Comparative Evaluation of Secreted Plant Carotenoid Cleavage Dioxygenase 1 (CCD1) Enzymes in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Media
2.2. CCD1s Gene Synthesis and Episomal Plasmid Construction
2.3. Construction of a CCD1 Phylogenetic Tree
2.4. LCMS Validation of Heterologous CCD1 Secretion
2.5. Spectrophotometry of β-Carotene Reduction in the Supernatant of β-Carotene-Containing Medium (BCM)
2.6. Head-Space Solid-Phase Micro-Extraction (HS-SPME) for Detection of β-Ionone by GCMS
3. Results and Discussion
3.1. Construction of a Phylogenetic Tree for Selection of Four CCD1s
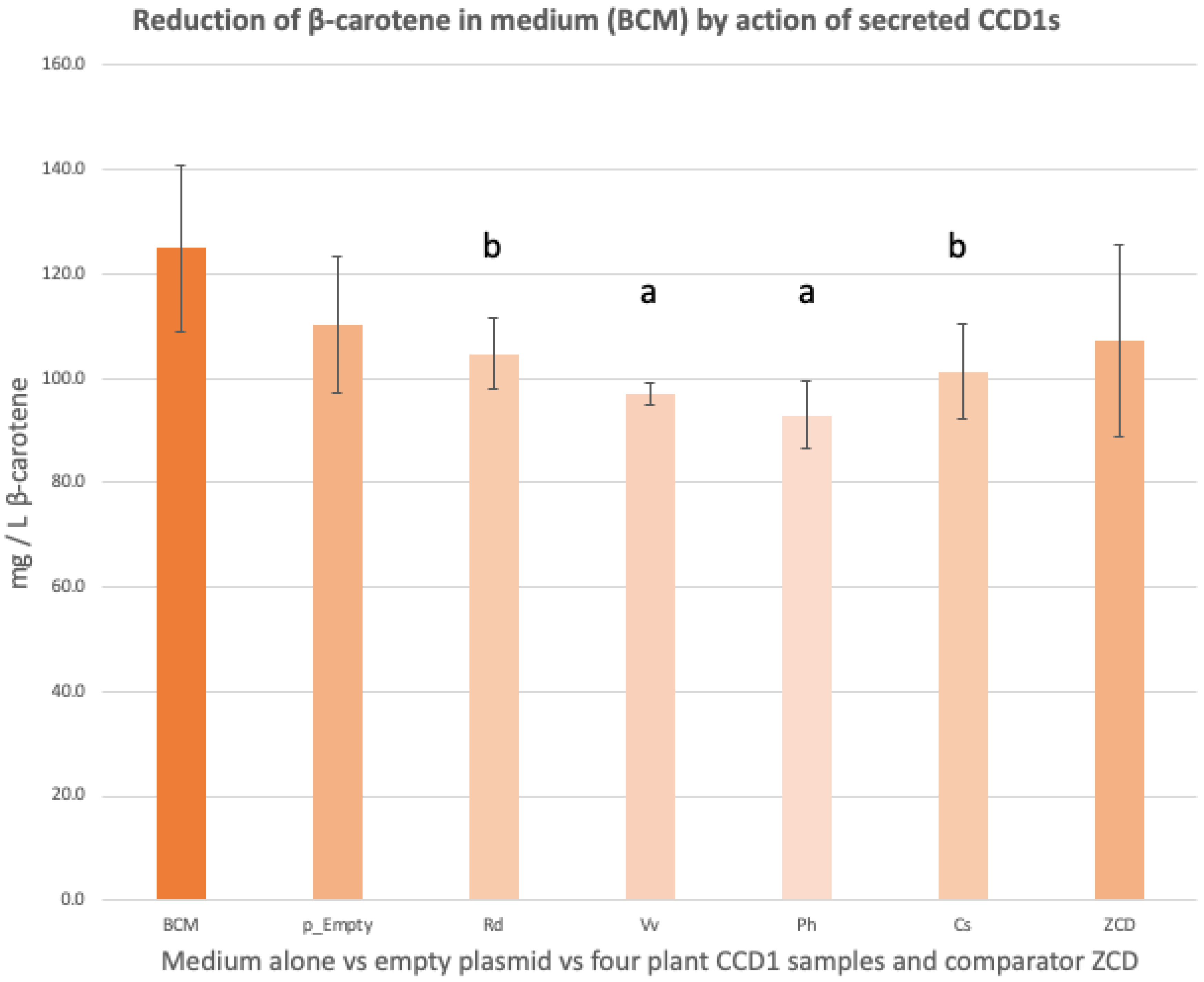
3.2. Spectrophotometry of β-Carotene Levels in the Supernatant of β-Carotene-Containing Medium (BCM)
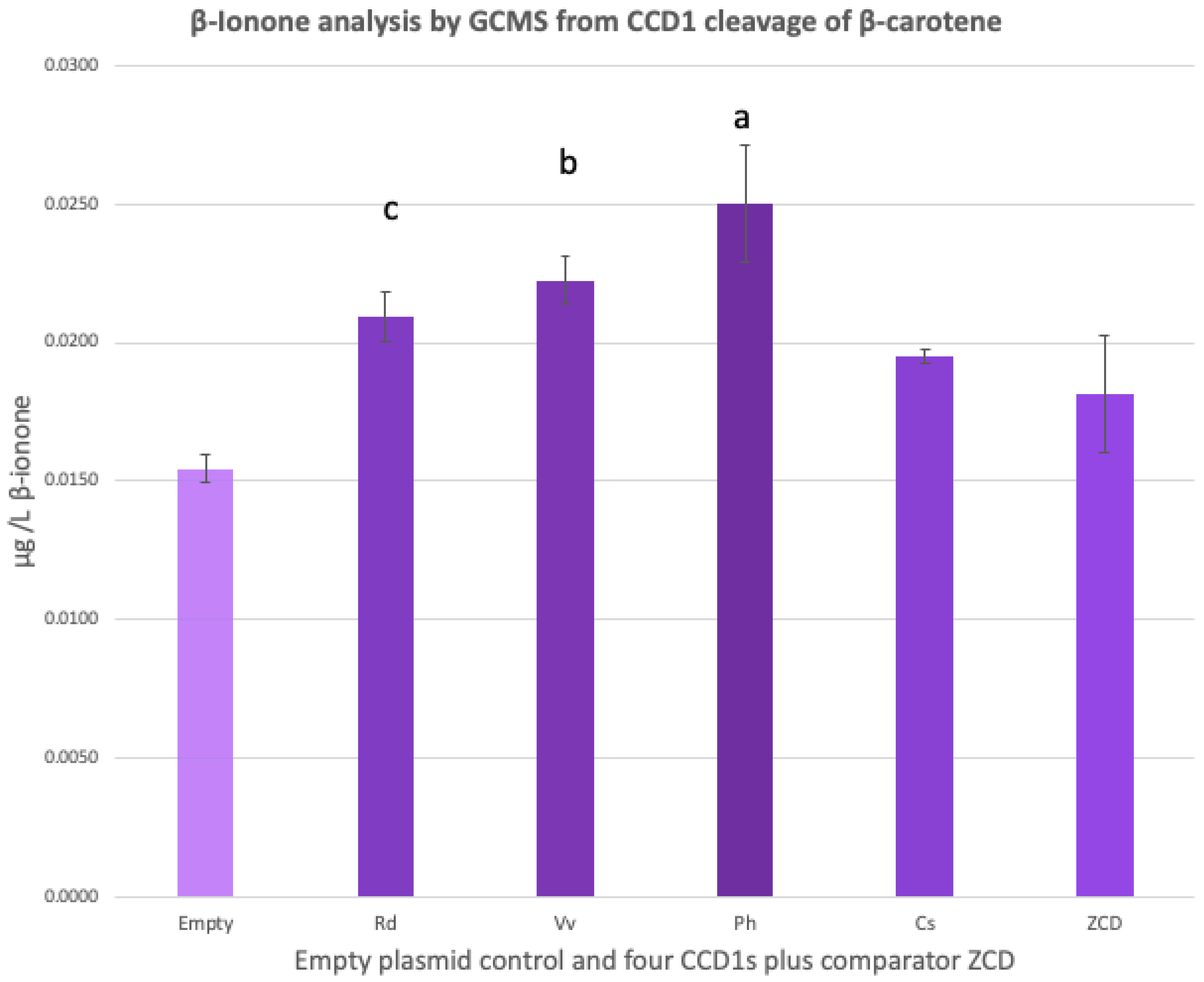
3.3. GCMS of β-Ionone from Head-Space Solid-Phase Micro-Extraction (HS-SPME)
3.4. Liquid Chromatography-Mass Spectrometry (LCMS) Analysis of Extracellularly Secreted CCD1s
3.5. Comparison of the Efficacy of the Four Secreted Plant CCD1s
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petronilho, S.; Lopez, R.; Ferreira, V.; Coimbra, M.A.; Rocha, S.M. Revealing the usefulness of aroma networks to explain wine aroma properties: A case study of Portuguese wines. Molecules 2020, 25, 272. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.L.; Boss, P.K.; Solomon, P.S.; Trengove, R.D.; Heymann, H.; Ebeler, S.E. Origins of grape and wine aroma. Part 1. Chemical components and viticultural impacts. Am. J. Enol. Vitic. 2014, 65, 1–24. [Google Scholar] [CrossRef]
- Cataldo, V.F.; Lopez, J.; Carcamo, M.; Agosin, E. Chemical vs. biotechnological synthesis of C13-apocarotenoids: Current methods, applications and perspectives. Appl. Microbiol. Biotechnol. 2016, 100, 5703–5718. [Google Scholar] [CrossRef]
- Timmins, J.J.B.; Kroukamp, H.; Paulsen, I.T.; Pretorius, I.S. The sensory significance of apocarotenoids in wine: Importance of carotenoid cleavage dioxygenase 1 (CCD1) in the production of β-ionone. Molecules 2020, 25, 2779–2793. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, S.; Terrier, N.; Procureur, J.; Bigey, F.; Gunata, Z. A carotenoid cleavage dioxygenase from Vitis vinifera L.: Functional characterisation and expression during grape berry development in relation to C13-norisoprenoid accumulation. J. Exp. Bot. 2005, 56, 2721–2731. [Google Scholar] [CrossRef] [PubMed]
- Baumes, R.; Wirth, J.; Bureau, S.; Gunata, Y.; Razungles, A. Biogeneration of C13-norisoprenoid compounds: Experiments supportive for an apo-carotenoid pathway in grapevines. Anal. Chim. Acta 2002, 458, 3–14. [Google Scholar] [CrossRef]
- Razungles, A.; Bayonove, C.L.; Cordonnier, R.E.; Baumes, R.L. Etude des caroténoïdes du raisin à maturité. Vitis 1987, 26, 183–191. [Google Scholar]
- Mendes-Pinto, M.M. Carotenoid breakdown products–the norisoprenoids–in wine aroma. Arch. Biochem. Biophys. 2009, 483, 236–245. [Google Scholar] [CrossRef]
- Razungles, A.; Bayonove, C.L.; Cordonnier, R.E.; Sapis, J.C. Grape carotenoids: Changes during the maturation period and localisation in mature berries. Am. J. Enol. Vitic. 1988, 39, 44–48. [Google Scholar]
- Saini, R.K.; Nile, S.H.; Park, S.W. Carotenoids from fruits and vegetables: Chemistry, analysis, occurrence, bioavailability and biological activities. Food Res. Int. 2015, 76, 735–750. [Google Scholar] [CrossRef]
- Chen, W.-K.; Yu, K.-J.; Liu, B.; Lan, Y.-B.; Sun, R.-Z.; Li, Q.; He, F.; Pan, Q.-H.; Duan, C.-Q.; Wang, J. Comparison of transcriptional expression patterns of carotenoid metabolism in ‘Cabernet Sauvignon’ grapes from two regions with distinct climate. J. Plant Physiol. 2017, 213, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Young, P.R.; Lashbrooke, J.G.; Alexandersson, E.; Jacobson, D.; Moser, C.; Velasco, R.; Vivier, M.A. The genes and enzymes of the carotenoid metabolic pathway in Vitis vinifera L. BMC Genom. 2012, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lashbrooke, J.G.; Young, P.R.; Dockrall, S.J.; Vasanth, K.; Vivier, M.A. Functional characterization of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 2013, 13, 156. [Google Scholar] [CrossRef]
- Kambiranda, D.; Basha, S.M.; Singh, R.K.; He, H.; Calvin, K.; Mercer, R. In depth proteome analysis of ripening Muscadine grape berry cv. Carlos reveals proteins associated with flavour and aroma compounds. J. Proteome Res. 2016, 15, 2910–2923. [Google Scholar] [CrossRef]
- Beekwilder, J.; van Rossum, H.M.; Koopman, F.; Sonntag, F.; Buchhaupt, M.; Schrader, J.; Hall, R.D.; Bosch, D.; Pronk, J.T.; van Maris, A.J.A.; et al. Polycistronic expression of a β-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to β-ionone production. J. Biotech. 2014, 192, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Ohloff, G. Importance of minor components in flavors and fragrances. Perfum. Flavor. 1978, 3, 11–22. [Google Scholar]
- Canuti, V.; Conversano, M.; Li Calzi, M.; Heymann, H.; Matthews, M.A.; Ebeler, S.E. Headspace solid-phase microextraction–gas chromatography–mass spectro- metry for profiling free volatile compounds in Cabernet Sauvignon grapes and wines. J. Chromatogr. A 2009, 1216, 3012–3022. [Google Scholar] [CrossRef]
- Asproudi, A.; Ferrandino, A.; Bonello, F.; Vaudano, E.; Pollon, M.; Petrozziello, M. Key norisoprenoid compounds in wines from early-harvested grapes in view of climate change. J. Food Chem. 2018, 268, 143–152. [Google Scholar] [CrossRef]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Pedroza, M.A.; Zalacain, A.; Lara, J.F.; Salinas, M.R. Global grape aroma potential and its individual analysis by SBSE-GC-MS. Food Res. Int. 2010, 43, 1003–1008. [Google Scholar] [CrossRef]
- Marais, J.; van Wyk, C.J.; Rapp, A. Carotenoid levels in maturing grapes as affected by climatic regions, sunlight and shade. S. Afr. J. Enol. Vitic. 1991, 12, 64–69. [Google Scholar] [CrossRef][Green Version]
- la Grange, D.C.; Pretorius, I.S.; van Zyl, W.H. Expression of a Trichoderma reesei β-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1996, 62, 1036–1044. [Google Scholar] [CrossRef]
- Nevalainen, H.; Peterson, R. Making recombinant proteins in filamentous fungi–are we expecting too much? Front. Microbiol. 2014, 5, 75. [Google Scholar] [PubMed]
- Brevnova, E.; McBride, J.; Wiswall, E.; Wenger, K.; Caiazza, N.; Hau, H.; Argyros, A.; Agbogbo, F.; Rice, C.F.; Barrett, T.; et al. Yeast Expressing Saccharolytic Enzymes for Consolidated Bioprocessing Using Starch and. Cellulose. Patent No. WO/2011/153516, 2013. [Google Scholar]
- Kroukamp, H.; den Haan, R.; la Grange, D.C.; Sibanda, N.; Foulquié-Moreno, M.R.; Thevelein, J.M.; van Zyl, W.H. Strain breeding enhanced heterologous cellobiohydrolase secretion by Saccharomyces cerevisiae in a protein specific manner. Biotechnol. J. 2017, 12, 1700346. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Tan, B.-C.; McCarty, D.R.; Klee, H.J. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef]
- Baldermann, S.; Kato, M.; Kurosawa, M.; Kurobayashi, Y.; Fujita, A.; Fleischmann, P.; Watanabe, N. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 2010, 61, 2967–2977. [Google Scholar] [CrossRef]
- Soria, A.C.; Sanz, J.; Villamiel, M. Analysis of volatiles in dehydrated carrot samples by solid-phase microextraction followed by GC-MS. J. Sep. Sci. 2008, 31, 3548–3555. [Google Scholar] [CrossRef]
- Yamamoto, M.; Baldermann, S.; Yoshikawa, K.; Fujita, A.; Mase, N.; Watanabe, N. Determination of volatile compounds in four commercial samples of Japanese green algae using solid phase microextraction gas chromatography mass spectrometry. Sci. World J. 2014, 2014, 289780. [Google Scholar] [CrossRef]
- Langen, J.; Wegmann-Herr, P.; Schmarr, H.-G. Quantitative determination of a-ionone, β-ionone and β-damascenone and enantiodifferentiation of a-ionone in wine for authenticity control using multidimensional gas chromatography with tandem mass spectrometric detection. Ann. Bioanal. Chem. 2016, 408, 6483–6496. [Google Scholar] [CrossRef]
- Maruti, A.; Duran-Guerrero, E.; Barroso, C.G.; Castro, R. Optimisation of a multiple headspace sorptive extraction method coupled to gas chromatography-mass spectrometry for the determination of volatile compounds in macroalgae. J. Chromatogr. A 2018, 1551, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.-C.; Horvath, G.; Molnar, P.; Turcsi, E.; Deli, J.; Schrader, J.; Sandmann, G.; Schmidt, H.; Schwab, W. Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from Rosa damascena. Phytochemistry 2009, 70, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Peng, S.; Xia, W.; Zheng, H.; Chen, X.; Yin, L. Analysis of five earthy-musty odorants in environmental water by HS-SPME/GC-MS. Int. J. Anal. Chem. 2014, 2014, 697260. [Google Scholar] [CrossRef] [PubMed]
- Nawade, B.; Shaltiel-Harpaz, L.; Yahyaa, M.; Bosamia, T.C.; Kabaha, A.; Kedoshim, R.; Zohar, M.; Isaacson, T.; Ibdah, M. Analysis of apocarotenoid volatiles during the development of Ficus carica fruits and characterisation of carotenoid cleavage dioxygenase genes. Plant Sci. 2020, 290, 110292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Lindley, N.D.; Too, H.-P. A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol. Bioeng. 2018, 115, 174–183. [Google Scholar] [CrossRef]
- Czajka, J.J.; Nathenson, J.A.; Benites, V.T.; Baidoo, E.E.K.; Cheng, Q.; Wang, Y.; Tang, Y.J. Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound β-ionone. Microb. Cell Factories 2018, 17, 136. [Google Scholar] [CrossRef]

| GenBank Accession Number | Gene Name | Gene Size | Plant Origin |
|---|---|---|---|
| EU327776.1 | Rd ssCCD1 | 1659 bp | Rosa × damascena |
| KF008001.1 | Vv ssCCD1 | 1629 bp | Vitis vinifera |
| AY576003.1 | Ph ssCCD1 | 1641 bp | Petunia × hybrida |
| AJ132927.1 | Cs ssCCD1 | 1686 bp | Crocus sativus |
| AJ489276.1 | Cs ssZCD | 1186 bp | Crocus sativus |
| Type of Primer | Sequence of Primer |
|---|---|
| Rd-ssCCD1 ORF_R | 5′-AAG AAC AAG CAA AGT TCT AA-3′ |
| Vv-ssCCD1 ORF_R | 5′-AAG AAC AAG CAA AAC TTT GA-3′ |
| Ph-ssCCD1 ORF_R | 5′-AAG AAC AAG CCA AAC TGT GA-3′ |
| Cs-ssCCD1 ORF_R | 5′-TGC TGG ATC ATT GCA GTA CC-3′ |
| Cs-ssZCD ORF_R | 5′-CCT CTC CAT ATT CGC TGC CA-3′ |
| ENO1p_all-CCD ORF_F | 5′-ACA CAA ACA CTA AAT CAA AG-3′ |
| GC Parameters | ||
| Start temperature: 50 °C | Injection temp: 270 °C | Sampling time: 1.0 to 1.5 min |
| Flow control: 69 cm/s | Pressure: 100 kPa | Purge flow: on at 3.0 mL/min |
| Linear velocity: 47 cm/s | Mode: Spitless | Split ratio: −1.0 |
| MS Parameters | ||
| Ion source temp: 200 °C | Interface temp: 270 °C | Solvent cut-off time: 1.5 min |
| Threshold: 1000 counts | Start m/z: 45.0 | End m/z: 350.0 |
| Start time: 2.0 min | End time: 12.0 min | Electron ionisation: 70 eV |
| Acquisition mode: scan | Interval: 0.15 s | Scan speed: 2500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timmins, J.J.B.; Kroukamp, H.; Walker, R.S.K.; Pretorius, I.S.; Paulsen, I.T. Comparative Evaluation of Secreted Plant Carotenoid Cleavage Dioxygenase 1 (CCD1) Enzymes in Saccharomyces cerevisiae. Fermentation 2022, 8, 395. https://doi.org/10.3390/fermentation8080395
Timmins JJB, Kroukamp H, Walker RSK, Pretorius IS, Paulsen IT. Comparative Evaluation of Secreted Plant Carotenoid Cleavage Dioxygenase 1 (CCD1) Enzymes in Saccharomyces cerevisiae. Fermentation. 2022; 8(8):395. https://doi.org/10.3390/fermentation8080395
Chicago/Turabian StyleTimmins, John J. B., Heinrich Kroukamp, Roy S. K. Walker, Isak S. Pretorius, and Ian T. Paulsen. 2022. "Comparative Evaluation of Secreted Plant Carotenoid Cleavage Dioxygenase 1 (CCD1) Enzymes in Saccharomyces cerevisiae" Fermentation 8, no. 8: 395. https://doi.org/10.3390/fermentation8080395
APA StyleTimmins, J. J. B., Kroukamp, H., Walker, R. S. K., Pretorius, I. S., & Paulsen, I. T. (2022). Comparative Evaluation of Secreted Plant Carotenoid Cleavage Dioxygenase 1 (CCD1) Enzymes in Saccharomyces cerevisiae. Fermentation, 8(8), 395. https://doi.org/10.3390/fermentation8080395







