Glycerol Utilization as a Sole Carbon Source Disrupts the Membrane Architecture and Solventogenesis in Clostridium beijerinckii NCIMB 8052
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Quantitative Gene Expression Profiling
2.2.1. Total RNA Isolation
2.2.2. Real-Time Quantitative PCR (RT-qPCR)
2.3. Enzyme Activity Assays
2.4. Analytical Methods
2.5. Statistical Analysis
3. Results
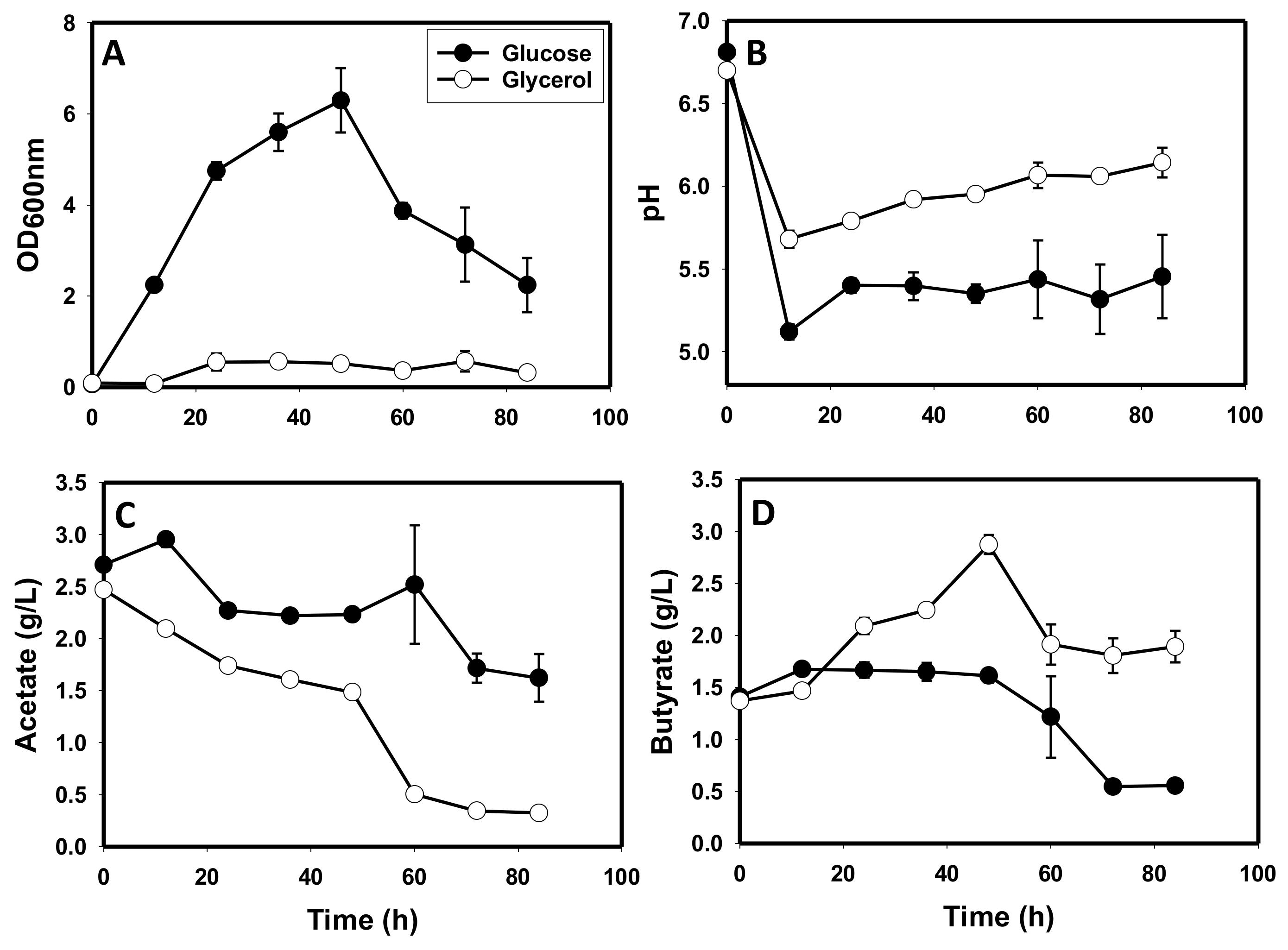
3.1. Comparative Fermentation Profiles of C. beijerinckii Grown on Glycerol versus Glucose
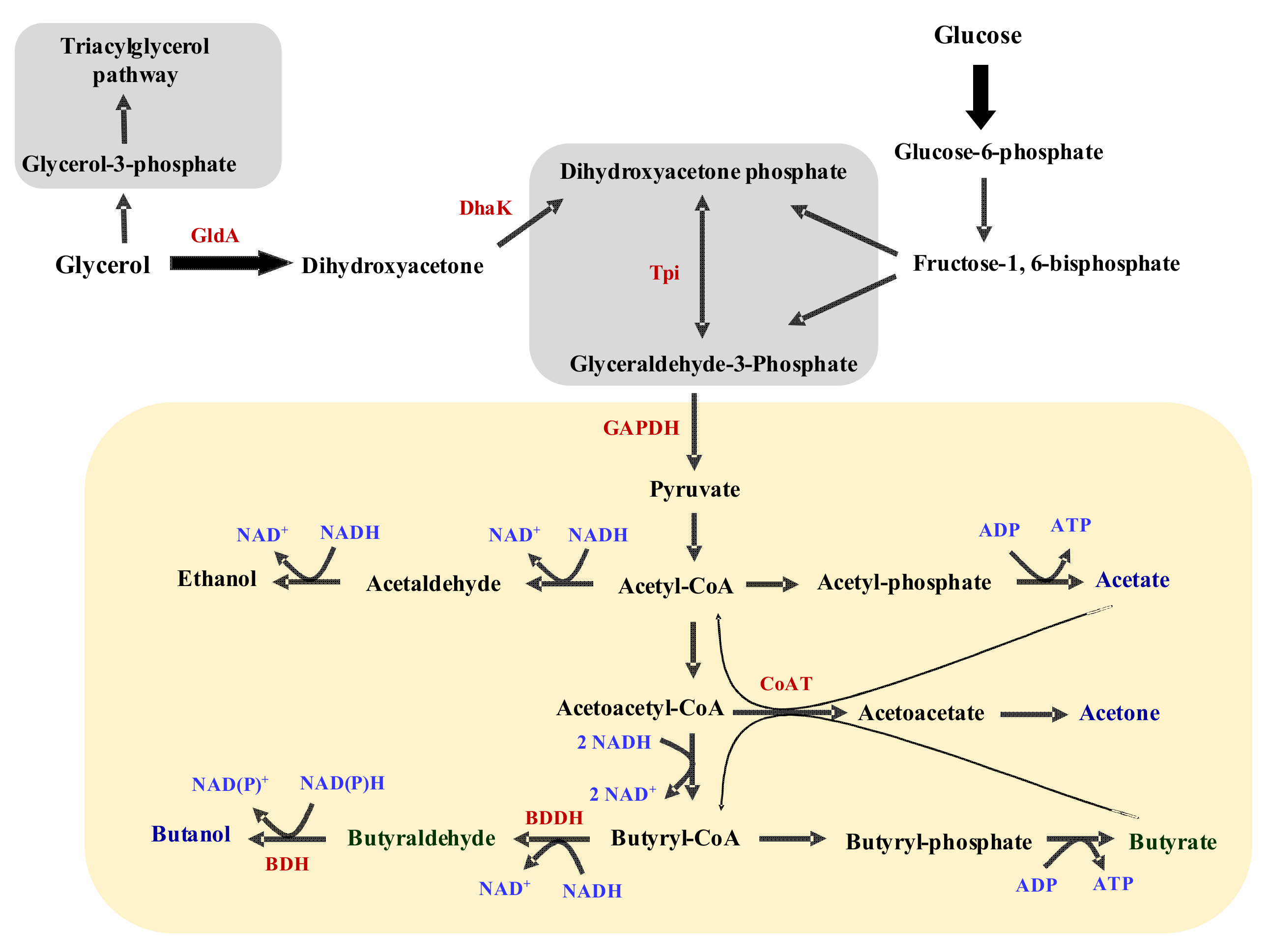
3.2. The Activities of Key Solventogenic Enzymes during Growth on Glycerol
3.3. Relative Expression of Important Metabolic Genes of C. beijerinckii Grown on Glycerol
3.4. Glycerol and Glucose Utilization Profiles of C. beijerinckii
3.5. The Metabolite Profiles of Glycerol-versus Glucose-Grown Cells of C. beijerinckii
4. Discussion
4.1. Production of Toxic Metabolites and Likely Impaired Construction of Membrane Architecture during Growth on Glycerol
4.2. Inefficient NADH Re-Oxidation
4.3. Glycerol-Mediated Gene and Enzyme Inhibitions and the Attendant Solvent Profiles
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Clomburg, J.M.; Ramon, G. Anaerobic fermentation of glycerol: A platform for renewable fuels and chemicals. Trends Biotechnol. 2013, 31, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, F.; Chen, T.; Jiang, Y.; Lu, J.; Dong, W.; Zhang, W.; Ma, J.; Zhang, M.; Jiang, M. Enhanced biobutanol production with high yield from crude glycerol by acetone uncoupled Clostridium sp. strain CT7. Bioresour. Technol. 2017, 244, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Murarka, A.; Dharmadi, Y.; Yazdani, S.S.; Gonzalez, R. Fermentative Utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 2008, 74, 1124–1135. [Google Scholar] [PubMed] [Green Version]
- Ujor, V.; Agu, C.V.; Gopalan, V.; Ezeji, T.C. Glycerol supplementation of the growth medium enhances in situ detoxification of furfural by Clostridium beijerinckii during butanol fermentation. Appl. Microbiol. Biotechnol. 2014, 98, 6511–6521. [Google Scholar] [CrossRef]
- Hirsch, R.L.; Bezdek, R.; Wendling, R. Peaking of World Oil Production and Its Mitigation. In Driving Climate Change; Sperling, D., Cannon, J.S., Eds.; Academic Press: Cambridge, MA, USA, 2007; pp. 9–27. [Google Scholar]
- Cheng, M.-H.; Huang, H.; Dien, B.S.; Singh, V. The costs of sugar production from different feedstocks and processing technologies. Biofuels Bioprod. Biorefining 2019, 13, 723–739. [Google Scholar] [CrossRef]
- Anitha, M.; Kamarudin, S.K.; Kofli, N.T. The potential of glycerol as a value-added commodity. Chem. Eng. J. 2016, 295, 119–130. [Google Scholar] [CrossRef]
- Mota, C.J.; Pinto, B.P.; de Lima, A.L. Glycerol Utilization. In Glycerol; Springer: New York, NY, USA, 2017; pp. 11–19. [Google Scholar]
- Vivek, N.; Sindhu, R.; Madhavan, A.; Anju, A.J.; Castro, E.; Faraco, F.; Pandey, A.; Binod, P. Recent advances in the production of value added chemicals and lipids utilizing biodiesel industry generated crude glycerol as a substrate–metabolic aspects, challenges and possibilities: An overview. Bioresour. Technol. 2017, 239, 507–517. [Google Scholar] [CrossRef]
- Yazdani, S.S.; Gonzalez, R. Anaerobic fermentation of glycerol: A path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 2007, 18, 213–219. [Google Scholar] [CrossRef]
- Neijssel, O.M.; Hueting, S.; Crabbendam, K.J.; Tempest, D.W. Dual pathways of glycerol assimilation in Klebsiella aerogenes NCIB 418. Arch. Microbiol. 1975, 104, 83–87. [Google Scholar] [CrossRef]
- Lin, E.C.C. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 1976, 30, 535–578. [Google Scholar] [CrossRef]
- Sarma, S.; Anand, A.; Dubey, V.K.; Moholka, V.S. Metabolic flux network analysis of hydrogen production from crude glycerol by Clostridium pasteurianum. Bioresour. Technol. 2017, 242, 169–177. [Google Scholar] [CrossRef]
- Palmqvist, E.; Almeida, J.S.; Hähn-Hägerdal, B. Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol. Bioeng. 1999, 62, 447–454. [Google Scholar] [CrossRef]
- Palmqvist, E.; Hähn-Hägerdal, B. Fermentation of lignocellulosic hydrolysate. II: Inhibitors and mechanisms of inhibition. Bioresour. Technol. 2000, 74, 25–33. [Google Scholar] [CrossRef]
- Sávári, H.I.; Franzén, C.J.; Taherzadeh, M.J.; Niklasson, C.; Lidén, G. Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-mediated chemostats. Appl. Environ. Microbiol. 2003, 69, 4076–4086. [Google Scholar]
- Bouvet, O.M.M.; Lenormand, P.; Ageron, E.; Grimont, P.A.D. Taxonomic diversity of anaerobic glycerol dissimilation in the Enterobacteriaceae. Res. Microbiol. 1995, 146, 279–290. [Google Scholar] [CrossRef]
- Booth, I.R. Glycerol and Methylglyoxal Metabolism. EcoSal Plus 2005, 1. [Google Scholar] [CrossRef] [PubMed]
- Agu, C.V.; Ujor, V.; Ezeji, T.C. Metabolic engineering of Clostridium beijerinckii to improve glycerol metabolism and furfural tolerance. Biotechnol. Biofuels 2019, 12, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujor, V.C.; Lai, L.B.; Okonkwo, C.C.; Gopalan, V.; Ezeji, T.C. Ribozyme-Mediated downregulation uncovers DNA integrity scanning protein A (DisA) as a solventogenesis determinant in Clostridium beijerinckii. Front. Bioeng. Biotechnol. 2021, 9, 669462. [Google Scholar] [CrossRef]
- Sauvageot, N.; Gouffi, K.; Laplace, J.-M.; Auffray, Y. Glycerol metabolism in Lactobacillus collinoides: Production of 3-hydroxypropionaldehyde, a precursor of acrolein. Int. J. Food Microbiol. 2000, 55, 167–170. [Google Scholar] [CrossRef]
- Claisse, O.; Lonvaud-Funel, A. Detection of lactic acid bacteria producing 3-hydroxypropionaldehyde (acrolein precursor) from glycerol by molecular tests. Int. J. Dairy Sci. 2001, 81, 173–181. [Google Scholar]
- Vivek, N.; Hazeena, S.H.; Rajesh ROGodan, T.K.; Anjali, K.B.; Nair, L.M.; Mohan, B.; Nair, S.C.; Sindhu, R.; Pandey, A.; Binod, P. Genomics of lactic acid bacteria for glycerol dissimilation. Mol. Biotechnol. 2019, 61, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, F.B. Formation of acrolein from glycerol by B. welchii. J. Infect. Dis. 1924, 35, 282–290. [Google Scholar] [CrossRef]
- Voisenet, M.E. Sur un ferment, contenu dans les eaux, agent de déshydratation de la glycérine. Ann. Inst. Pasteur 1914, 28, 807–818. [Google Scholar]
- Hao, J.; Lin, R.; Zheng, Z.; Sun, Y.; Liu, D. 3-Hydroxypropionaldehyde guided glycerol feeding strategy in aerobic 1,3-propanediol production by Klebsiella pneumoniae. J. Ind. Microbiol. Biotechnol. 2008, 35, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Liyanage, H.; Kashket, S.; Young, M.; Kashket, E.R. Clostridium beijerinckii and Clostridium difficile detoxify methylglyoxal by a novel mechanism involving glycerol dehydrogenase. Appl. Environ. Microbiol. 2001, 67, 2004–2010. [Google Scholar] [CrossRef] [Green Version]
- Inoue, Y.; Kimura, A. Methylglyoxal and regulation of its metabolism in microorganisms. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 1995; Volume 37, pp. 177–227. [Google Scholar]
- Kalapos, M.P. Methylglyoxal in living organisms: Chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 1999, 110, 145–175. [Google Scholar] [CrossRef]
- Gangwar, B.; Kumar, S.; Darokar, M.P. Glabridin averts biofilms formation in methicillin-resistant Staphylococcus aureus by modulation of the surfaceome. Front. Microbiol. 2020, 11, 1779. [Google Scholar] [CrossRef]
- Inui, M.; Suda, M.; Kimura, S.; Yasuda, K.; Suzuki, H.; Toda, H.; Yamamoto, S.; Okino, S.; Suzuki, N.; Yukawa, H. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl. Microbiol. Biotechnol. 2008, 77, 1305–1316. [Google Scholar] [CrossRef]
- Clark, S.W.; Bennett, G.N.; Rudolph, F.B. Isolation and characterization of mutants of Clostridium acetobutylicum ATCC 824 deficient in acetoacetyl-coenzyme A:acetate/butyrate:Coenzyme A-transferase (EC 2.8.3.9) and in other solvent pathway enzymes. Appl. Environ. Microbiol. 1989, 55, 970–976. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Ujor, V.; Lai, L.B.; Gopalan, V.; Ezeji, T.C. Use of proteomic analysis to elucidate the role of calcium in acetone-butanol-ethanol fermentation by Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 2012, 79, 282–293. [Google Scholar] [CrossRef] [Green Version]
- Galindo-Cuspinera, V.; Lubran, M.B.; Rankin, S.A. Comparison of volatile compounds in water- and oil-soluble annatto (Bixa orellana L.) Extracts. J. Agric. Food Chem. 2002, 50, 2010–2015. [Google Scholar] [CrossRef] [PubMed]
- Vollenweider, S.; Evers, S.; Zurbriggen, K.; Lacroix, C. Unraveling the hydroxypropionaldehyde (HPA) system: An active antimicrobial agent against human pathogens. J. Agric. Food Chem. 2010, 58, 10315–10322. [Google Scholar] [CrossRef] [PubMed]
- Schuler, D.; Eder, E. Detection of 1,N2-propanodeoxyguanosine adducts of 2-hexenal in organs of Fischer 344 rats by a 32P-post-labeling technique. Carcinogenesis 1999, 20, 1345–1350. [Google Scholar] [CrossRef] [Green Version]
- Auerbach, C.; Moutschen-Dahmen, M.; Moutschen, J. Genetic and cytogenetical effects of formaldehyde and related compounds. Mutat. Res. Genet. Toxicol. 1977, 39, 317–361. [Google Scholar] [CrossRef]
- Ikeda, M. H. Greim (ed): DFG Essential MAK value documentations. Int. Arch. Occup. Environ. Health 2008, 81, 933–934. [Google Scholar] [CrossRef]
- Schaefer, L.; Auchtung, T.; Hermans, K.E.; Whitehead, D.; Borhan, B.; Britton, R.A. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 2010, 156, 1589–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, P.H.; Kanuri, M.; Nechev, L.V.; Harris, T.M.; Lloyd, R.S. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ. Mol. Mutagen. 2005, 45, 455–459. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Z.; Pan, X.; Zhu, M.; Xie, J. Gene expression profile and cytotoxicity of human bronchial epithelial cells exposed to crotonaldehyde. Toxicol. Lett. 2010, 197, 113–122. [Google Scholar] [CrossRef]
- Kaneda, T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef]
- Carey, A.B.; Ashenden, A.; Köper, I. Model architectures for bacterial membranes. Biophys. Rev. 2022, 14, 111–143. [Google Scholar] [CrossRef]
- Lepage, C.; Fayolle, F.; Hermann, M.; Vandecasteele, J. Changes in membrane lipid composition of Clostridium acetobutylicum during acetone-butanol fermentation: Effects of solvents, growth temperature and pH. J. Gen. Microbiol. 1987, 133, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Isar, J.; Rangaswamy, V. Improved n-butanol production by solvent tolerant Clostridium beijerinckii. Biomass-Bioenergy 2012, 37, 9–15. [Google Scholar] [CrossRef]
- Ingram, L.O. Adaptation of membrane lipids to alcohols. J. Bacteriol. 1976, 125, 670–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, F.J.; de Bont, J.A.M. Adaptation mechanisms of microorganisms to the toxic effects of organic solvent on membranes. Biochim. Biophys. Acta. 1996, 1286, 225–245. [Google Scholar] [PubMed]
- Torres, S.; Pandey, A.; Castro, G.R. Organic solvent adaptation of Gram positive bacteria: Applications and biotechnological potentials. Biotechnol. Adv. 2011, 29, 442e52. [Google Scholar] [CrossRef] [PubMed]
- Cray, J.A.; Stevenson, A.; Ball, P.; Bankar, S.B.; Eleutherio, E.C.; Ezeji, T.C.; Singhal, R.S.; Thevelein, J.M.; Timson, D.J.; Hallsworth, J.E. Chaotropicity: A key factor in product tolerance of biofuel-producing microorganisms. Curr. Opin. Biotechnol. 2015, 33, 228–259. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, H.; Yang, F.; Ma, Y. Current progress on butyric acid production by fermentation. Curr. Microbiol. 2009, 59, 656–663. [Google Scholar] [CrossRef]
- Gu, Y.; Hu, S.; Chen, J.; Shao, L.; He, H.; Yang, Y.; Yang, S.; Jiang, W. Ammonium acetate enhances solvent production by Clostridium acetobutylicum EA 2018 using cassava as a fermentation medium. J. Ind. Microbiol. Biotechnol. 2009, 36, 1225–1232. [Google Scholar] [CrossRef]
- Chen, C.-K.; Blaschek, H.P. Effect of acetate on molecular and physiological aspects of Clostridium beijerinckii NCIMB 8052 solvent production and strain degeneration. Appl. Environ. Microbiol. 1999, 65, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Wiesenborn, D.P.; Rudolph, F.B.; Papoutsakis, E.T. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl. Environ. Microbiol. 1989, 55, 323–329. [Google Scholar] [CrossRef] [Green Version]







| S.N. | Locus Tag | Gene | Primer Sequence 5′ to 3′ | |
|---|---|---|---|---|
| 1 | Cbei_0305 | adhE | Forward | GACCTGTGGAGTTATAGAGAGAGA |
| 2 | Cbei_0305 | adhE | Reverse | ATGGCTGTTGATGTTGGATTTG |
| 3 | Cbei_2421 | bdhA | Forward | CGAAGGAGATCCATGGGATTTAG |
| 4 | Cbei_2421 | bdhA | Reverse | CATGTGGATCCATCTCAGAACC |
| 5 | Cbei_2753 | gldA | Forward | ACACTGATGATGGCGTCTTT |
| 6 | Cbei_2753 | gldA | Reverse | GGCATCACCCATACCAGATAC |
| 7 | Cbei_2148 | dhaK | Forward | GAGCAGTGTTTACATCACCTACA |
| 8 | Cbei_2148 | dhaK | Reverse | TCCCTTCCATTTCAGCCATATC |
| 9 | Cbei_0599 | tpiA | Forward | AAGGCTGTAGCTGGTTCAAATA |
| 10 | Cbei_0599 | tpiA | Reverse | TCCCATAGCTTCAAGCATTCC |
| 11 | Cbei_0597 | gapdh-1 | Forward | ACTTTAGATGCTCCACACAGAA |
| 12 | Cbei_0597 | gapdh-1 | Reverse | CTCTTTGAGCACCACCATCT |
| 13 | Cbei_2282 | gapdh | Forward | GGGAACCATTAGGAGTTGTACTT |
| 14 | Cbei_2282 | gapdh | Reverse | AATCCACATAAGCTCCCTTGAG |
| 15 | Cbei_2572 | gapdh | Forward | TTGTAGCAGGTGGTTACTCATAC |
| 16 | Cbei_2572 | gapdh | Reverse | GGATTTCCGACCACAAGTTTATTC |
| 17 | Cbei_2654 | ctfA | Forward | ATCAGCTCAACCAGGAGAAAC |
| 18 | Cbei_2654 | ctfA | Reverse | GCTTCAGGGCAAGTACCAATA |
| 19 | Cbei_2653 | ctfB | Forward | AGGTGGATCTCCAATCACTTTAC |
| 20 | Cbei_2653 | ctfB | Reverse | ATCTACCTCTAAAGCGCCTAATG |
| 21 | Cbei_3833 | ctfA | Forward | ACCTTCTATAGGGCTGCTACT |
| 22 | Cbei_3833 | ctfA | Reverse | CCAGGAGTCATTATGGCATCTC |
| 23 | Cbei_3834 | ctfB | Forward | GGGTGGCGCTATGGATTTA |
| 24 | Cbei_3834 | cftB | Reverse | CCACTTGAGCCTTAGCAGTAA |
| 25 | Cbei_R0024 | 16SrRNA | Forward | CTCTGTCTTCAGGGACGATAATG |
| 26 | Cbei_R0024 | 16SrRNA | Reverse | TATCCACCTACGCTCCCTTTA |
| Gene ID | Function of Protein Product | Relative Expression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | ||||||||
| Glu + A | Gly + A | Glu − A | Gly − A | Glu + A | Gly + A | Glu − A | Gly − A | ||
| Cbei_0305 | Bifunctional acetaldehyde-CoA/alcohol dehydrogenase (adhE) | 84.44 ± 8.5 | 4.00 ± 0.42 | 1.02 ± 0.21a | 1.13 ± 0.06 a | 1.00 ± 0.14 | 1.44 ± 0.18 | 1.00 ± 0.18 | 0.30 ± 0.53 |
| Cbei_2421 | NADH-dependent butanol dehydrogenase A (bdhA) | 0.26 ± 0.04 | 0.51 ± 0.03 | 1.01 ± 0.12 | 0.33 ± 0.01 | 0.37 ± 0.02 | 1.00 ± 0.18 | 1.00 ± 0.07 | 0.13 ± 0.02 |
| Cbei_2753 | Glycerol dehydrogenase (gldA) | 0.27 ± 0.03 | 1.36 ± 0.12 | 1.00 ± 0.04 | 1.4 ± 0.15 | 0.48 ± 0.04 | 1.00 ± 0.17 | 1.00 ± 0.09 | 0.30 ± 0.03 |
| Cbei_2148 | Dihydroxyacetone kinase (subunit dhaK) | 13.83 ± 1.38 | 880 ± 71.36 | 1.02 ± 0.25 | 899 ± 71.36 | 0.01 ± 0.00 | 1.27 ± 0.07 | 1.04 ± 0.34 | 2.3 ± 0.44 |
| Cbei_0599 | Triosephosphate isomerase (A subunit; tpiA) | 0.23 ± 0.02 | 0.4 ± 0.05 | 1.01 ± 0.14 | 0.33 ± 0.03 | 1.70 ± 0.05 | 1.03 ± 0.12 | 1.02 ± 0.25 | 0.13 ± 0.02 |
| Cbei_0597 | Glyceraldehyde-3-phosphate dehydrogenase (type I) | 0.26 ± 0.03 a | 0.30 ± 0.04 a | 1.00 ± 0.11 | 0.30 ± 0.02 | 1.50 ± 0.07 | 1.00 ± 0.12 | 1.01 ± 0.14 | 0.10 ± 0.01 |
| Cbei_2282 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (gapdh-N1) | 26.3 ± 7.00 | 1.60 ± 0.31 | 1.04 ± 0.35 a | 0.70 ± 0.24 a | 1.00 ± 0.40 | 1.5 ± 0.07 | 1.00 ± 0.10 | 0.20 ± 0.07 |
| Cbei_2572 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (gapdh-N2) | 2.12 ± 0.02 | 0.50 ± 0.10 | 1.00 ± 0.03 | 0.40 ± 0.06 | 0.60 ± 0.11 | 1.13 ± 0.05 | 1.00 ± 0.09 a | 0.80 ± 0.04 a |
| Cbei_2654 | Acetate CoA-transferase, alpha subunit (ctfA) | 189 ± 15.00 | 1.40 ± 0.05 | 1.00 ± 0.04 | 0.30 ± 0.05 | 0.61 ± 0.05 | 1.70 ± 0.05 | 1.00 ± 0.10 | 0.01 ± 0.00 |
| Cbei_2653 | Butyrate-acetoacetate CoA-transferase, subunit B (ctfB) | 89 ± 1.30 | 0.90 ± 0.09 | 1.01 ± 0.14 | 0.17 ± 0.04 | 0.60 ± 0.11 | 1.65 ± 0.13 | 1.00 ± 0.12 | 0.01 ± 0.00 |
| Cbei_3833 | Butyrate-acetoacetate CoA-transferase, subunit A (ctfA) | 1.6 ± 0.03 | 0.04 ± 0.00 | 1.00 ± 0.11 | 0.02 ± 0.00 | 0.2 ± 0.01 | 0.8 ± 0.09 | 1.00 ± 0.12 | 0.00 ± 0.00 |
| Cbei_3834 | Acyl-CoA:acetate CoA transferase, beta subunit (ctfB) | 1.0 ± 0.08 | 0.02 ± 0.00 | 1.00 ± 0.07 | 0.00 ± 0.00 | 0.14 ± 0.01 | 0.72 ± 0.11 | 1.00 ± 0.08 | 0.00 ± 0.00 |
| Substrate | Treatment | Initial Concentration (g/L) | Final Concentration (g/L) | Substrate Consumed (g/L) |
|---|---|---|---|---|
| Glucose | 60 g/L in P2 medium * | 57.4 ± 0.00 | 17.10 ± 3.79 | 39.10 ± 2.23 |
| Glycerol | 60 g/L in P2 medium | 55.10 ± 0.00 | 49.80 ± 0.46 | 5.30 ± 0.46 |
| Glycerol | 60 g/L without acetate | 59.80 ± 3.32 | 55.10 ± 3.60 | 4.70 ± 0.40 |
| Glycerol | 12 g/L without acetate | 12.60 ± 0.00 | 9.32 ± 0.65 | 3.28 ± 0.65 |
| Glycerol | 20 g/L without acetate | 21.11 ± 0.00 | 17.20 ± 0.36 | 3.91 ± 0.36 |
| Glycerol | 36.1 g/L without acetate | 37.00 ± 0.00 | 32.90 ± 0.36 | 4.10 ± 0.36 |
| Glycerol | 60 g/L in P2 medium + butyraldehyde (3 g/L) | 61.00 ± 0.00 | 57.20 ± 1.13 | 3.80 ± 1.03 |
| Glycerol | 60 g/L + acetate (6 g/L) + butyrate (3.5 g/L) | 62.33 ± 0.00 | 57.59 ± 0.49 | 4.74 ± 0.49 |
| Glucose | 60 g/L + acetate (6 g/L) + butyrate (3.5 g/L) | 59.06 ± 0.00 | 13.33 ± 0.80 | 45.73 ± 0.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agyeman-Duah, E.; Kumar, S.; Gangwar, B.; Ujor, V.C. Glycerol Utilization as a Sole Carbon Source Disrupts the Membrane Architecture and Solventogenesis in Clostridium beijerinckii NCIMB 8052. Fermentation 2022, 8, 339. https://doi.org/10.3390/fermentation8070339
Agyeman-Duah E, Kumar S, Gangwar B, Ujor VC. Glycerol Utilization as a Sole Carbon Source Disrupts the Membrane Architecture and Solventogenesis in Clostridium beijerinckii NCIMB 8052. Fermentation. 2022; 8(7):339. https://doi.org/10.3390/fermentation8070339
Chicago/Turabian StyleAgyeman-Duah, Eric, Santosh Kumar, Bhavana Gangwar, and Victor C. Ujor. 2022. "Glycerol Utilization as a Sole Carbon Source Disrupts the Membrane Architecture and Solventogenesis in Clostridium beijerinckii NCIMB 8052" Fermentation 8, no. 7: 339. https://doi.org/10.3390/fermentation8070339
APA StyleAgyeman-Duah, E., Kumar, S., Gangwar, B., & Ujor, V. C. (2022). Glycerol Utilization as a Sole Carbon Source Disrupts the Membrane Architecture and Solventogenesis in Clostridium beijerinckii NCIMB 8052. Fermentation, 8(7), 339. https://doi.org/10.3390/fermentation8070339







