Research Progress on the Construction of Artificial Pathways for the Biosynthesis of Adipic Acid by Engineered Microbes
Abstract
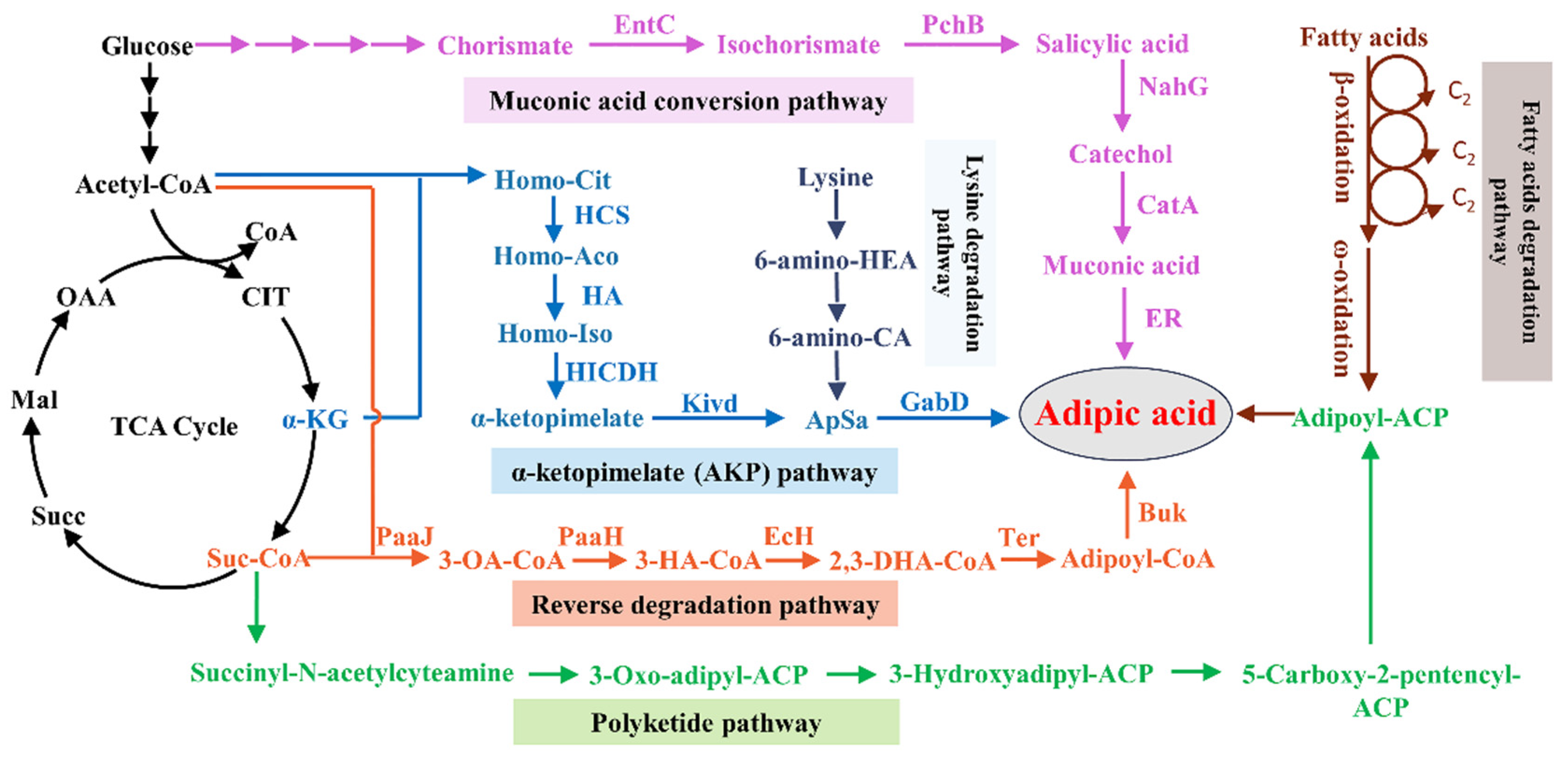
:1. Introduction
2. Biosynthesis of Adipic Acid via the Reverse Degradation Pathway
2.1. The Artificial Design of the Reverse Degradation Pathway
2.2. The Identification of the Reverse Degradation Pathway in Thermobifida fusca
3. Other Feasible Artificial Pathways for Adipic Acid Production
3.1. The Fatty Acid Degradation Pathway
3.2. The Muconic Acid Conversion Pathway
3.3. Some Potential Pathways for Adipic Acid Production
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vescan, A. Environmental Protection and Social Entrepreneurship Activities: The Vision of the Young People. Environ. Sci. Proc. 2021, 9, 15. [Google Scholar]
- Liu, E.K.; He, W.Q.; Yan, C.R. ‘White revolution’ to ‘white pollution’ Agricultural plastic film mulch in China. Environ. Res. Lett. 2014, 9, 091001. [Google Scholar] [CrossRef]
- Mohanty, S.; Nayak, S.K. Biodegradable Nanocomposites of Poly(butylene adipate-co-terephthalate) (PBAT) and Organically Modified Layered Silicates. J. Polym. Environ. 2012, 20, 195–207. [Google Scholar] [CrossRef]
- Signori, F.; Coltelli, M.B.; Bronco, S.J.P.D. Stability, Thermal degradation of poly(lactic acid) (PLA) and poly(butylene adipate-co-terephthalate) (PBAT) and their blends upon melt processing. Polym. Degrad. Stabil. 2009, 94, 74–82. [Google Scholar] [CrossRef]
- Skoog, E.; Shin, J.H.; Saez-Jimenez, V.; Mapelli, V.; Olsson, L. Biobased adipic acid—The challenge of developing the production host. Biotechnol. Adv. 2018, 36, 2248–2263. [Google Scholar] [CrossRef]
- Turk, S.C.H.J.; Kloosterman, W.P.; Ninaber, D.K.; Kolen, K.P.A.M.; Knutova, J.; Suir, E.; Schürmann, M.; Raemakersfranken, P.C.; Müller, M.; De Wildeman, S.M.A.; et al. Metabolic Engineering toward Sustainable Production of Nylon-6. ACS Synth. Biol. 2016, 5, 65–73. [Google Scholar] [CrossRef]
- Cui, X.M.J.F.; Chemicals, S. Market status and development prospect of adiopic acid at home and abroad. Fine Spec. Chem. 2013, 21, 6–16. [Google Scholar]
- Polen, T.; Spelberg, M.; Bott, M. Toward biotechnological production of adipic acid and precursors from biorenewables. J. Biotechnol. 2013, 167, 75–84. [Google Scholar] [CrossRef]
- Jou, W.S.; Chen, K.N.; Chao, D.Y.J.P.D. Stability, Flame retardant and dielectric properties of glass fibre reinforced nylon-66 filled with red phosphorous. Polym. Degrad. Stabil. 2001, 74, 239–245. [Google Scholar] [CrossRef]
- Niu, W.; Draths, K.M.; Frost, J.W.J.B.P. Benzene-free synthesis of adipic acid. Biotechnol. Prog. 2002, 18, 201–211. [Google Scholar] [CrossRef]
- Sato, K.; Aoki, M.; Noyori, R.J.S. A ‘Green’ Route to Adipic Acid: Direct Oxidation of Cyclohexenes with 30 Percent Hydrogen Peroxide. Science 1998, 281, 1646–1647. [Google Scholar] [CrossRef]
- Alini, S.; Basile, F.; Blasioli, S.; Rinaldi, C.; Vaccari, A. Development of new catalysts for N2O-decomposition from adipic acid plant. Appl. Catal. B-Environ. 2007, 70, 323–329. [Google Scholar] [CrossRef]
- Aryapratama, R.; Janssen, M. Prospective life cycle assessment of bio-based adipic acid production from forest residues. J. Clean. Prod. 2017, 164, 434–443. [Google Scholar] [CrossRef]
- Diamond, G.M.; Murphy, V.; Boussie, T.R. Application of high throughput experimentation to the production of commodity chemicals from renewable feedstocks. Mod. Appl. High Throughput R D Heterog. Catal. 2014, 288–309. [Google Scholar] [CrossRef]
- Yu, J.L.; Xia, X.X.; Zhong, J.J.; Qian, Z.G. Direct biosynthesis of adipic acid from a synthetic pathway in recombinant Escherichia coli. Biotechnol. Bioeng. 2014, 111, 2580–2586. [Google Scholar] [CrossRef]
- Babu, T.; Yun, E.J.; Kim, S.; Kim, D.H.; Liu, K.H.; Kim, S.R.; Kim, K.H. Engineering Escherichia coli for the production of adipic acid through the reversed β-oxidation pathway. Process. Biochem. 2015, 50, 2066–2071. [Google Scholar] [CrossRef]
- Cheong, S.; Clomburg, J.M.; Gonzalez, R.J.N.B. Energy- and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 2016, 34, 556–561. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Gatgens, J.; Lubcke, M.; Pietruszka, J.; Bott, M.; Polen, T. Improved production of adipate with Escherichia coli by reversal of beta-oxidation. Appl. Microbiol. Biotechnol. 2017, 101, 2371–2382. [Google Scholar] [CrossRef]
- Zhao, M.; Huang, D.; Zhang, X.; Koffas, M.A.; Zhou, J.; Deng, Y. Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metab. Eng. 2018, 47, 254–262. [Google Scholar] [CrossRef]
- Liu, H.; Song, R.; Liang, Y.; Zhang, T.; Deng, L.; Wang, F.; Tan, T. Genetic manipulation of Escherichia coli central carbon metabolism for efficient production of fumaric acid. Bioresour. Technol. 2018, 270, 96–102. [Google Scholar] [CrossRef]
- Liu, H.; Marsafari, M.; Wang, F.; Deng, L.; Xu, P. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab. Eng. 2019, 56, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Thykaer, J.; Christensen, B.; Nielsen, J.J.M.E. Metabolic network analysis of an adipoyl-7-ADCA-producing strain of Penicillium chrysogenum: Elucidation of adipate degradation. Metab. Eng. 2002, 4, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Macchi, R.; Franchi, F.; Barzaghi, D.; Fernández, C.; García, J.L.; Bertoni, G.; Díaz, E. Characterization of the last step of the aerobic phenylacetic acid degradation pathway. Microbiology 2007, 153, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Dekishima, Y.; Lan, E.I.; Shen, C.R.; Cho, K.M.; Liao, J.C. Extending Carbon Chain Length of 1-Butanol Pathway for 1-Hexanol Synthesis from Glucose by Engineered Escherichia coli. J. Am. Chem. Soc. 2011, 133, 11399–11401. [Google Scholar] [CrossRef]
- Hartmanis, M.G. Butyrate kinase from Clostridium acetobutylicum. J. Biol. Chem. 1987, 262, 617–621. [Google Scholar] [CrossRef]
- Westin, M.A.; Hunt, M.C.; Alexson, S.E. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. J. Biol. Chem. 2005, 280, 38125–38132. [Google Scholar] [CrossRef]
- Parke, D.; Garcia, M.A.; Ornston, L.N.J.A.; Microbiology, E. Cloning and genetic characterization of dca genes required for beta-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1. Appl. Environ. Microb. 2001, 67, 4817–4827. [Google Scholar] [CrossRef]
- Deng, Y.; Mao, Y. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J. Appl. Microbiol. 2015, 119, 1057–1063. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Wang, J.; Zhao, Y.; Deng, Y. Biosynthesis of adipic acid in metabolically engineered Saccharomyces cerevisiae. J. Microbiol. 2020, 58, 1065–1075. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, M.; Zhou, S.; Zhao, Y.; Deng, Y.J. Biosynthesis of adipic acid by a highly efficient induction-free system in Escherichia coli. J. Biotechnol. 2020, 314–315, 8–13. [Google Scholar] [CrossRef]
- Hao, T.; Li, G.; Zhou, S.; Deng, Y. Engineering the Reductive TCA Pathway to Dynamically Regulate the Biosynthesis of Adipic Acid in Escherichia coli. ACS Synth. Biol. 2021, 10, 632–639. [Google Scholar] [CrossRef]
- Xu, P.; Vansiri, A.; Bhan, N.; Koffas, M.A. ePathBrick: A synthetic biology platform for engineering metabolic pathways in E. Coli. ACS Synth. Biol. 2012, 1, 256–266. [Google Scholar] [CrossRef]
- Edwards, J.; Cole, L.J.; Green, J.B.; Thomson, M.J.; Wood, A.J.; Whittingham, J.L.; Moir, J.W.B. Binding to DNA protects Neisseria meningitidis fumarate and nitrate reductase regulator (FNR) from oxygen. J. Biol. Chem. 2010, 285, 1105–1112. [Google Scholar] [CrossRef]
- Constantinidou, C.; Hobman, J.L.; Griffiths, L.; Patel, M.D.; Penn, C.W.; Cole, J.A.; Overton, T.W. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 2006, 281, 4802–4815. [Google Scholar] [CrossRef]
- Beardslee, T.; Picataggio, S.J.L.T. Bio-based adipic acid from renewable oils. Lipid Technol. 2012, 24, 223–225. [Google Scholar] [CrossRef]
- Picataggio, S.; Mielenz, J.J.M.; Biology, C. Determination of Candida tropicalis acyl coenzyme A oxidase isozyme function by sequential gene disruption. Mol. Cell. Biol. 1991, 11, 4333–4339. [Google Scholar]
- Ju, J.H.; Oh, B.R.; Heo, S.Y.; Lee, Y.U.; Shon, J.H.; Kim, C.H.; Kim, Y.M.; Seo, J.W.; Hong, W.K. Production of adipic acid by short- and long-chain fatty acid acyl-CoA oxidase engineered in yeast Candida tropicalis. Bioproc. Biosyst. Eng. 2020, 43, 33–43. [Google Scholar] [CrossRef]
- Sun, J.; Raza, M.; Sun, X.; Yuan, Q. Biosynthesis of adipic acid via microaerobic hydrogenation of cis, cis-muconic acid by oxygen-sensitive enoate reductase. J. Biotechnol. 2018, 280, 49–54. [Google Scholar] [CrossRef]
- Hagen, A.; Poust, S.; Rond, T.; Fortman, J.L.; Katz, L.; Petzold, C.J.; Keasling, J.D. Engineering a Polyketide Synthase for In Vitro Production of Adipic Acid. ACS Synth. Biol. 2016, 5, 21–27. [Google Scholar] [CrossRef]
- Salvachúa, D.; Johnson, C.W.; Singer, C.A.; Rohrer, H.; Peterson, D.J.; Black, B.A.; Knapp, A.; Beckham, G.T. Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 2018, 20, 5007–5019. [Google Scholar] [CrossRef]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Choi, S.; Lee, H.-N.; Park, E.; Lee, S.-J.; Kim, E.-S. Recent advances in microbial production of cis, cis-muconic acid. Biomolecules 2020, 10, 1238. [Google Scholar] [CrossRef] [PubMed]
- Draths, K.M.; Frost, J.W. Environmentally compatible synthesis of adipic acid from D-glucose. J. Am. Chem. Soc. 1994, 116, 399–400. [Google Scholar] [CrossRef]
- Frost, J.; Draths, K. Biocatalytic syntheses or aromatics from D-glucose: Renewable microbial sources of aromatic compounds. Annu. Rev. Microb. 1995, 49, 557–580. [Google Scholar] [CrossRef] [PubMed]
- Raj, K.; Partow, S.; Correia, K.; Khusnutdinova, A.N.; Yakunin, A.F.; Mahadevan, R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. 2018, 6, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.C.; Khusnutdinova., A.N.; Flick., R.; Kim., T.; Bornscheuer, U.T.; Yakunin, A.F.; Mahadevan, R. Alkene hydrogenation activity of enoate reductases for an environmentally benign biosynthesis of adipic acid. Chem. Sci. 2017, 8, 1406. [Google Scholar] [CrossRef] [PubMed]
- Khosla, C. Structures and Mechanisms of Polyketide Synthases. J. Org. Chem. 2009, 74, 6416–6420. [Google Scholar] [CrossRef]
- Donadio, S.; Staver, M.; McAlpine, J.; Swanson, S.; Katz, L. Modular Organization of Genes Required for Complex Polyketide Biosynthesis. Science 1991, 252, 675–679. [Google Scholar] [CrossRef]
- Olano, C.; Wilkinson, B.; Sanchez, C.; Moss, S.J.; Sheridan, R.; Math, V.; Weston, A.J.; Braña, A.F.; Martin, C.J.; Oliynyk, M.; et al. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: Cluster analysis and assignment of functions. Chem. Biol. 2004, 11, 87–97. [Google Scholar]
- Hagen, A.; Poust, S.; de Rond, T.; Yuzawa, S.; Katz, L.; Adams, P.D.; Petzold, C.J.; Keasling, J.D. In vitro analysis of carboxyacyl substrate tolerance in the loading and first extension modules of borrelidin polyketide synthase. Biochemistry 2014, 53, 5975–5977. [Google Scholar] [CrossRef]
- Howell, D.M.; Graupner, M.; Xu, H.; White, R.H. Identification of Enzymes Homologous to Isocitrate Dehydrogenase That Are Involved in Coenzyme B and Leucine Biosynthesis in Methanoarchaea. J. Bacteriol. 2000, 182, 5013–5016. [Google Scholar] [CrossRef]
- Drevland, R.M.; Waheed, A.; Graham, D.E. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 2007, 189, 4391–4400. [Google Scholar] [CrossRef]
- Drevland, R.M.; Jia, Y.; Palmer, D.R.; Graham, D.E. Methanogen homoaconitase catalyzes both hydrolyase reactions in coenzyme B biosynthesis. J. Biol. Chem. 2008, 283, 28888–28896. [Google Scholar] [CrossRef]
- Howell, D.M.; Harich, K.; Xu, H.; White, R.H. α-Keto Acid Chain Elongation Reactions Involved in the Biosynthesis of Coenzyme B (7-Mercaptoheptanoyl Threonine Phosphate) in Methanogenic Archaea. Biochemistry 1998, 37, 10108–10117. [Google Scholar] [CrossRef]
- Smit, B.A.; Van, H.; Engels, W.; Meijer, L.; Wouters, J.T.; Smit, G. Identification, Cloning, and Characterization of a Lactococcus lactis Branched-Chain α-Keto Acid Decarboxylase Involved in Flavor Formation. J. Bacteriol. 2005, 71, 303–311. [Google Scholar] [CrossRef]
- Taylor, P.P.; Pantaleone, D.P.; Senkpeil, R.F. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 1998, 16, 412. [Google Scholar] [CrossRef]
- Emma, K.; Ho, S.J.; Gunnar, W.; Eriksson, L.A.; Olsson, L.; Mapelli, V. In silico and in vitro studies of the reduction of unsaturated α,β bonds of trans-2-hexenedioic acid and 6-amino-trans-2-hexenoic acid—Important steps towards biobased production of adipic acid. PLoS ONE 2018, 13, e0193503. [Google Scholar]
- Becker, J.; Zelder, O.; Hafner, S.; Schroder, H.; Wittmann, C. From zero to hero—Design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab. Eng. 2011, 13, 159–168. [Google Scholar] [CrossRef]
- Becker, J.; Wittmann, C. Bio-based production of chemicals, materials and fuels-Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 2012, 23, 631–640. [Google Scholar] [CrossRef]
- Becker, J.; Wittmann, C. Systems and synthetic metabolic engineering for amino acid production—The heartbeat of industrial strain development. Curr. Opin. Biotechnol. 2012, 23, 718–726. [Google Scholar] [CrossRef]
| Strains | Phenotype | Substrates | Production of Adipic Acid | Reference |
|---|---|---|---|---|
| E. Coli | PTrc-paaJ (β-ketoadipyl-CoA thiolase from E. coli), PCMV-Hbd (3-hydroxybutyryl-CoA dehydrogenase from C. acetobutylicum), PCMV-Crt (crotonase from C. acetobutylicum), PTrc-Ter (trans-enoyl-CoA reductase from E. gracilis), PCMV-Ptb (phosphate butyryltransferase from C. acetobutylicum), PCMV-Buk1(butyryl kinase from C. acetobutylicum); ▲ptsG (Glucose phosphotransferase system),▲poxB (py-ruvate oxidase), ▲pta (phosphotransacetylase), ▲sdhA (succinate dehydrogenase), ▲iclR (isocitrate lyase regulator) | glucose | 0.64 mg/L | [15] |
| PT7-paaJ (thiolase from E. coli), PT7-paaH (3-hydroxyacyl-CoA reductase from E. coli), PT7-paaZ (3-oxoadipyl-CoA dehydratase from E. coli), PT7-Bcd (butyryl-CoA dehydrogenase from C. acetobutylicum), PT7-Bcd (peroxisomal acyl-CoA thioesterase from M. musculus) | glucose | 12 ug/L | [16] | |
| PT7-paaJ (thiolase from E. coli), PT7-paaH (3-hydroxyacyl-CoA reductase from E. coli), PT7-paaZ (3-oxoadipyl-CoA dehydratase from E. coli), PT7-DcaA (adipyl-CoA dehydrogenase from A. baylyi), PT7-TesB (acyl-CoA thioesterase from A. baylyi) | glucose | 36 mg/L | [18] | |
| PT7-Tfu_0875 (β-ketothiolase from T. fusca), PT7-Tfu_2399 (3-hydroxyacyl-CoA dehydrogenase from T. fusca), PTrc-Tfu_0067 (3-hydroxyadipyl-CoA dehydrogenase from T. fusca), PTrc-Tfu_1647 (5-Carboxy-2-pentenoyl-CoA reductase), PT7-Tfu_2576-7 (adipyl-CoA synthetase from T. fusca); ▲ldhA (L-lactate dehydrogenase), ▲sucD (succinate coenzyme A ligase), ▲atoB (acetyl coenzyme A acetyltransferase). | glycerin | 68 g/L | [19] | |
| PUTRrpst-Tfu_0875 (β-ketothiolase from T. fusca), PUTRrpst-Tfu_2399 (3-hydroxyacyl-CoA dehydrogenase from T. fusca), PUTRrpst-Tfu_0067 (3-hydroxyadipyl-CoA dehydrogenase from T. fusca), PUTRrpst-Tfu_1647 (5-Carboxy-2-pentenoyl-CoA reductase), PUTRlpp-Tfu_2576-7 (adipyl-CoA synthetase from T. fusca);▲ldhA (L-lactate dehydrogenase), ▲sucD (succinate coenzyme A ligase), ▲atoB (acetyl coenzyme A acetyltransferase) | glycerin | 57.6 g/L | [20] | |
| T. fusca | PT7-Tfu_1647 (5-Carboxy-2-pentenoyl-CoA reductase from T. fusca) | glucose | 2.23 g/L | [28] |
| S. cerevisiae | PTEF1-Tfu_0875 (β-ketothiolase from T. fusca), PGPD1-Tfu_2399(3-hydroxyacyl-CoA dehydrogenase from T. fusca), PADH1-Tfu_0067 (3-hydroxyadipyl-CoA dehydrogenase from T. fusca), PPGK1-Tfu_1647 (5-Carboxy-2-pentenoyl-CoA reductase), PTDH3-Tfu_2576-7 (adipyl-CoA synthetase from T. fusca); ▲LSC1 (succinyl-CoA ligase) | glucose | 3.83 mg/L | [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, Y.; Liu, H.; Zhang, R.; Jin, Y.; Yu, Y.; Deng, L.; Wang, F. Research Progress on the Construction of Artificial Pathways for the Biosynthesis of Adipic Acid by Engineered Microbes. Fermentation 2022, 8, 393. https://doi.org/10.3390/fermentation8080393
Ning Y, Liu H, Zhang R, Jin Y, Yu Y, Deng L, Wang F. Research Progress on the Construction of Artificial Pathways for the Biosynthesis of Adipic Acid by Engineered Microbes. Fermentation. 2022; 8(8):393. https://doi.org/10.3390/fermentation8080393
Chicago/Turabian StyleNing, Yuchen, Huan Liu, Renwei Zhang, Yuhan Jin, Yue Yu, Li Deng, and Fang Wang. 2022. "Research Progress on the Construction of Artificial Pathways for the Biosynthesis of Adipic Acid by Engineered Microbes" Fermentation 8, no. 8: 393. https://doi.org/10.3390/fermentation8080393
APA StyleNing, Y., Liu, H., Zhang, R., Jin, Y., Yu, Y., Deng, L., & Wang, F. (2022). Research Progress on the Construction of Artificial Pathways for the Biosynthesis of Adipic Acid by Engineered Microbes. Fermentation, 8(8), 393. https://doi.org/10.3390/fermentation8080393






