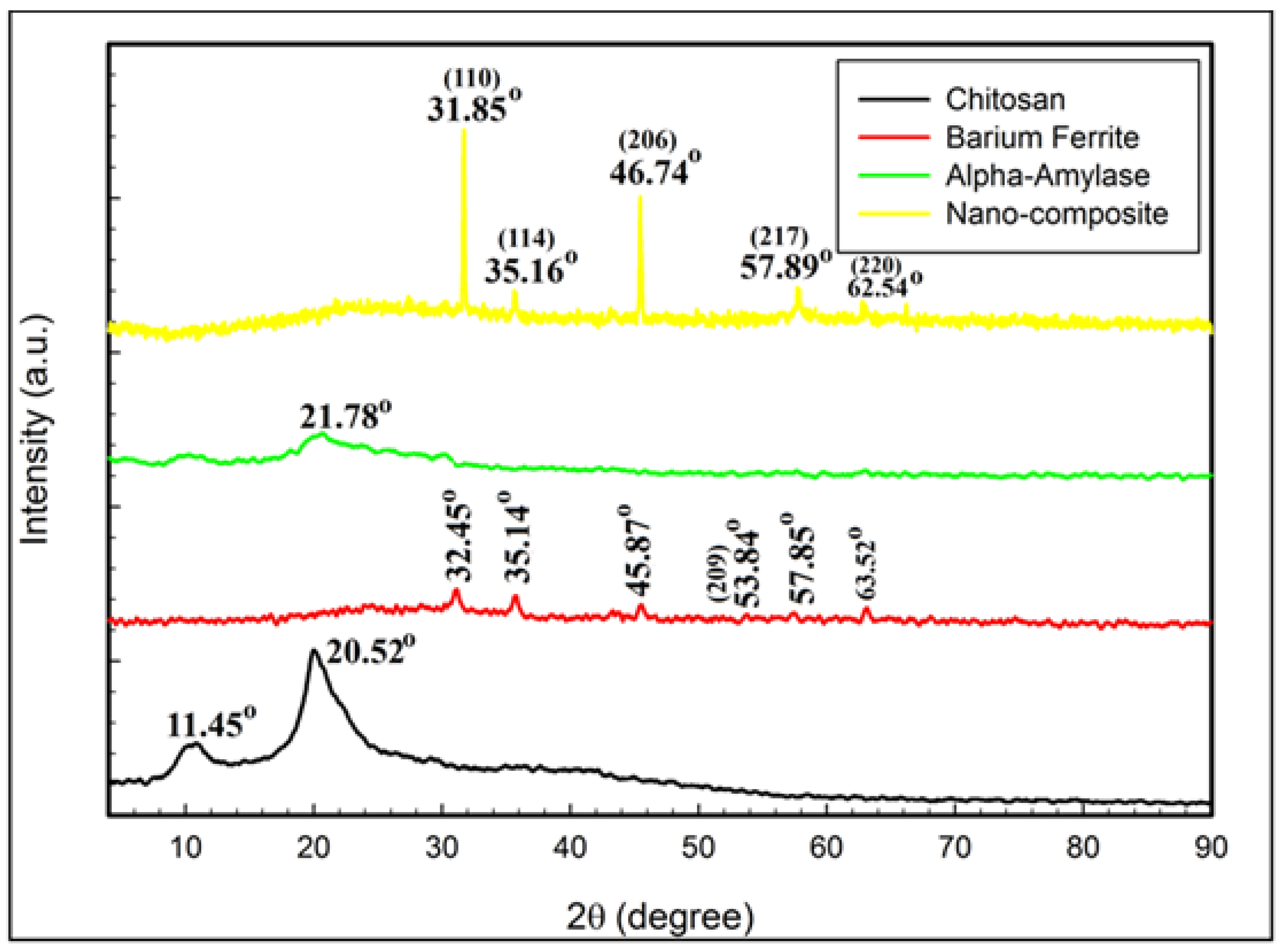
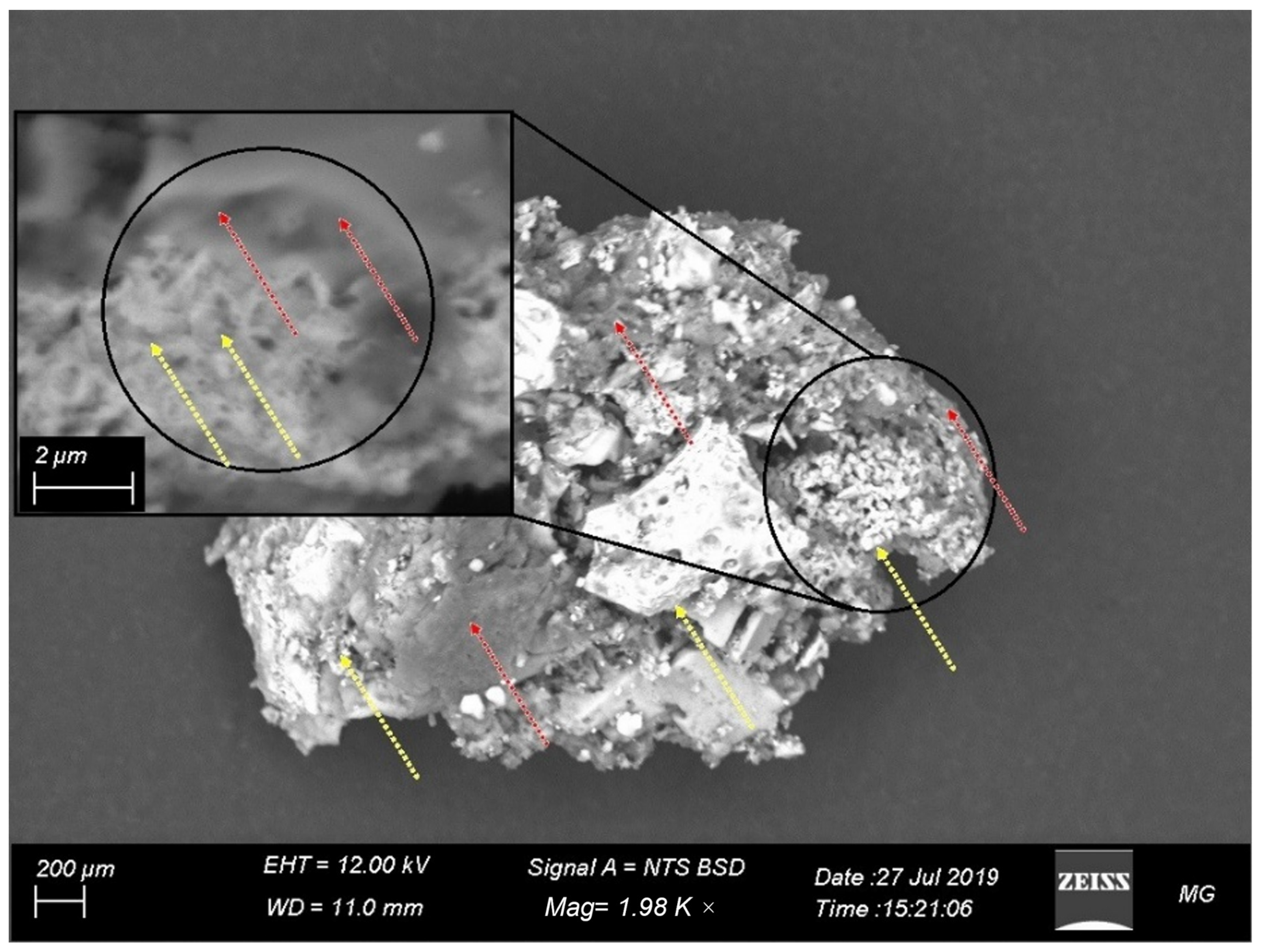
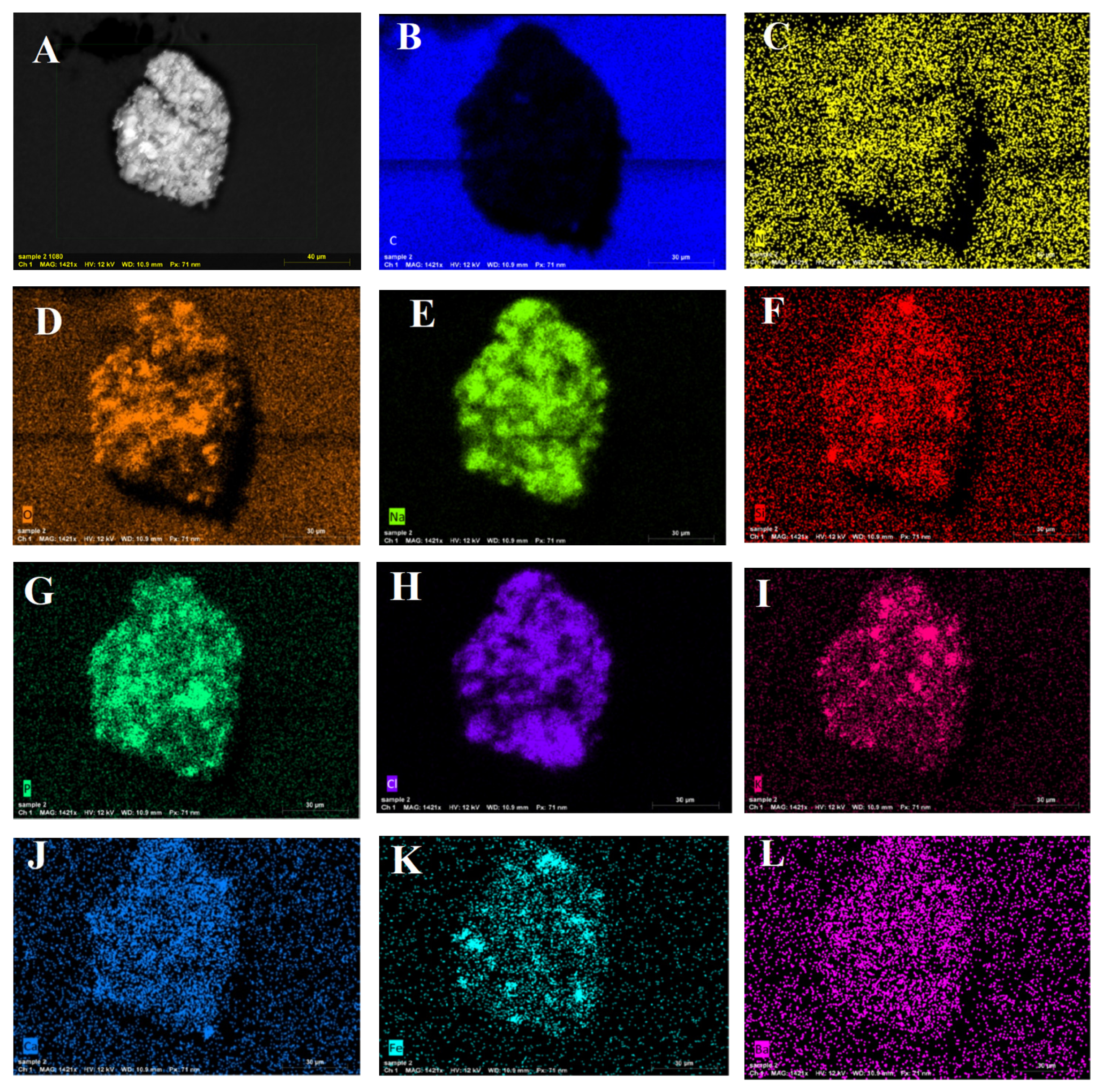
2.8. Characterization of the Prepared α-Amylase-CLBFNPs
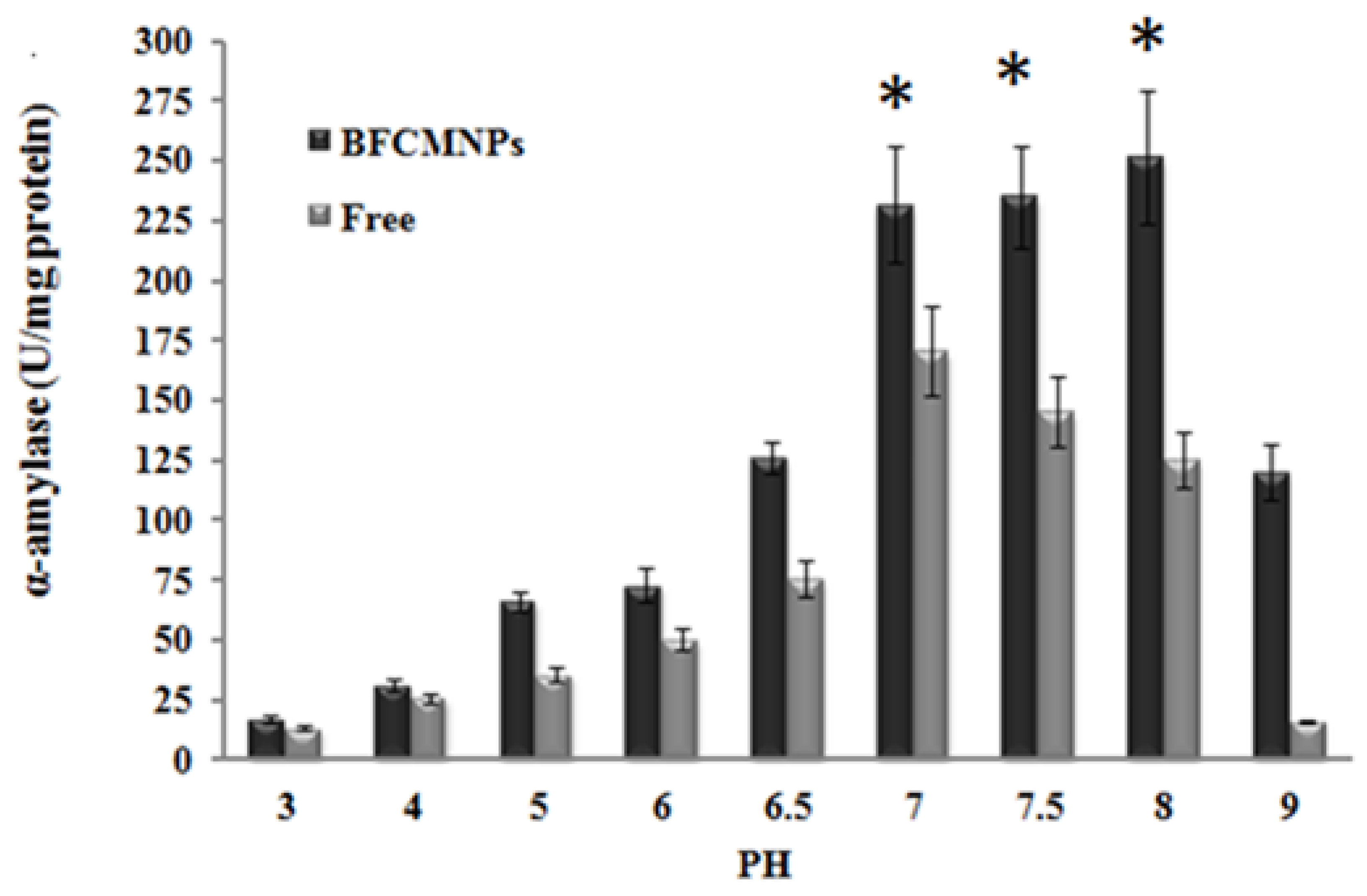
2.9. Effects of Different Working Temperatures and pH on the Activity of the Prepared α-Amylase-CLBFNPs
2.10. Reusability of the Prepared α-Amylase-CLBFNPs
2.11. Statistical Analysis
3. Results
3.1. Isolation of Alpha-Amylase Producers from Soil
3.2. Identification of MS009
3.3. Plackett–Burman Design to Optimize Alpha-Amylase Production
3.4. Central Composite Design to Optimize Alpha-Amylase Production
3.5. Gamma Irradiation Enhanced the Production of Alpha-Amylase
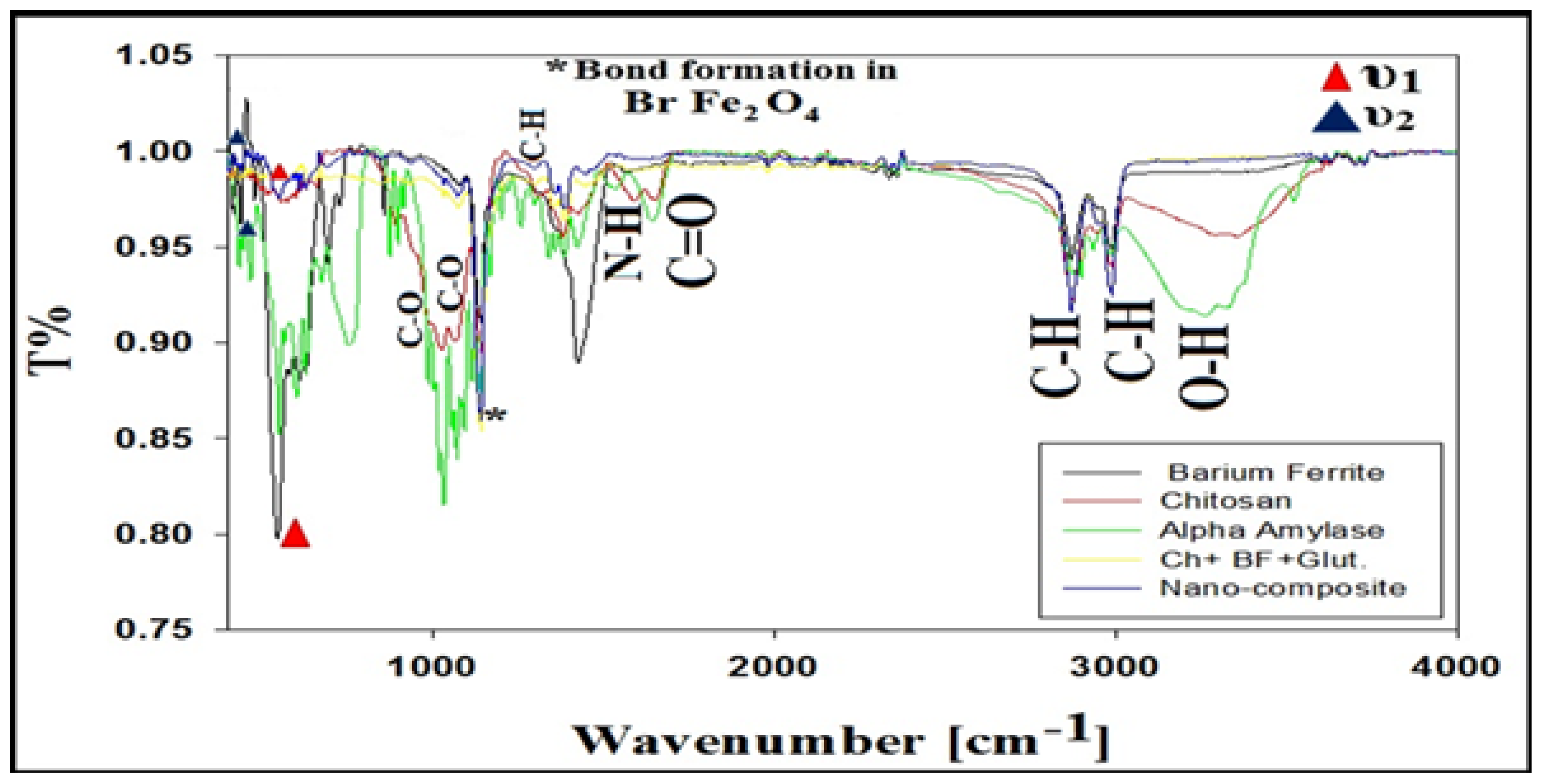
3.6. XRD, FTIR, and SEM/EDAX Characterization of Alpha-Amylase-CLBFNPs
3.7. The Synthesized Alpha-Amylase-CLBFNPs Are Active and Reusable
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumari, N.; Sushil; Rani, B.; Malik, K.; Avtar, R. Microbial amylases: An overview on recent advancement. J. Entomol. Zool. Stud. 2019, 7, 198–205. [Google Scholar]
- Hussain, I.; Siddique, F.; Mahmood, M.S.; Ahmed, S.I. A Review of the Microbiological Aspect of α-amylase Production. Int. J. Agric. Biol. 2013, 15, 1029–1034. [Google Scholar]
- Mahajan, R.; Manhas, M.; Priya, B.; Kaur, K.; Kaur, G. Isolation and optimization of amylase producing bacteria from different soils of Jammu province. Res. J. Pharm. Biol. Chem. Sci. 2011, 2, 239–246. [Google Scholar]
- Ram, K.T.; Ramkumar, J.; Karthikeyan, S.; Ramesh, B.N.G.; Manivasagan, V. Isolation, optimization, production and purification of alpha amylase from soil bacteria. Int. Res. J. Eng. Technol. 2017, 4, 2290–2293. [Google Scholar]
- Ire, F.S.; Okolo, B.N.; Moneke, A.N.; Odibo, F.J.C. Influence of cultivation conditions on the production of a protease from Aspergillus carbonarius using submerged fermentation. Afr. J. Food Sci. 2011, 5, 353–365. [Google Scholar]
- Singh, S.; Sharma, V.; Soni, M.L. Biotechnological applications of industrially important amylase enzyme. Int. J. Pharm. Biol. Sci. 2011, 2, 487–496. [Google Scholar]
- Krishnamoorthy, K.; Landge, A.V. Statistical Optimisation of Fermentation Media for Novel Clostridium novyi-NT Using Response Surface Methodology. Braz. Arch. Biol. Technol. 2018, 61, 18–26. [Google Scholar] [CrossRef]
- Ahmed, K.; Valeem, E.E. Biosynthesis, purification and characterization of commercial enzyme by Penicillium expansum link. Pak. J. Bot. 2015, 47, 1521–1526. [Google Scholar]
- Djekrif-Dakhmouche, S.; Gheribi-Aoulmi, Z.; Meraihi, Z.; Bennamoun, L. Application of a Statistical Design to the Optimization of Culture Medium for α-Amylase Production by Aspergillus niger ATCC 16404 Grown on Orange Waste Powder. J. Food Eng. 2006, 73, 190–197. [Google Scholar] [CrossRef]
- Jha, S.; Modi, H.A. Statistical optimization of chitinase production by Streptomyces rubiginosus SP24 and efficacy of purifed chitinase to control root-knot nematode infection in Vigna radiata under controlled conditions. Chem. Biol. Technol. Agric. 2018, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Haddar, A.; Fakhfakh-Zouari, N.; Hmidet, N.; Frikha, F.; Nasri, M.; Kamoun, A.S. Low-cost fermentation medium for alkaline protease production by Bacillus mojavensis A21 using hulled grain of wheat and sardinella peptone. J. Biosci. Bioeng. 2010, 10, 288–294. [Google Scholar] [CrossRef]
- Niyongabo, N.F. Production of Microbial Industrial Enzymes. Acta Sci. Microbiol. 2019, 2, 75–89. [Google Scholar] [CrossRef]
- Yahya, S.; Jahangir, S.; Shaukat, S.S.; Sohail, M.; Khan, S.A. Production optimization by using plackett-burman design and partial characterization of amylase from aspergillus tubingensis sy. Pak. J. Bot. 2016, 48, 2557–2561. [Google Scholar]
- Diep, T.B.; Thom, N.T.; Sang, H.D.; Binh, N.V.; An, T.X.; Thao, H.P.; Duong, P.D.; Quynh, T.M.; Thuan, T.B.; Lan, V.T.T. Study on Enhancement Protease-Producing of Bacillus Subtilis by Combining Ribosome Engineering and Gamma Irradiation; Lan, N.T.P., Ed.; (VINATOM-Annual Report—2016); Vietnam Atomic Energy Institute: Hanoi, Vietnam, 2017; pp. 16–19. [Google Scholar]
- Kodym, A.; Afza, R. Physical and chemical mutagenesis. Methods Mol. Biol. 2003, 236, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Moussa, H.R. Effect of gamma radiation on antioxidant enzymes and G6PDH activities in vicia faba plants. J. New Seeds 2008, 9, 89–99. [Google Scholar] [CrossRef]
- Awan, S.D.; Tabbasam, N.; Ayub, N.; Babar, M.E.; Rahman, M.; Ran, S.M.; Rajoka, M.I. Gamma radiation induced mutagenesis in Aspergillus niger to enhance its microbial fermentation activity for industrial enzyme production. Mol. Biol. Rep. 2011, 38, 1367–1374. [Google Scholar] [CrossRef]
- Gomaa, O.M.; Momtaz, O.A. 16S rRNA characterization of a Bacillus isolate and its tolerance profile after subsequent subculturing. Arab J. Biotech. 2007, 10, 107–116. [Google Scholar]
- Arabaci, G.; Usluoglu, A. Catalytic properties and immobilization studies of catalase from Malva sylvestris L. J. Chem. 2013, 2013, 686185. [Google Scholar] [CrossRef] [Green Version]
- Danial, E.N.; Elnashar, M.M.; Awad, G.E. Immobilized Inulinase on Grafted Alginate Beads Prepared by the One-Step and the Two-Steps Methods Indus. Chem. Eng. Res. 2010, 49, 3120–3125. [Google Scholar] [CrossRef]
- El-nashar, M.M.M. Immobilized Molecules Using Biomaterials and Nano biotechnology. J. Biomater. Nanobiotechnol. 2010, 1, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Cetinus, S.A.; Sahin, E.; Saraydin, D. Preparation of Cu (II) adsorbed chitosan beads for catalase immobilization. Food Chem. 2009, 114, 962–969. [Google Scholar] [CrossRef]
- Eberhardt, A.M.; Pedroni, V.; Volpe, M.; Ferreira, M.L. Immobilization of catalase from Aspergillus niger on inorganic and biopolymeric supports for H2O2 decomposition. Appl. Catal. B Environ. 2004, 47, 153–163. [Google Scholar] [CrossRef]
- Cetinus, S.A.; Oztop, H.N. Immobilization of catalase on chitosan film. Enzym. Microb. Technol. 2000, 26, 497–501. [Google Scholar] [CrossRef]
- Kaushal, J.; Seema; Singh, G.; Arya, S.K. Immobilization of catalase onto chitosan and chitosan–bentonite complex: A comparative study. Biotechnol. Rep. 2018, 18, e00258. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Y.; Wang, Y.; Pan, Q.; Ding, X.; Xu, K.; Li, N.; Wen, Q. Ionic liquid-coated Fe3O4/APTES/graphene oxide nanocomposites: Synthesis, characterization and evaluation in protein extraction processes. RSC Adv. 2016, 6, 5718–5728. [Google Scholar] [CrossRef]
- Singh, V.; Rakshit, K.; Rathee, S.; Angmo, S.; Kaushal, S.; Garg, P.; Chung, J.H.; Sandhir, R.; Sangwan, R.S.; Singhal, N. Metallic/bimetallic magnetic nanoparticle functionalization for immobilization of α-amylase for enhanced reusability in bio-catalytic processes. Bioresour. Technol. 2016, 214, 528–533. [Google Scholar] [CrossRef]
- Galli, M.; Guerrini, A.; Cauteruccio, S.; Thakare, P.; Dova, D.; Orsini, F.; Arosio, P.; Carrara, C.; Sangregoria, C.; Lascialfari, A.; et al. Superparamagnetic iron oxide nanoparticles functionalized by peptide nucleic acids. RSC Adv. 2017, 7, 15500–15512. [Google Scholar] [CrossRef] [Green Version]
- Waifalkar, P.P.; Parit, S.B.; Chougale, A.D.; Sahoo, S.C.; Patil, P.S.; Patil, P.B. Immobilization of invertase on chitosan coated γ-Fe2O3 magnetic nanoparticles to facilitate magnetic separation. J. Colloid Interface Sci. 2016, 482, 159–164. [Google Scholar] [CrossRef]
- Thangaraj, B.; Jia, Z.; Dai, L.; Liu, D.; Du, W. Effect of silica coating on Fe3O4 magnetic nanoparticles for lipase immobilization and their application for biodiesel production. Arab. J. Chem. 2016, 12, 4694–4706. [Google Scholar] [CrossRef] [Green Version]
- Leitgeb, M.; Knez, Ž.; Vasić, K. Micro- and Nanocarriers for Immobilization of Enzymes. In Micro and Nanotechnologies for Biotechnology; Stanciu, S.G., Ed.; IntechOpen: London, UK, 2016; Available online: https://www.intechopen.com/chapters/50581 (accessed on 31 March 2022). [CrossRef] [Green Version]
- Wallyn, J.; Anton, N.; Vandamme, T.F. Synthesis, Principles, and Properties of Magnetite Nanoparticles for In Vivo Imaging Applications—A Review. Pharmaceutics 2019, 11, 601. [Google Scholar] [CrossRef] [Green Version]
- El-Batal, A.I.; Farrag, A.A.; Elsayed, M.A.; El-Khawaga, A.M. Biodiesel Production by Aspergillus niger Lipase Immobilized on Barium Ferrite Magnetic Nanoparticles. Bioenginerring 2016, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Du, J.; Lai, Q.; Zeng, R.; Ye, D.; Xu, J.; Shao, Z. Proposal of nine novel species of the Bacillus cereus group. Int. J. Syst. Evol. Microbiol. 2017, 67, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kohler, N.; Zhang, M. Surface modification of superparamagnetic magnetite nanoparticles and their intracellular uptake. Biomaterials 2002, 23, 1553–1561. [Google Scholar] [CrossRef]
- Gawande, M.B.; Goswami, A.; Asefa, T.; Guo, H.; Biradar, A.V.; Beng, D.-L.; Zboril, R.; Varma, R.S. Core–shell nanoparticles: Synthesis and applications in catalysis and electrocatalysis. Chem. Soc. Rev. 2015, 44, 7540–7590. [Google Scholar] [CrossRef]
- Ominyi, M.C.; Ogbonna, J.C.; Nwoba, E.G.; Nwagu, K.E.; Ukachi, R. Isolation and Screening of α-amylase and Glucoamylase Producing Fungi and Their Application in Bioethanol Production. Int. J. Sci. Nat. 2013, 4, 44–55. [Google Scholar]
- Beyler-Çiğil, A.; Çakmakçı, E.; Danış, Ö.; Demir, S.; Kahraman, M.V. Alpha-amylase Immobilization on Modified Polyimide Material. Chem. Eng. Trans. 2013, 32, 1687–1692. [Google Scholar]
- Padhiar, A.R.; Kommu, S. Isolation, Characterization and Optimization of Bacteria producing Amylase. Int. J. Adv. Res. Biol. Sci. 2016, 3, 1–7. [Google Scholar]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; S Kaushik, K.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in Chemical and Biological Methods to Identify Microorganisms—From Past to Present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [Green Version]
- Safari, M.S.; Keyhanfar, M.; Shafiei, R. Investigating the antibacterial effects of some Lactobacillus, Bifidobacterium and acetobacter strains killed by different methods on Streptococcus mutans and Escherichia coli. Mol. Biol. Res. Commun. 2019, 8, 103–111. [Google Scholar]
- Cihan, A.C.; Tekin, N.; Ozcan, B.; Cokmus, C. The Genetic Diversity OF Genus Bacillus and the Related Genera Revealed by 16S rRNA Gene Sequences and ARDRA Analysis Isolated from Geothermal Regions Of Turkey. Braz. J. Microbiol. 2012, 43, 309–324. [Google Scholar] [CrossRef]
- Helmy, O.M.; Kashef, M.T. Different phenotypic and molecular mechanisms associated with multidrug resistance in Gram-negative clinical isolates from Egypt. Infect. Drug Resist. 2017, 10, 479–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheherazed, D.D.; Leila, B.; Amel, A.K.; Kenza, L.; Tahar, N.; Zoubida, G.A.; Zahia, M. An Optimization Study of α-Amylase Production by Aspergillus niger ATCC 16404 Grown on Orange Waste Powder. Adv. Biosci. Biotechnol. 2016, 7, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Ali, E.H.; El-Nagdy, M.A.; Al-Garni, S.M.; Ahmed, M.S.; Rawaa, A.M. Enhancement of alpha amylase production by Aspergillus. Eur. J. Biol. Res. 2017, 7, 154–164. [Google Scholar]
- Iftikhar, T.; Niaz, M.; Hussain, Y.; Abbas, S.Q.; Ashraf, I.; Anjum, Z.M. Improvement of Selected Strains through Gamma Irradiation for Enhanced Lipolytic Potential. Pak. J. Bot. 2010, 42, 2257–2267. [Google Scholar]
- Tallapragada, P.; Dikshit, R.; Jadhav, A.; Sarah, U. Partial purification and characterization of amylase enzyme under solid state fermentation from Monascus sanguineus. J. Genet. Eng. Biotechnol. 2017, 15, 95–101. [Google Scholar] [CrossRef]
- Dhavale, R.P.; Parit, S.B.; Sahoo, S.C.; Kollu, P.; Patil, P.S.; Patil, P.B.; Chougale, A.D. α-amylase immobilized on magnetic nanoparticles:reusable robust nano-biocatalyst for starch hydrolysis. Mater. Res. Express. 2018, 5, 075403. [Google Scholar] [CrossRef]
- Prakash, O.; Jaiswal, N. Immobilization of a Thermostable—Amylase on Agarose and Agar Matrices and its Application in Starch Stain Removal. World Appl. Sci. J. 2011, 13, 572–577. [Google Scholar]
- Díaz-Hernández, A.; Gracida, J.; García-Almendárez, B.E.; Regalado, C.; Núñez, R.; Amaro-Reyes, A. Characterization of Magnetic Nanoparticles Coated with Chitosan: A Potential Approach for Enzyme Immobilization. J. Nanomater. 2018, 2018, 9468574. [Google Scholar] [CrossRef]
- Kumar, S.; Morya, V.; Gadhavi, J.; Vishnoi, A.; Singh, J.; Datta, B. Investigation of nanoparticle immobilized cellulase: Nanoparticle identity, linker length and polyphenol hydrolysis. Heliyon 2019, 5, e01702. [Google Scholar] [CrossRef] [Green Version]
- Vengadaramana, A.; Balakumar, S.; Arasaratnam, V. Effect of temperature, pH, substrate (Starch) and glucose on stability of α-amylase from Bacillus licheniformis ATCC 6346. Sch. Acad. J. Pharm. 2014, 3, 492–495. [Google Scholar]
- Tanyildizi, M.S.; Ozer, D.; Elibol, M. Optimization of Amylase production by Bacillus sp. using Response surface methodology. Process Biochem. 2005, 40, 2291–2296. [Google Scholar] [CrossRef]
- Fossi, B.T.; Taveaand, F.; Ndjonenkeu, T. Production and Partial Characterization of a Themostable amylase from Ascomycetes yeast strain isolated from starchy soils. Afr. J. Biotechnol. 2005, 4, 14–18. [Google Scholar]
- Gopinath, S.C.B.; Anbu, P.; Arshad, M.K.M.; Lakshmipriya, T.; Voon, C.H.; Hashim, U.; Chinni, S.V. Biotechnological Processes in Microbial Amylase Production. Biomed. Res. Int. 2017, 2017, 1272193. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.X.; Chang, Y.P. Valorizing guava (Psidium guajava L.) seeds through germination-induced carbohydrate changes. J. Food Sci. Technol. 2017, 54, 2041–2049. [Google Scholar] [CrossRef]
- Keharom, S.; Mahachai, R.; Chanthai, S. The optimization study of α-amylase activity based on central composite design-response surface methodology by dinitrosalicylic acid method. Int. Food Res. J. 2016, 23, 10–17. [Google Scholar]
- Cordeiro, C.A.M.; Martinas, M.L.L.; Lucaino, A. Production and Properties of alpha amylase from thermophylic Bacillus species. Braz. J. Microbiol. 2003, 33, 57–61. [Google Scholar]
- Far, B.E.; Ahmadi, Y.; Khosroshahi, A.Y.; Dilmaghani, A. Microbial Alpha-Amylase Production: Progress, Challenges and Perspectives. Adv. Pharm. Bull. 2020, 10, 350–358. [Google Scholar]
- Panda, S.H.; Swain, M.R.; Kar, S.; Ray, R.C.; Montet, D. Statistical Optimization of Alpha Amylase Production by Probiotic Lactobacillus Plantarum MTCC 1407 in Submerged Fermentation. Pol. J. Microbiol. 2008, 57, 149–155. [Google Scholar]
- Aly, E.E.; Abd-El-Hamid, G.; Abo-El-Soued, M.A.; Khalil, N.M.; Mostafa, H.S. Mutagenic Effect of Gamma Irradiation on Phenotype and Enzyme Activities in Aspergillus niger. Egypt. J. Bot. 2016, 56, 507–526. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, H.M.M.; Bashandy, A.S. Effect of Gamma Radiation on Alpha-Amylase Production by Bacillus Subtilis Utilizing Potato Peel Waste. Isot. Rad. Res. 2010, 42, 1071–1082. [Google Scholar]
- El-Batal, A.; Karem, H.A. Phytase production and phytic acid reduction in rapeseed meal by Aspergillus niger during solid state fermentation. Int. Food Res. J. 2001, 34, 715–720. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Reisz, J.A.; Bansal, N.; Qian, J.; Zhao, W.; Furdui, C.M. Effects of ionizing radiation on biological molecules—Mechanisms of damage and emerging methods of detection. Antioxid. Redox Signal. 2014, 21, 260–292. [Google Scholar] [CrossRef] [PubMed]
- Swarnalatha, V.; Aluri Esther, R.; Dhamodharan, R. Immobilization of α-amylase on gum acacia stabilized magnetite nanoparticles, an easily recoverable and reusable support. J. Mol. Catal. B Enzym. 2013, 96, 6–13. [Google Scholar] [CrossRef]
- Long, J.; Li, X.; Zhan, X.; Xu, X.; Tian, Y.; Xie, Z.; Jin, Z. Sol–gel encapsulation of pullulanase in the presence of hybrid magnetic (Fe3O4–chitosan) nanoparticles improves thermal and operational stability. Bioprocess Biosyst. Eng. 2017, 40, 821–831. [Google Scholar] [CrossRef]
- El-Shishtawy, R.M.; Ahmed, N.S.E.; Almulaiky, Y.Q. Immobilization of Catalase on Chitosan/ZnO and Chitosan/ZnO/Fe2O3 Nanocomposites: A Comparative Study. Catalysts 2021, 11, 820. [Google Scholar] [CrossRef]
- Baskar, G.; Banu, N.; Leuca, G.H.; Gayathri, V.; Jeyashree, N. Magnetic immobilization and characterization of α-amylase as nanobiocatalyst for hydrolysis of sweet potato starch. Biochem. Eng. J. 2015, 102, 18–23. [Google Scholar] [CrossRef]
- Saif, B.; Wang, C.; Chuan, D.; Shuang, S. Synthesis and characterization of Fe3O4 coated on APTES as carriers for morin-anticancer drug. J. Biomater. Nanobiotechnol. 2015, 6, 267–275. [Google Scholar] [CrossRef]
- Unsoy, G.; Yalcin, S.; Khodadust, R.; Gunduz, G.; Gunduz, U. Synthesis optimization and characterization of chitosan-coated iron oxide nanoparticles produced for biomedical applications. J. Nanoparticle Res. 2012, 14, 964. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, X.; Sun, H.; Chi, G.; Xu, J.; Sun, Y. Control synthesis of magnetic Fe3O4–chitosan nanoparticles under UV irradiation in aqueous system. Curr. Appl. Phys. 2010, 10, 828–862. [Google Scholar] [CrossRef]
- Ma, W.; Ya, F.-Q.; Han, M.; Wang, R. Characteristics of equilibrium, kinetics studies for adsorption of fluoride on magnetic chitosan particle. J. Hazard. Mater. 2007, 143, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, M.; Khani, A.; Najafzadeh, K. α-amylase immobilization on the silica nanoparticles for cleaning performance towards starch soils in laundry detergents. J. Mol. Catal. B Enzym. 2012, 74, 1–5. [Google Scholar] [CrossRef]
- Jain, M.; Sebatini, M.; Sharmila, G.; Muthukumaran, C.; Baskar, G.; Tamilarasan, K. Fabrication of a chitosan-coated magnetic nanobiocatalyst for starch hydrolysis. Chem. Eng. Technol. 2015, 38, 1444–1451. [Google Scholar] [CrossRef]
- Sohrabi, N.; Rasouli, N.; Torkzadeh, M. Enhanced stability and catalytic activity of immobilized α-amylase on modified Fe3O4 nanoparticles. Chem. Eng. J. 2014, 33, 240–426. [Google Scholar] [CrossRef]
- Milani, Z.M.; Jalal, R.; Goharshadi, E.K. Carbodiimide for covalent α-amylase immobilization onto magnetic nanoparticles. Int. J. Nanosci. 2017, 16, 175–190. [Google Scholar] [CrossRef]




| Tested Variables | Level 1 | Level 2 |
|---|---|---|
| Starch g% | 0.5 | 1 |
| Peptone g% | 0.3 | 0.6 |
| Yeast extract g% | 0.3 | 0.6 |
| Culture volume/total flask volume mL | 1/2.5 | 1/5.0 |
| Tween 80% (v/v) | 0.1 | 0.2 |
| CaCl2 g% | 1 | 2 |
| Incubation period (h) | 48 | 72 |
| Incubation temperature (°C) | 35 | 40 |
| Shaking speed (rpm) | 150 | 180 |
| pH | 7 | 8 |
| Inoculum size mL * | 2.5 | 5 |
| Variable | Peptone (g%) | Culture Volume/250 mL Flask | CaCl2 (g%) | Incubation Period (h) |
|---|---|---|---|---|
| Level 1 | 0.15 | 25 | 1 | 48 |
| Level 2 | 0.45 | 50 | 1.5 | 60 |
| Level 3 | 0.3 | 75 | 2 | 72 |
| Level 4 | 0.6 | 100 | 2.5 | 84 |
| Isolate | Source | Classification | Diameter of the Clear Zone (mm) | α Amylase Specific Activity (U/mg) |
|---|---|---|---|---|
| MA003 | AEAG | Bacteria | 8 | ND |
| MA011 | AEAG | Fungi | 8 | ND |
| MA016 | AEAG | Bacteria | 17 | ND |
| MA023 | AEAG | Fungi | 1 | ND |
| MA035 | AEAG | Bacteria | 28 | 19.22 ± 0.94 |
| MA040 | AEAG | Fungi | 12 | ND |
| MA054 | AEAG | Bacteria | 58 | 147.32 ± 5.32 |
| MA063 | AEAG | Fungi | 12 | 12.23 ± 0.67 |
| MS001 | ESFG | Bacteria | 50 | 98.99 ± 3.45 |
| MS009 | ESFG | Bacteria | 60 | 177.12 ± 6.12 |
| MS011 | ESFG | Fungi | 39 | 50.08 ± 2.05 |
| MS017 | ESFG | Fungi | 33 | 28.43 ± 1.34 |
| MS020 | ESFG | Bacteria | 5 | ND |
| MS023 | ESFG | Fungi | 45 | ND |
| MS029 | ESFG | Bacteria | 31 | 25.04 ± 1.09 |
| MS032 | ESFG | Fungi | 27 | 18.63 ± 0.88 |
| MS038 | ESFG | Bacteria | 25 | 14.56 ± 0.89 |
| MS043 | ESFG | Fungi | 58 | 140.08 ± 2.98 |
| MS057 | ESFG | Bacteria | 29 | 20.36 ± 1.02 |
| MS061 | ESFG | Fungi | 10 | ND |
| MS070 | ESFG | Bacteria | 20 | 136.862 ± 3.67 |
| MS076 | ESFG | Fungi | 30 | 21.47 ± 0.98 |
| MS079 | ESFG | Fungi | 4 | ND |
| MS083 | ESFG | Bacteria | 15 | ND |
| MS086 | ESFG | Fungi | 5 | ND |
| MS089 | ESFG | Bacteria | 5 | ND |
| MS091 | ESFG | Fungi | 10 | ND |
| MS095 | ESFG | Bacteria | 1 | ND |
| MM003 | EMVG | Fungi | 8 | ND |
| MM007 | EMVG | Bacteria | 10 | ND |
| MM012 | EMVG | Fungi | 3 | ND |
| MM017 | EMVG | Bacteria | 15 | ND |
| MM029 | EMVG | Fungi | 8 | ND |
| MM033 | EMVG | Bacteria | 45 | 9.12 ± 0.04 |
| MM036 | EMVG | Fungi | 5 | ND |
| MM040 | EMVG | Bacteria | 15 | ND |
| MM043 | EMVG | Fungi | 3 | ND |
| MM050 | EMVG | Bacteria | 10 | ND |
| MM054 | EMVG | Fungi | 2 | ND |
| MM061 | EMVG | Bacteria | 2 | ND |
| MM066 | EMVG | Fungi | 5 | ND |
| MM070 | EMVG | Bacteria | 56 | 133.11 ± 3.54 |
| MM075 | EMVG | Fungi | 10 | ND |
| MM084 | EMVG | Bacteria | 24 | 13.39 ± 0.87 |
| Run | Starch (g%) | Peptone (g%) | Yeast Extract (g%) | Culture Volume/Total Flask Volume (mL) | Tween 80 % (v/v) | CaCl2 (g%) | Incubation Period (h) | Temperature (°C) | Shaking (rpm) | pH | Inoculum Size (mL) | Amylase-Specific Activity (U/mg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0.6 | 0.3 | 1/2.5 | 0.2 | 2 | 48 | 35 | 150 | 8 | 2.5 | 10.616 ± 0.54 |
| 2 | 0.5 | 0.3 | 0.3 | 1/5.0 | 0.1 | 1 | 48 | 35 | 150 | 7 | 2.5 | 35.93 ± 1.32 |
| 3 | 0.5 | 0.3 | 0.6 | 1/5.0 | 0.2 | 2 | 48 | 40 | 180 | 8 | 2.5 | 25.586 ± 0.98 |
| 4 | 0.5 | 0.6 | 0.6 | 1/2.5 | 0.1 | 1 | 48 | 40 | 150 | 8 | 5 | 153.795 ± 3.03 |
| 5 | 1 | 0.3 | 0.6 | 1/2.5 | 0.1 | 2 | 72 | 40 | 150 | 7 | 2.5 | 53.03 ± 1.78 |
| 6 | 1 | 0.6 | 0.3 | 1/5.0 | 0.1 | 2 | 48 | 40 | 180 | 7 | 5 | 52.126 ± 1.56 |
| 7 | 0.5 | 0.6 | 0.3 | 1/5.0 | 0.2 | 2 | 72 | 35 | 150 | 7 | 5 | 187.685 ± 4.69 |
| 8 | 0.5 | 0.6 | 0.3 | 1/2.5 | 0.2 | 1 | 72 | 40 | 180 | 7 | 2.5 | 190.27 ± 4.69 |
| 9 | 0.5 | 0.3 | 0.3 | 1/2.5 | 0.1 | 2 | 72 | 35 | 180 | 8 | 5 | 26.812 ± 0.98 |
| 10 | 1 | 0.3 | 0.3 | 1/5.0 | 0.2 | 1 | 72 | 40 | 150 | 8 | 5 | 186.186 ± 4.78 |
| 11 | 1 | 0.6 | 0.6 | 1/5.0 | 0.1 | 1 | 72 | 35 | 180 | 8 | 2.5 | 222.254 ± 5.07 |
| 12 | 1 | 0.3 | 0.6 | 1/2.5 | 0.2 | 1 | 48 | 35 | 180 | 7 | 5 | 33.072 ± 1.69 |
| Run | Peptone (g%) | Aeration (Culture Volume/250 mL Flask) (mL) | CaCl2 (g%) | Incubation Period (h) | Amylase-Specific Activity (U/mg) |
|---|---|---|---|---|---|
| 1 | 0.6 | 100 | 2 | 48 | 124.72 ± 3.01 |
| 2 | 0.6 | 50 | 2 | 48 | 90.427 ± 2.35 |
| 3 | 0.3 | 100 | 2 | 72 | 225.79 ± 5.08 |
| 4 | 0.6 | 50 | 2 | 72 | 221.548 ± 4.96 |
| 5 | 0.45 | 25 | 1.5 | 60 | 108.137 ± 2.75 |
| 6 | 0.15 | 75 | 1.5 | 60 | 166.74 ± 3.58 |
| 7 | 0.3 | 50 | 2 | 48 | 128.84 ± 3.26 |
| 8 | 0.3 | 100 | 1 | 48 | 81.36 ± 2.43 |
| 9 | 0.3 | 50 | 1 | 48 | 112.693 ± 2.97 |
| 10 | 0.6 | 100 | 1 | 48 | 118.691 ± 2.47 |
| 11 | 0.45 | 75 | 1.5 | 60 | 151.833 ± 3.48 |
| 12 | 0.45 | 75 | 1.5 | 60 | 155.76 ± 3.91 |
| 14 | 0.3 | 100 | 1 | 72 | 153.01 ± 2.99 |
| 15 | 0.6 | 50 | 1 | 48 | 116.705 ± 2.98 |
| 16 | 0.45 | 75 | 1.5 | 60 | 127.023 ± 3.01 |
| 17 | 0.45 | 75 | 2.5 | 60 | 161.495 ± 4.23 |
| 18 | 0.45 | 125 | 1.5 | 60 | 102.909 ± 2.74 |
| 19 | 0.6 | 100 | 1 | 72 | 226.426 ± 5.67 |
| 20 | 0.45 | 75 | 1.5 | 84 | 232.456 ± 5.98 |
| 21 | 0.45 | 75 | 1.5 | 60 | 176.324 ± 3.79 |
| 22 | 0.45 | 75 | 0.5 | 60 | 154.256 ± 2.77 |
| 23 | 0.3 | 50 | 1 | 72 | 203.134 ± 4.58 |
| 24 | 0.6 | 100 | 2 | 72 | 190.135 ± 3.54 |
| 25 | 0.75 | 75 | 1.5 | 60 | 167.646 ± 3.47 |
| 26 | 0.3 | 100 | 2 | 48 | 101.44 ± 2.32 |
| 27 | 0.6 | 50 | 1 | 72 | 205.286 ± 4.02 |
| 28 | 0.45 | 75 | 1.5 | 60 | 171.822 ± 3.26 |
| 29 | 0.45 | 75 | 1.5 | 60 | 147.745 ± 3.26 |
| 30 | 0.3 | 50 | 2 | 72 | 214.525 ± 3.79 |
| Purification Method | Amylase-Specific Activity (U/mg) | Purification Factor |
|---|---|---|
| Crude enzyme | 68.59 ± 2.97 | 1 |
| (NH4)2SO4 (40%) | 70.985 ± 3.68 | 1.03 |
| (NH4)2SO4 (50%) | 74.42 ± 4.46 | 1.08 |
| (NH4)2SO4 (60%) | 82.5 ± 4.14 | 1.2 |
| (NH4)2SO4 (70%) | 94.75 ± 5.14 | 1.4 |
| (NH4)2SO4 (80%) | 75.1 ± 4.38 | 1.1 |
| Acetone | 100.88 ± 5.64 | 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hallol, M.; Helmy, O.; Shawky, A.-E.; El-Batal, A.; Ramadan, M. Optimization of Alpha-Amylase Production by a Local Bacillus paramycoides Isolate and Immobilization on Chitosan-Loaded Barium Ferrite Nanoparticles. Fermentation 2022, 8, 241. https://doi.org/10.3390/fermentation8050241
Hallol M, Helmy O, Shawky A-E, El-Batal A, Ramadan M. Optimization of Alpha-Amylase Production by a Local Bacillus paramycoides Isolate and Immobilization on Chitosan-Loaded Barium Ferrite Nanoparticles. Fermentation. 2022; 8(5):241. https://doi.org/10.3390/fermentation8050241
Chicago/Turabian StyleHallol, Merehan, Omneya Helmy, Alla-Eldien Shawky, Ahmed El-Batal, and Mohamed Ramadan. 2022. "Optimization of Alpha-Amylase Production by a Local Bacillus paramycoides Isolate and Immobilization on Chitosan-Loaded Barium Ferrite Nanoparticles" Fermentation 8, no. 5: 241. https://doi.org/10.3390/fermentation8050241
APA StyleHallol, M., Helmy, O., Shawky, A.-E., El-Batal, A., & Ramadan, M. (2022). Optimization of Alpha-Amylase Production by a Local Bacillus paramycoides Isolate and Immobilization on Chitosan-Loaded Barium Ferrite Nanoparticles. Fermentation, 8(5), 241. https://doi.org/10.3390/fermentation8050241







