Optimization of cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Oleaginous Yeast Strain
2.2. Culture Conditions
2.2.1. Growth Media Composition
2.2.2. Yeast Culture in Erlenmeyer Flasks (Design of Experiment)
2.2.3. Yeast Culture in Bioreactor
2.3. Determination of Substrate Consumption during Fermentation
2.4. Lipid Extraction and Fatty Acids Analysis
2.4.1. Lipid Extraction from Y. lipolytica Biomass
2.4.2. Preparation and Quantification of Fatty Acid Methyl Esters (FAMEs)
2.5. Statistical Analyses
3. Results and Discussion
3.1. Impact of the Fermentation Conditions on Yeast Growth and C17:1 Biosynthesis
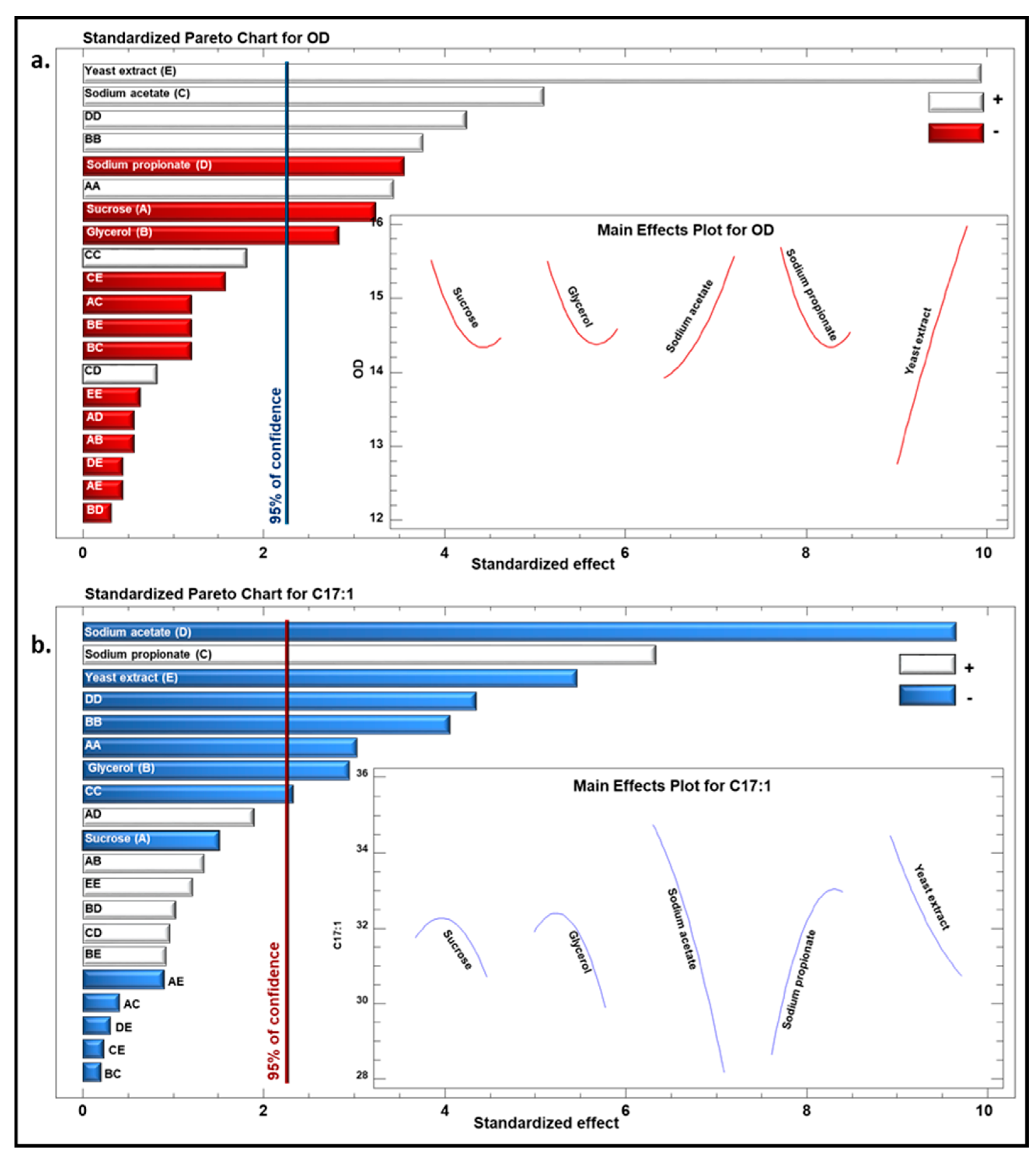
3.1.1. Models’ Validation and Fitting
X1 × X5 − 0.25 X2 × X3 − 0.07 X2 × X4 − 0.24 X2 × X5 + 0.15 X3 × X4 − 0.32 X3 × X5 − 0.08 X4 × X5 + 0.62* (X1)2 + 0.68 (X2)2 + 0.39
(X3)2 + 0.74 (X4)2 + 0.02 (X5)2
0.11 X2 × 3 + 0.41 X2 × X4 + 0.37 X2 × X5 + 0.39 X3 × X4 − 0.1 X3 × X5 − 0.11 X4 × X5 − 0.95 (X1)2 − 1.25 (X2)2 − 0.68 (X3)2 − 1.40
(X4)2 + 0.17 (X5)2
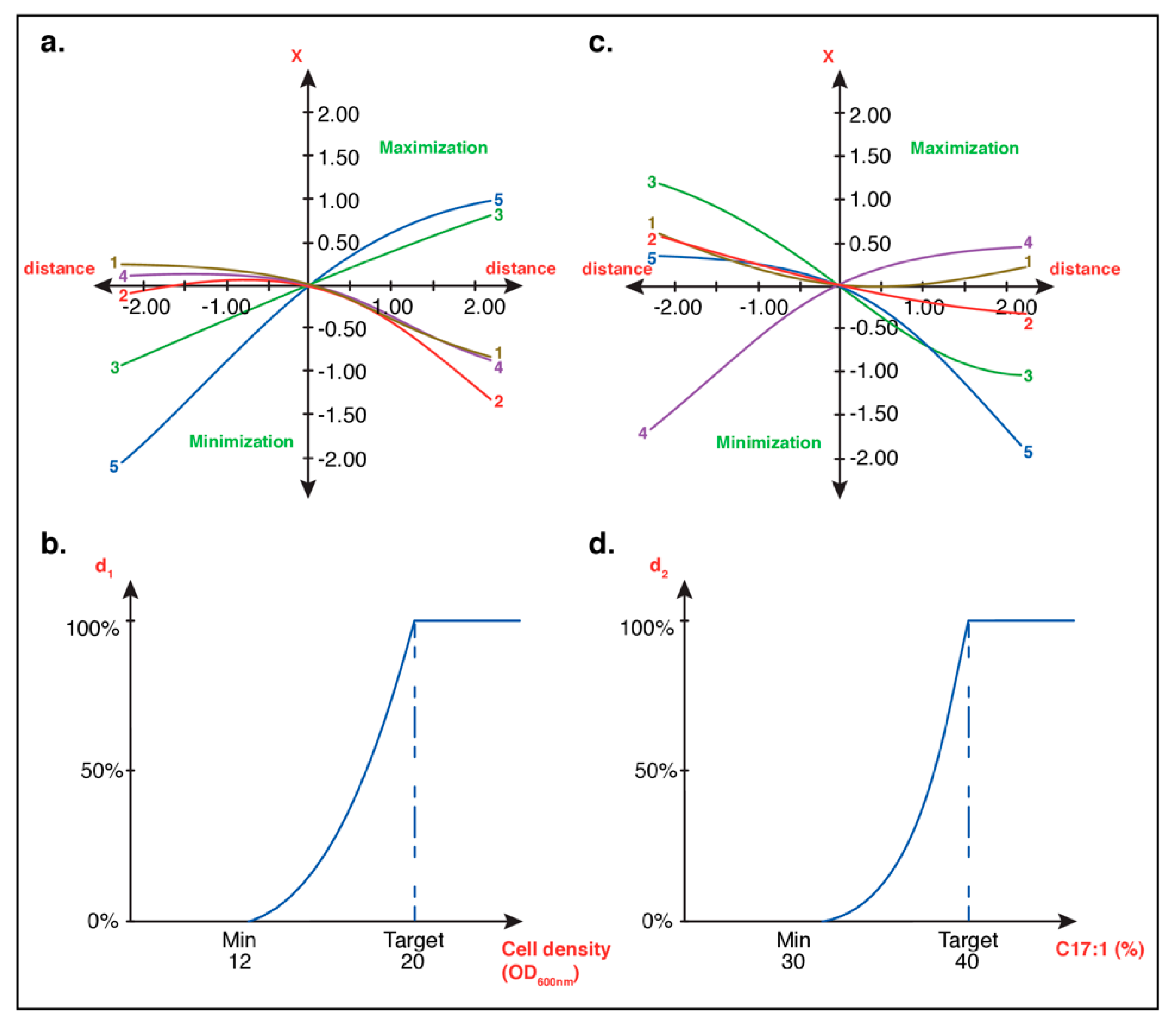
3.1.2. Optimal Culture Conditions Analyses
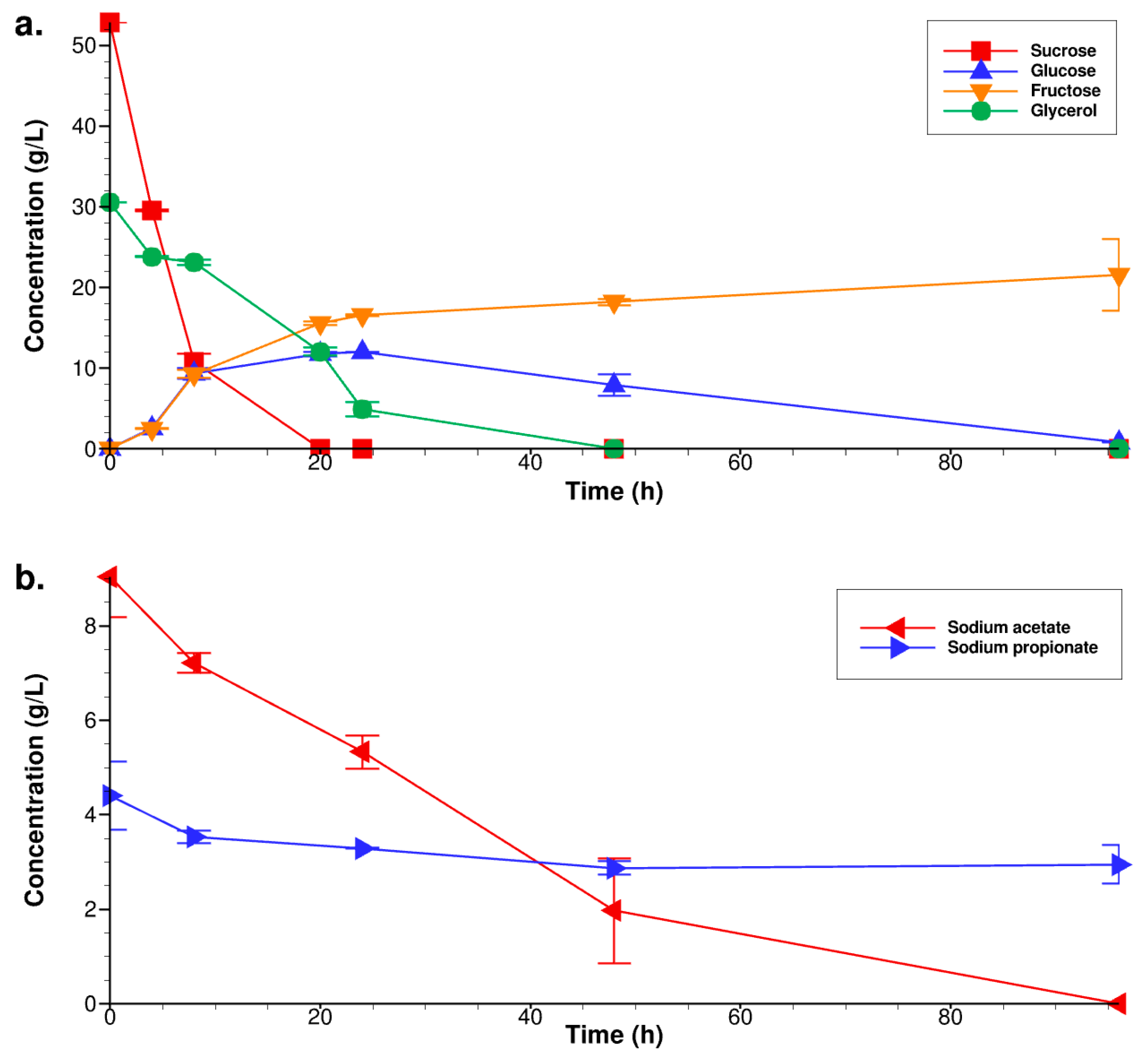
3.2. Fermentation under the Optimal Culture Conditions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Abghari, A.; Chen, S. Yarrowia lipolytica as an Oleaginous Cell Factory Platform for Production of Fatty Acid-Based Biofuel and Bioproducts. Front. Energy Res. 2014, 2, 21. [Google Scholar] [CrossRef]
- Koubaa, M.; Imatoukene, N.; Drévillon, L.; Vorobiev, E. Current Insights in Yeast Cell Disruption Technologies for Oil Recovery: A Review. Chem. Eng. Process.-Process Intensif. 2020, 150, 107868. [Google Scholar] [CrossRef]
- Li, Q.; Du, W.; Liu, D. Perspectives of Microbial Oils for Biodiesel Production. Appl. Microbiol. Biotechnol. 2008, 80, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Z.; Ratledge, C. Single Cell Oils: Microbial and Algal Oils; AOCS Press: Champaign, IL, USA, 2010; ISBN 978-1-893997-73-8. [Google Scholar]
- Thevenieau, F.; Nicaud, J.-M. Microorganisms as Sources of Oils. OCL 2013, 20, D603. [Google Scholar] [CrossRef]
- Li, Y.; Horsman, M.; Wu, N.; Lan, C.Q.; Dubois-Calero, N. Biofuels from Microalgae. Biotechnol. Prog. 2008, 24, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Balamurugan, S.; Li, T.; Cai, J.X.; Chen, T.T.; Wang, X.; Yang, W.D.; Li, H.Y. Biotechnological Approaches to Enhance Biofuel Producing Potential of Microalgae. Fuel 2021, 302, 121169. [Google Scholar] [CrossRef]
- Meng, X.; Yang, J.; Xu, X.; Zhang, L.; Nie, Q.; Xian, M. Biodiesel Production from Oleaginous Microorganisms. Renew. Energy 2009, 34, 1–5. [Google Scholar] [CrossRef]
- Amalia, L.; Zhang, Y.-H.; Ju, Y.-H.; Tsai, S.-L. Enhanced Lipid Production in Yarrowia lipolytica Po1g by over-Expressing Lro1 Gene under Two Different Promoters. Appl. Biochem. Biotechnol. 2020, 191, 104–111. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Muniglia, L.; Chevalot, I.; Aggelis, G.; Marc, I. Yarrowia lipolytica as a Potential Producer of Citric Acid from Raw Glycerol. J. Appl. Microbiol. 2002, 92, 737–744. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Knight, R.; Alper, H.S. Heterologous Production of Pentane in the Oleaginous Yeast Yarrowia lipolytica. J. Biotechnol. 2013, 165, 184–194. [Google Scholar] [CrossRef]
- Rakicka, M.; Rywińska, A.; Cybulski, K.; Rymowicz, W. Enhanced Production of Erythritol and Mannitol by Yarrowia lipolytica in Media Containing Surfactants. Braz. J. Microbiol. 2016, 47, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledesma-Amaro, R.; Dulermo, T.; Nicaud, J.M. Engineering Yarrowia lipolytica to Produce Biodiesel from Raw Starch. Biotechnol. Biofuels 2015, 8, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicaud, J.-M. Yarrowia lipolytica. Yeast 2012, 29, 409–418. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Nicaud, J.-M. Yarrowia lipolytica as a Biotechnological Chassis to Produce Usual and Unusual Fatty Acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef]
- Park, Y.-K.; Dulermo, T.; Ledesma-Amaro, R.; Nicaud, J.-M. Optimization of Odd Chain Fatty Acid Production by Yarrowia lipolytica. Biotechnol. Biofuels 2018, 11, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degwert, J.; Jacob, J.; Steckel, F. Use of Cis-9-Heptadecenoic Acid for Treating Psoriasis and Allergies. International Patent Application No. WO1994021247A1, 13 January 1998. [Google Scholar]
- Clausen, C.A.; Coleman, R.D.; Yang, V.W. Fatty Acid-Based Formulations for Wood Protection against Mold and Sapstain. For. Prod. J. 2010, 60, 301–304. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Liang, S.; Zong, M.-H.; Yang, J.-G.; Lou, W.-Y. Microbial Synthesis of Functional Odd-Chain Fatty Acids: A Review. World J. Microbiol. Biotechnol. 2020, 36, 35. [Google Scholar] [CrossRef]
- Jenkins, B.; West, J.A.; Koulman, A. A Review of Odd-Chain Fatty Acid Metabolism and the Role of Pentadecanoic Acid (C15:0) and Heptadecanoic Acid (C17:0) in Health and Disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.-K.; Bordes, F.; Letisse, F.; Nicaud, J.-M. Engineering Precursor Pools for Increasing Production of Odd-Chain Fatty Acids in Yarrowia lipolytica. Metab. Eng. Commun. 2021, 12, e00158. [Google Scholar] [CrossRef]
- Fickers, P.; Le Dall, M.T.; Gaillardin, C.; Thonart, P.; Nicaud, J.M. New Disruption Cassettes for Rapid Gene Disruption and Marker Rescue in the Yeast Yarrowia lipolytica. J. Microbiol. Methods 2003, 55, 727–737. [Google Scholar] [CrossRef]
- Lazar, Z.; Rossignol, T.; Verbeke, J.; Crutz-Le Coq, A.-M.; Nicaud, J.-M.; Robak, M. Optimized Invertase Expression and Secretion Cassette for Improving Yarrowia lipolytica Growth on Sucrose for Industrial Applications. J. Ind. Microbiol. Biotechnol. 2013, 40, 1273–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.-M.; Nicaud, J.-M. Hexokinase—A Limiting Factor in Lipid Production from Fructose in Yarrowia lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef] [PubMed]
- AFNOR. Céréales et Produits Céréaliers: Détermination de la Teneur en Matières Grasses Totales; Paris La Défense: Pairs, France, 1986. [Google Scholar]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.-M. Yarrowia lipolytica as a Model for Bio-Oil Production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef]
- Rios, L.F.; Klein, B.C.; Luz, L.F.; Maciel Filho, R.; Wolf Maciel, M.R. Nitrogen Starvation for Lipid Accumulation in the Microalga Species Desmodesmus sp. Appl. Biochem. Biotechnol. 2015, 175, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Calvey, C.H.; Su, Y.-K.; Willis, L.B.; McGee, M.; Jeffries, T.W. Nitrogen Limitation, Oxygen Limitation, and Lipid Accumulation in Lipomyces starkeyi. Bioresour. Technol. 2016, 200, 780–788. [Google Scholar] [CrossRef] [Green Version]
- González-García, Y.; Rábago-Panduro, L.M.; French, T.; Camacho-Córdova, D.I.; Gutiérrez-González, P.; Córdova, J. High Lipids Accumulation in Rhodosporidium toruloides by Applying Single and Multiple Nutrients Limitation in a Simple Chemically Defined Medium. Ann. Microbiol. 2017, 67, 519–527. [Google Scholar] [CrossRef]
- Fakankun, I.; Mirzaei, M.; Levin, D.B. Impact of Culture Conditions on Neutral Lipid Production by Oleaginous Yeast. Methods Mol. Biol. 2019, 1995, 311–325. [Google Scholar]
- Wang, X.; Fosse, H.K.; Li, K.; Chauton, M.S.; Vadstein, O.; Reitan, K.I. Influence of Nitrogen Limitation on Lipid Accumulation and EPA and DHA Content in Four Marine Microalgae for Possible Use in Aqauafeed. Front. Mar. Sci. 2019, 6, 95. [Google Scholar] [CrossRef]
- Boulton, C.A.; Ratledge, C. 1981 Correlation of Lipid Accumulation in Yeasts with Possession of ATP: Citrate Lyase. Microbiology 1981, 127, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Dulermo, T.; Lazar, Z.; Dulermo, R.; Rakicka, M.; Haddouche, R.; Nicaud, J.-M. Analysis of ATP-Citrate Lyase and Malic Enzyme Mutants of Yarrowia lipolytica Points out the Importance of Mannitol Metabolism in Fatty Acid Synthesis. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 1107–1117. [Google Scholar] [CrossRef] [Green Version]
- Meesters, P.A.E.P.; Huijberts, G.N.M.; Eggink, G. High-Cell-Density Cultivation of the Lipid Accumulating Yeast Cryptococcus curvatus Using Glycerol as a Carbon Source. Appl. Microbiol. Biotechnol. 1996, 45, 575–579. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Bai, F. High-Density Cultivation of Oleaginous Yeast Rhodosporidium toruloides Y4 in Fed-Batch Culture. Enzym. Microb. Technol. 2007, 41, 312–317. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Fakas, S.; Fick, M.; Chevalot, I.; Galiotou-Panayotou, M.; Komaitis, M.; Marc, I.; Aggelis, G. Biotechnological Valorisation of Raw Glycerol Discharged after Bio-Diesel (Fatty Acid Methyl Esters) Manufacturing Process: Production of 1,3-Propanediol, Citric Acid and Single Cell Oil. Biomass Bioenergy 2008, 32, 60–71. [Google Scholar] [CrossRef]
- Liang, Y.; Cui, Y.; Trushenski, J.; Blackburn, J.W. Converting Crude Glycerol Derived from Yellow Grease to Lipids through Yeast Fermentation. Bioresour. Technol. 2010, 101, 7581–7586. [Google Scholar] [CrossRef]
- Back, A.; Rossignol, T.; Krier, F.; Nicaud, J.-M.; Dhulster, P. High-Throughput Fermentation Screening for the Yeast Yarrowia lipolytica with Real-Time Monitoring of Biomass and Lipid Production. Microb. Cell Factories 2016, 15, 147. [Google Scholar] [CrossRef]
- D’Amato, D.; Corbo, M.R.; Nobile, M.A.D.; Sinigaglia, M. Effects of Temperature, Ammonium and Glucose Concentrations on Yeast Growth in a Model Wine System. Int. J. Food Sci. Technol. 2006, 41, 1152–1157. [Google Scholar] [CrossRef]
- Zhu, L.Y.; Zong, M.H.; Wu, H. Efficient Lipid Production with Trichosporon fermentans and Its Use for Biodiesel Preparation. Bioresour. Technol. 2008, 99, 7881–7885. [Google Scholar] [CrossRef]
- Tsigie, Y.A.; Wang, C.-Y.; Kasim, N.S.; Diem, Q.-D.; Huynh, L.-H.; Ho, Q.-P.; Truong, C.-T.; Ju, Y.-H. Oil Production from Yarrowia lipolytica Po1g Using Rice Bran Hydrolysate. J. Biomed. Biotechnol. 2012, 2012, e378384. [Google Scholar] [CrossRef]
- Guerfali, M.; Ayadi, I.; Sassi, H.-E.; Belhassen, A.; Gargouri, A.; Belghith, H. Biodiesel-Derived Crude Glycerol as Alternative Feedstock for Single Cell Oil Production by the Oleaginous Yeast Candida viswanathii Y-E4. Ind. Crops Prod. 2020, 145, 112103. [Google Scholar] [CrossRef]
- Gao, R.; Li, Z.; Zhou, X.; Cheng, S.; Zheng, L. Oleaginous Yeast Yarrowia lipolytica Culture with Synthetic and Food Waste-Derived Volatile Fatty Acids for Lipid Production. Biotechnol. Biofuels 2017, 10, 247. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Xu, P.; Chu, M.-Y.; Zong, M.-H.; Yang, J.-G.; Lou, W.-Y. Using 1-Propanol to Significantly Enhance the Production of Valuable Odd-Chain Fatty Acids by Rhodococcus opacus PD630. World J. Microbiol. Biotechnol. 2019, 35, 164. [Google Scholar] [CrossRef] [PubMed]
- Kolouchová, I.; Schreiberová, O.; Sigler, K.; Masák, J.; Řezanka, T. Biotransformation of Volatile Fatty Acids by Oleaginous and Non-Oleaginous Yeast Species. FEMS Yeast Res. 2015, 15, fov076. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain | Description | Reference |
|---|---|---|
| E. coli | ||
| JME547 | pUB-Cre-Hygro | [22] |
| JME2103 | JMP62-URA3 ex-pTEF-YlHXK1-Tlip2 | [24] |
| JME2347 | JMP62-LEU2 ex-pTEF-ScSUC2-Tlip2 | [23] |
| Y. lipolytica | ||
| JMY7780 | Δphd1 Δmfe1 Δtgl4 pTEF-DGA2 pTEF-GPD1 hp4d-LDP1-URA3 ex pTEF-Repct-LEU2 ex | [21] |
| JMY7829 | Δphd1 Δmfe1 Δtgl4 pTEF-DGA2 pTEF-GPD1 hp4d-LDP1 pTEF-Repct | This study |
| JMY7877 | JMY7829 pTEF-ScSUC2-LEU2 ex pTEF-YlHXK1-URA3 ex | This study |
| Name | Sequence |
|---|---|
| pTEF-internal-Fw | TCTGGAATCTACGCTTGTTCA |
| SUC2-internal-Rev | GCAGATTCTAGCTTCCAGGAC |
| HXK1-internal-Rev | CTCATCTTCTCGAAGGTCTGCTG |
| Experiment | Sucrose (g/L) | Glycerol (g/L) | Sodium Acetate (g/L) | Sodium Propionate (g/L) | Yeast Extract (g/L) | Cell Density (OD600nm) | C17:1 Content (%) |
|---|---|---|---|---|---|---|---|
| 1 | 40 (−1) | 20 (−1) | 15 (−1) | 4 (−1) | 4 (+ 1) | 19.2 | 31.7 |
| 2 | 80 (+1) | 20 (−1) | 15 (−1) | 4 (−1) | 2 (−1) | 14.0 | 31.2 |
| 3 | 40 (−1) | 40 (+1) | 15 (−1) | 4 (−1) | 2 (−1) | 14.4 | 29.3 |
| 4 | 80 (+1) | 40 (+1) | 15 (−1) | 4 (−1) | 4 (+1) | 17.8 | 26.4 |
| 5 | 40 (−1) | 20 (−1) | 25 (+1) | 4 (−1) | 2 (−1) | 16.8 | 27.4 |
| 6 | 80 (+1) | 20 (−1) | 25 (+1) | 4 (−1) | 4 (+1) | 19.4 | 19.7 |
| 7 | 40 (−1) | 40 (+1) | 25 (+1) | 4 (−1) | 4 (+1) | 19.2 | 21.3 |
| 8 | 80 (+1) | 40 (+1) | 25 (+1) | 4 (−1) | 2 (−1) | 14.8 | 21.1 |
| 9 | 40 (−1) | 20 (−1) | 15 (−1) | 6 (+1) | 2 (−1) | 13.0 | 34.7 |
| 10 | 80 (+1) | 20 (−1) | 15 (−1) | 6 (+1) | 4 (+1) | 17.0 | 30.7 |
| 11 | 40 (−1) | 40 (+1) | 15 (−1) | 6 (+1) | 4 (+1) | 17.0 | 30.4 |
| 12 | 80 (+1) | 40 (+1) | 15 (−1) | 6 (+1) | 2 (−1) | 12.2 | 34.2 |
| 13 | 40 (−1) | 20 (−1) | 25 (+1) | 6 (+1) | 4 (+1) | 20.0 | 26.5 |
| 14 | 80 (+1) | 20 (−1) | 25 (+1) | 6 (+1) | 2 (−1) | 15.0 | 29.3 |
| 15 | 40 (−1) | 40 (+1) | 25 (+1) | 6 (+1) | 2 (−1) | 15.6 | 26.4 |
| 16 | 80 (+1) | 40 (+1) | 25 (+1) | 6 (+1) | 4 (+1) | 16.0 | 25.1 |
| 17 | 20 (−2) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 17.6 | 29.5 |
| 18 | 100 (+2) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 15.8 | 28.3 |
| 19 | 60 (0) | 10 (−2) | 20 (0) | 5 (0) | 3 (0) | 17.8 | 29.4 |
| 20 | 60 (0) | 50 (+2) | 20 (0) | 5 (0) | 3 (0) | 16.0 | 25.8 |
| 21 | 60 (0) | 30 (0) | 10 (−2) | 5 (0) | 3 (0) | 13.8 | 36.6 |
| 22 | 60 (0) | 30 (0) | 30 (+2) | 5 (0) | 3 (0) | 17.6 | 23.1 |
| 23 | 60 (0) | 30 (0) | 20 (0) | 3 (−2) | 3 (0) | 18.2 | 21.5 |
| 24 | 60 (0) | 30 (0) | 20 (0) | 7 (+2) | 3 (0) | 16.2 | 32.9 |
| 25 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 1 (−2) | 12.0 | 38.2 |
| 26 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 5 (+2) | 16.4 | 28.7 |
| 27 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 15.0 | 32.1 |
| 28 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 14.6 | 32.2 |
| 29 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 14.8 | 31.2 |
| 30 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 3 (0) | 13.2 | 32.0 |
| 31 | 42.7 (−0.866) | 25 (−0.5) | 18.2 (−0.3535) | 4.7 (−0.2739) | 2.8 (−0.2236) | 14.0 | 32.3 |
| 32 | 77.3 (+0.866) | 25 (−0.5) | 18.2 (−0.3535) | 4.7 (−0.2739) | 2.8 (−0.2236) | 13.2 | 32.4 |
| 33 | 60 (0) | 40 (+1) | 18.2 (−0.3535) | 4.7 (−0.2739) | 2.8 (−0.2236) | 13.6 | 31.6 |
| 34 | 60 (0) | 30 (0) | 25.3 (+1.0607) | 4.7 (−0.2739) | 2.8 (−0.2236) | 15.0 | 28.4 |
| 35 | 60 (0) | 30 (0) | 20 (0) | 6.1 (+1.0955) | 2.8 (−0.2236) | 12.8 | 32.4 |
| 36 | 60 (0) | 30 (0) | 20 (0) | 5 (0) | 4.1 (+1.118) | 15.8 | 30.1 |
| Cell Density (OD600nm) | % C17:1 | |||
|---|---|---|---|---|
| Experiment | Ymeasured | Ypredicted | Ymeasured | Ypredicted |
| 31 | 14 | 15.02 | 32.29 | 33.27 |
| 32 | 13.2 | 14.44 | 32.41 | 31.76 |
| 33 | 13.6 | 14.31 | 31.63 | 30.49 |
| 34 | 15 | 15.51 | 28.39 | 27.45 |
| 35 | 12.8 | 14.27 | 32.43 | 33.30 |
| 36 | 15.8 | 16.14 | 30.07 | 30.49 |
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F-Ratio | p-Value |
|---|---|---|---|---|---|
| Regression | 149.27 | 20 | 7.46 | 12.22 | <0.01 * |
| Residue | 9.16 | 15 | 0.61 | ||
| Lack of fit | 7.16 | 12 | 0.59 | 0.89 | 61.9 |
| Pure error | 2.0 | 3 | 0.66 | ||
| Total | 158.43 | 35 |
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F-Ratio | p-Value |
|---|---|---|---|---|---|
| Regression | 637.21 | 20 | 31.86 | 23.34 | <0.01 * |
| Residue | 20.47 | 15 | 1.36 | ||
| Lack of fit | 19.87 | 12 | 1.65 | 8.37 | 5.3 |
| Pure error | 0.59 | 3 | 0.19 | ||
| Total | 657.68 | 35 |
| FAME | % in Erlenmeyer | % in 5 L Bioreactors |
|---|---|---|
| C15:0 | 5.03 ± 0.19 | 5.37 ± 0.34 |
| C16:0 | 7.78 ± 0.15 | 9.37 ± 1.59 |
| C16:1 | 1.00 ± 0.06 | 1.29 ± 0.30 |
| C17:0 | 19.71 ± 0.42 | 18.00 ± 1.71 |
| C17:1 | 31.62 ± 1.15 | 31.62 ± 0.94 |
| C18:0 | 10.02 ± 0.24 | 9.85 ± 0.27 |
| C18:1 | 17.30 ± 0.27 | 17.87 ± 0.56 |
| C18:2 | 2.59 ± 0.18 | 2.61 ± 0.17 |
| C19:0 | 2.72 ± 0.06 | 2.64 ± 0.10 |
| C19:1 | 1.41 ± 0.07 | 1.39 ± 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Sahyouni, W.; El Kantar, S.; Khelfa, A.; Park, Y.-K.; Nicaud, J.-M.; Louka, N.; Koubaa, M. Optimization of cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica. Fermentation 2022, 8, 245. https://doi.org/10.3390/fermentation8060245
Al Sahyouni W, El Kantar S, Khelfa A, Park Y-K, Nicaud J-M, Louka N, Koubaa M. Optimization of cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica. Fermentation. 2022; 8(6):245. https://doi.org/10.3390/fermentation8060245
Chicago/Turabian StyleAl Sahyouni, Wendy, Sally El Kantar, Anissa Khelfa, Young-Kyoung Park, Jean-Marc Nicaud, Nicolas Louka, and Mohamed Koubaa. 2022. "Optimization of cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica" Fermentation 8, no. 6: 245. https://doi.org/10.3390/fermentation8060245
APA StyleAl Sahyouni, W., El Kantar, S., Khelfa, A., Park, Y. -K., Nicaud, J. -M., Louka, N., & Koubaa, M. (2022). Optimization of cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica. Fermentation, 8(6), 245. https://doi.org/10.3390/fermentation8060245








