Kinetic Model for Enzymatic Hydrolysis of Cellulose from Pre-Treated Rice Husks
Abstract
:1. Introduction
2. Materials and Methods
2.1. H2O2–Aqueous Ammonia Pretreatment (Previous Author Modification)
2.2. Cellulase Enzyme Preparation from Aspergillus niger (Modification of Previous Studies)
2.3. Enzymatic Hydrolysis
2.4. Analytical Methods
2.4.1. Evaluation of Endoglucanase Activity
2.4.2. Evaluation of Exoglucanase Activity
3. Results
3.1. Effect of Hydrogen Peroxide–Aqueous Ammonia Pretreatment on the Composition of Rice Husks
3.2. Characterization of Cellulase Enzyme Extract
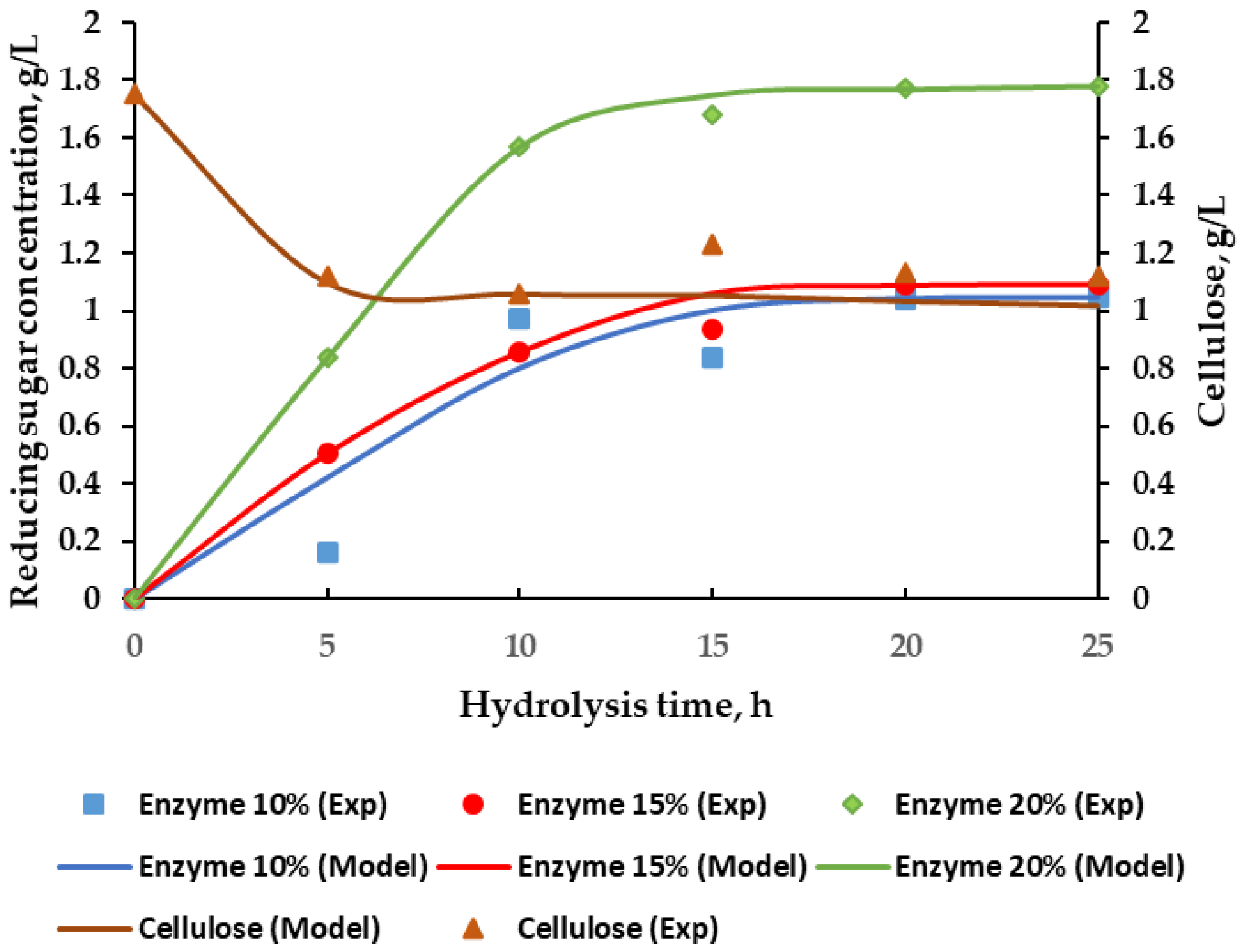
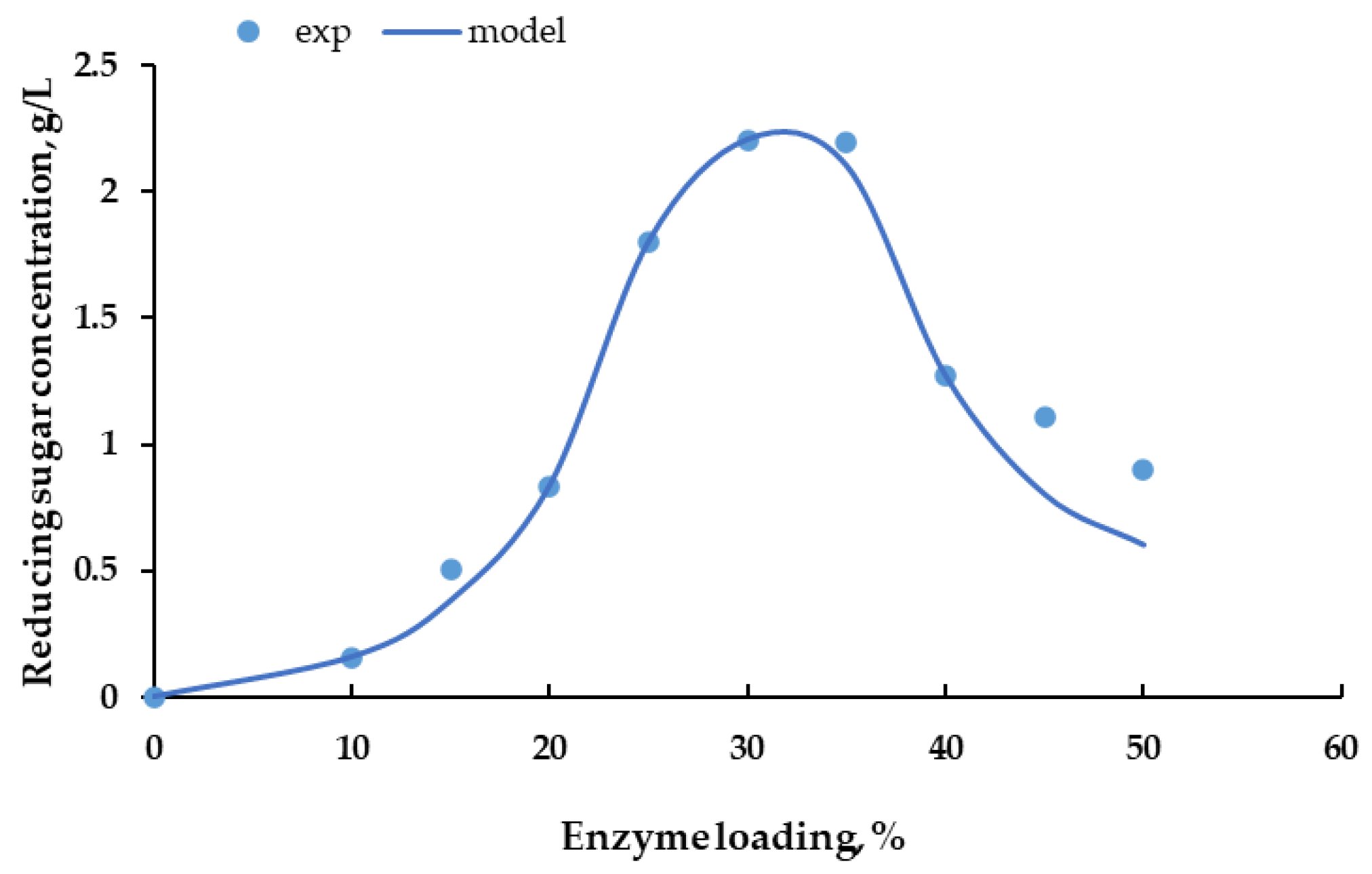
3.3. Effect of Hydrolysis Time and Enzyme Concentration on the Reducing Sugar Concentration
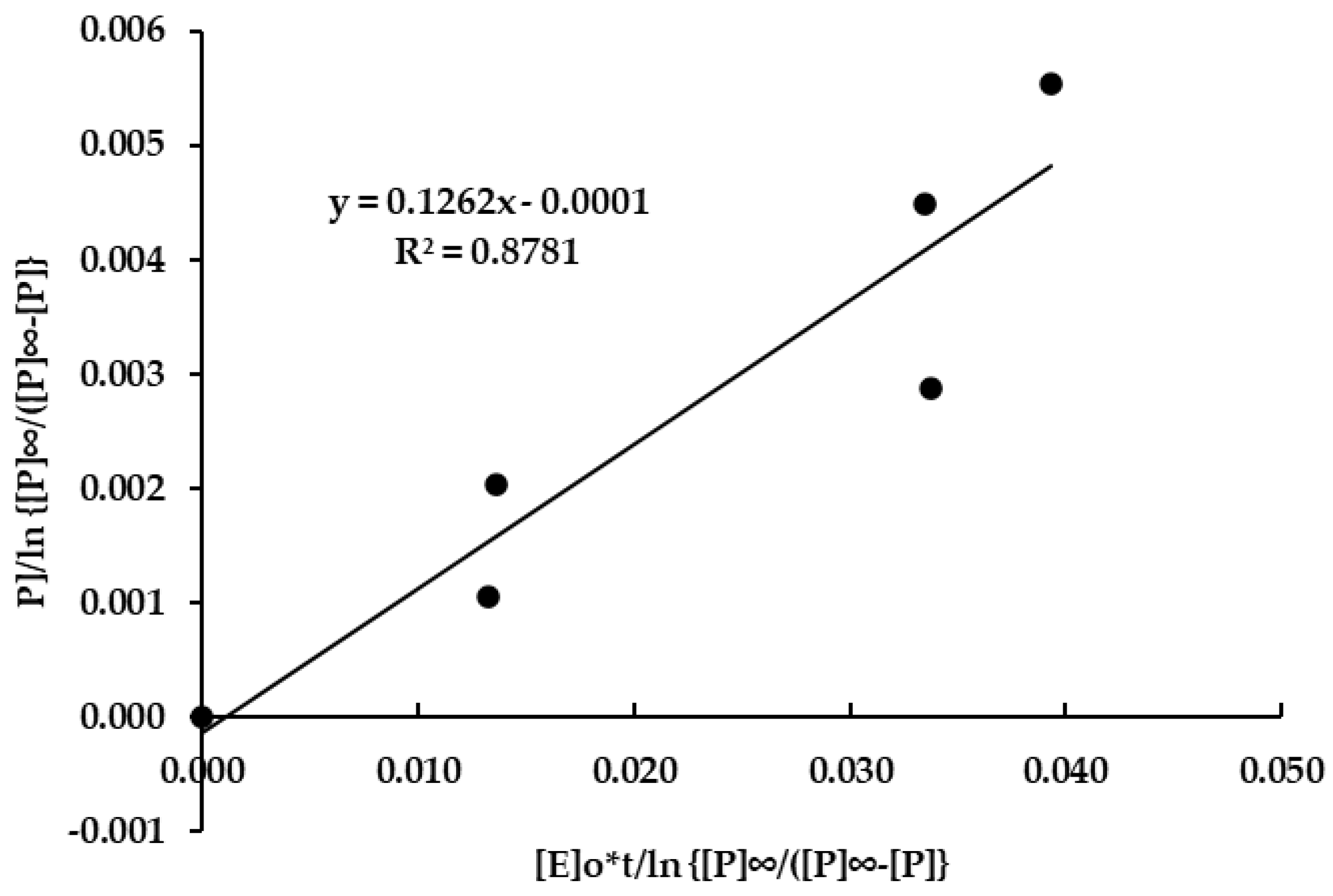
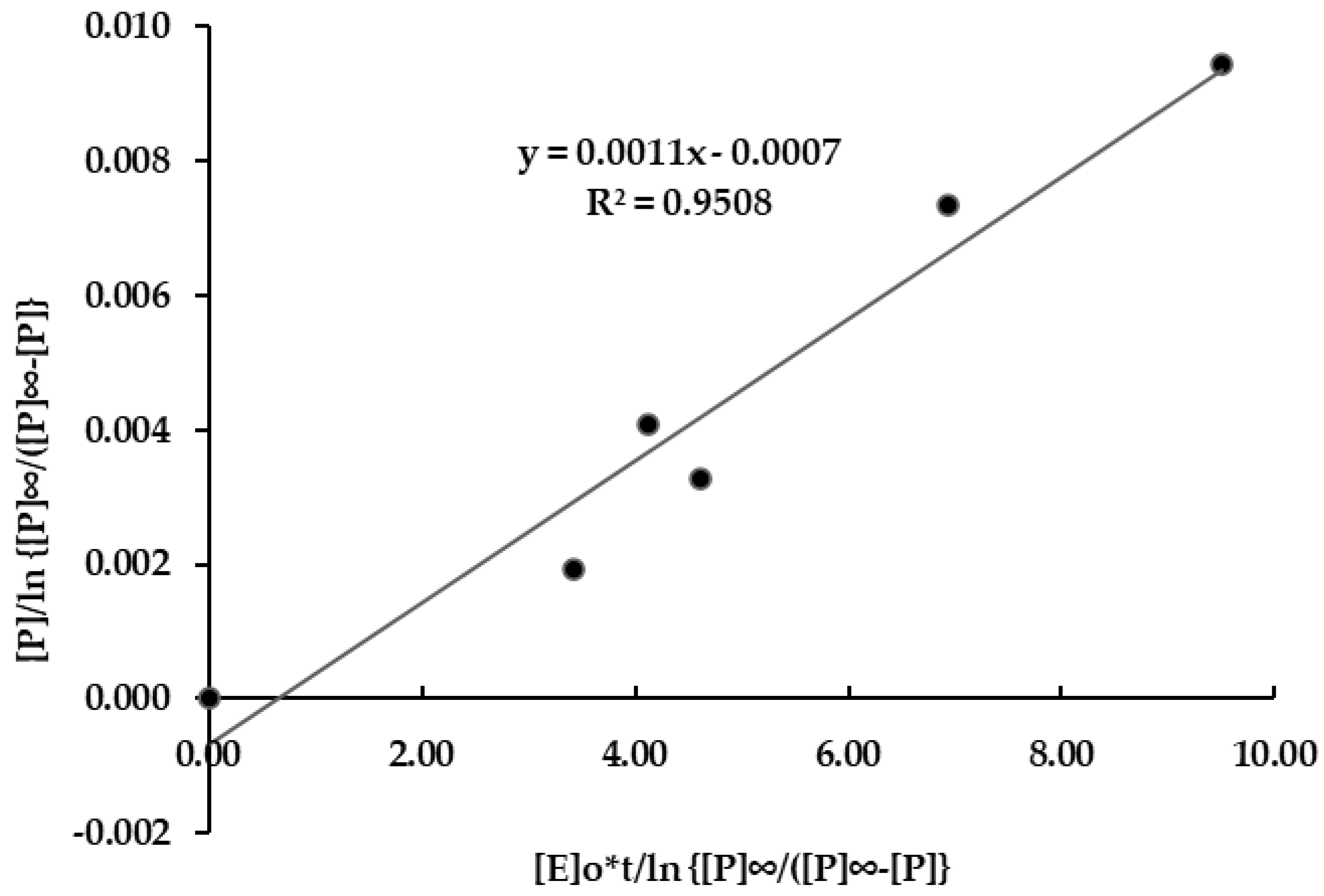
3.4. Cellulase Enzyme Reaction Kinetics
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rosita, N. Book 5: Province of South Sumatra. In Business Directory/Companies of Rice Milling Industry; Bahagia, M.Q., Ed.; Central Bureau of Statistics: Jakarta, Indonesia, 2020; pp. 1–318. [Google Scholar]
- Cacua, A.; Gelvez, J.J.; Rodríguez, D.C.; Parra, J.W. Production of bioethanol from rice husk pre-treated with alkalis and hydrolyzed with acid cellulase at pilot scale. J. Phys. Conf. Ser. 2018, 1126, 12034. [Google Scholar] [CrossRef]
- Brémond, U.; de Buyer, R.; Steyer, J.P.; Bernet, N.; Carrere, H. Biological pre-treatments of biomass for improving biogas production: An overview from lab scale to full-scale. Renew. Sustain. Energy Rev. 2018, 90, 583–604. [Google Scholar] [CrossRef]
- Plácido, J.; Capareda, S. Ligninolytic enzymes: A biotechnological alternative for bioethanol production. Bioresour. Bioprocess. 2015, 2, 23. [Google Scholar] [CrossRef] [Green Version]
- Zabed, H.; Sahu, J.N.; Boyce, A.N.; Faruq, G. Fuel ethanol production from lignocellulosic biomass: An overview on feedstocks and technological approaches. Renew. Sustain. Energy Rev. 2016, 66, 751–774. [Google Scholar] [CrossRef]
- Gonçalves, F.A.; Ruiz, H.A.; Nogueira, C.D.C.; Dos Santos, E.S.; Teixeira, J.A.; De Macedo, G.R. Comparison of delignified coconuts waste and cactus for fuel-ethanol production by the simultaneous and semi-simultaneous saccharification and fermentation strategies. Fuel 2014, 131, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Chukwuma, O.B.; Rafatullah, M.; Tajarudin, H.A.; Ismail, N. Lignocellulolytic enzymes in biotechnological and industrial processes: A review. Sustainability 2020, 12, 7282. [Google Scholar] [CrossRef]
- Axelsson, J. Separate Hydrolysis and Fermentation of Pretreated Spruce. Master’ Thesis, Linköping University, Örnsköldsvik Sweden, 2011. [Google Scholar]
- Jȩdrzejczyk, M.; Soszka, E.; Czapnik, M.; Ruppert, A.M.; Grams, J. Physical and chemical pre-treatment of lignocellulosic biomass. In Second and Third Generation of Feedstocks: The Evolution of Biofuels; Elsevier Inc.: Amsterdam, The Netherlands, 2019; pp. 143–196. [Google Scholar]
- Albuquerque, E.D.; Torres, F.A.G.; Fernandes, A.A.R.; Fernandes, P.M.B. Combined effects of high hydrostatic pressure and specific fungal cellulase improve coconut husk hydrolysis. Process. Biochem. 2016, 51, 1767–1775. [Google Scholar] [CrossRef]
- Novia, N.; Said, M.; Jannah, A.M.; Pebriantoni, P.; Bayu, M. Aqueous Ammonia Soaking-Dilute Acid Pretreatment to Produce Bioethanol from Rice Husk. Technol. Reports Kansai Univ. 2020, 62, 891–900. [Google Scholar]
- Kim, J.S.; Lee, Y.Y.; Kim, T.H. A review on alkaline pre-treatment technology for bioconversion of lignocellulosic biomass. Bioresour. Technol. 2016, 199, 42–48. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, Q.; Hu, X.; Li, X.; Huang, Y.; Li, W.; Ma, L. Structural evolution during corn stalk acidic and alkaline hydrogen peroxide pre-treatment. Ind. Crop. Prod. 2022, 176, 114386. [Google Scholar] [CrossRef]
- Ali, N.; Giwa, A.S.; Abdalla, M.; Liu, X. Alkaline hydrogen peroxide pre-treatment of bamboo culm for improved enzymatic release of reducing sugars using recombinant cellulases. Cellulose 2020, 27, 769–779. [Google Scholar] [CrossRef]
- Novia, N.; Hasanudin, H.; Hermansyah, H.; Fudholi, A. Kinetics of Lignin Removal from Rice Husk Using Hydrogen Peroxide and Combined Hydrogen Peroxide–Aqueous Ammonia Pretreatments. Fermentation 2022, 8, 157. [Google Scholar] [CrossRef]
- Jeoh, T.; Cardona, M.J.; Karuna, N.; Mudinoor, A.R.; Nill, J. Mechanistic Kinetic Models of Enzymatic Cellulose Hydrolysis — A Review. Biotechnol. Bioeng. 2017, 114, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- Niglio, S.; Procentese, A.; Russo, M.E.; Piscitelli, A.; Marzocchella, A. Integrated enzymatic pre-treatment and hydrolysis of apple pomace in a bubble column bioreactor. Biochem. Eng. J. 2019, 150, 107306. [Google Scholar] [CrossRef]
- Abdella, A.; Mazeed, T.E.; El-baz, A.F.; Yang, S. Production of β-glucosidase from wheat bran and glycerol by Aspergillus niger in stirred tank and rotating fibrous bed bioreactors. Process. Biochem. 2016, 51, 1331–1337. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Isak, S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [Green Version]
- Bychkov, A.; Podgorbunskikh, E.; Bychkova, E.; Lomovsky, O. Current achievements in the mechanically pre-treated conversion of plant biomass. Biotechnol. Bioeng. 2019, 116, 1231–1244. [Google Scholar] [CrossRef]
- Nill, J.; Karuna, N.; Jeoh, T. The impact of kinetic parameters on cellulose hydrolysis rates. Process. Biochem. 2018, 74, 108–117. [Google Scholar] [CrossRef]
- Fabbrizi, G.; Giannoni, T.; Lorenzi, L.; Nicolini, A.; Iodice, P.; Coccia, V.; Cavalaglio, G.; Gelosia, M. High Solid and Low Cellulase Enzymatic Hydrolysis of Cardoon Stems Pretreated by Acidified γ-Valerolactone/Water Solution. Energies 2022, 15, 2600. [Google Scholar] [CrossRef]
- Policastro, G.; Luongo, V.; Frunzo, L.; Fabbricino, M. A comprehensive review of mathematical models of photo fermentation. Crit. Rev. Biotechnol. 2021, 41, 628–648. [Google Scholar] [CrossRef]
- Bisswanger, H. Enzyme Kinetics: Principles and Methods. In Psychology in the Brain, 3rd ed.; Wiley-VCH: Weinheim, Germany, 2017; pp. 1–21. [Google Scholar]
- Bezerra, R.M.F.; Dias, A.A. Discrimination among eight modified Michaelis-Menten kinetics models of cellulose hydrolysis with a large range of substrate/enzyme ratios: Inhibition by cellobiose. Appl. Biochem. Biotechnol. Part A Enzym. Eng. Biotechnol. 2004, 112, 173–184. [Google Scholar] [CrossRef]
- Miao, Y.; Chen, J.Y.; Jiang, X.; Huang, Z. Kinetic studies on the product inhibition of enzymatic lignocellulose hydrolysis. Appl. Biochem. Biotechnol. 2012, 167, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.E.; Procentese, A.; Montagnaro, F.; Marzocchella, A. Effect of enzymes adsorption on enzymatic hydrolysis of coffee silverskin: Kinetic characterization and validation. Biochem. Eng. J. 2022, 180, 108364. [Google Scholar] [CrossRef]
- Carvalho, M.L.; Sousa, R.; Rodríguez-Zúñiga, U.F.; Suarez, C.A.G.; Rodrigues, D.S.; Giordano, R.C.; Giordano, R.L.C. Kinetic study of the enzymatic hydrolysis of sugarcane bagasse. Braz. J. Chem. Eng. 2013, 30, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, Y.; Song, M.; Topakas, E.; Yu, Q.; Yuan, Z.; Wang, Z.; Guo, Y. Combining Michaelis-Menten theory and enzyme deactivation reactions for the kinetic study of enzymatic hydrolysis by different pre-treated sugarcane bagasse. Process. Biochem. 2021, 105, 72–78. [Google Scholar] [CrossRef]
- Angarita, J.D.; Souza, R.B.A.; Cruz, A.J.G.; Biscaia, E.C.; Secchi, A.R. Kinetic modeling for enzymatic hydrolysis of pre-treated sugarcane straw. Biochem. Eng. J. 2015, 104, 10–19. [Google Scholar] [CrossRef]
- Nnaemeka, I.C.; Egbuna Samuel, O.; Onoh Maxwell, I.; Christain, A.O.; Onyekwulu Chinelo, S. Optimization and kinetic studies for enzymatic hydrolysis and fermentation of colocynthis vulgaris Shrad seeds shell for bioethanol production. J. Bioresour. Bioprod. 2021, 6, 45–64. [Google Scholar] [CrossRef]
- Ude, M.U.; Oluka, I.; Eze, P.C. Optimization and kinetics of glucose production via enzymatic hydrolysis of mixed peels. J. Bioresour. Bioprod. 2020, 5, 283–290. [Google Scholar] [CrossRef]
- Radeva, G.; Valchev, I.; Petrin, S.; Valcheva, E.; Tsekova, P. Kinetic model of enzymatic hydrolysis of steam-exploded wheat straw. Carbohydr. Polym. 2012, 87, 1280–1285. [Google Scholar] [CrossRef]
- Zheng, Y.; Pan, Z.; Zhang, R.; Jenkins, B.M. Kinetic modeling for enzymatic hydrolysis of pre-treated creeping wild ryegrass. Biotechnol. Bioeng. 2009, 102, 1558–1569. [Google Scholar] [CrossRef]
- Makarova, E.I.; Budaeva, V.V.; Kukhlenko, A.A.; Orlov, S.E. Enzyme kinetics of cellulose hydrolysis of Miscanthus and oat hulls. 3 Biotech. 2017, 7, 317. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Chen, X.; Shekiro, J.; Kuhn, E.; Wang, W.; Ji, Y.; Kozliak, E.; Himmel, M.E.; Tucker, M.P. Kinetic Modelling and Experimental Studies for the Effects of Fe2+ Ions on Xylan Hydrolysis with Dilute-Acid Pre-treatment and Subsequent Enzymatic Hydrolysis. Catalysts 2018, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Nikzad, M.; Talebnia, F.; Movagharnejad, K.; Najafpour, G.; Esfahanian, M. Kinetic modeling of enzymatic hydrolysis of pre-treated sorghum bicolor and rice husk. Int. J. Eng. Trans. B Appl. 2017, 30, 1622–1630. [Google Scholar] [CrossRef]
- Datta, R. Acidogenic fermentation of lignocellulose–acid yield and conversion of components. Biotechnol. Bioeng. 1981, 23, 2167–2170. [Google Scholar] [CrossRef]
- Yao, F.; Tian, D.; Shen, F.; Hu, J.; Zeng, Y.; Yang, G.; Zhang, Y.; Deng, S.; Zhang, J. Recycling solvent system in phosphoric acid plus hydrogen peroxide pre-treatment towards a more sustainable lignocellulose biorefinery for bioethanol. Bioresour. Technol. 2019, 275, 19–26. [Google Scholar] [CrossRef]
- Kim, T.H.; Lee, Y.Y. Pretreatment of corn stover by soaking in aqueous ammonia at moderate temperatures. Appl. Biochem. Biotechnol. 2007, 137–140, 81–92. [Google Scholar] [CrossRef]
- Nathan, V.K.; Rani, M.E.; Rathinasamy, G.; Dhiraviam, K.N.; Jayavel, S. Process optimization and production kinetics for cellulase production by Trichoderma viride VKF3. Springerplus 2014, 3, 92. [Google Scholar] [CrossRef] [Green Version]
- Darwesh, O.M.; El-Maraghy, S.H.; Abdel-Rahman, H.M.; Zaghloul, R.A. Improvement of paper wastes conversion to bioethanol using novel cellulose degrading fungal isolate. Fuel 2020, 262, 116518. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Kim, T.H.; Taylor, F.; Hicks, K.B. Bioethanol production from barley hull using SAA (soaking in aqueous ammonia) pretreatment. Bioresour. Technol. 2008, 99, 5694–5702. [Google Scholar] [CrossRef]
- Moreira, B.R.; Breitkreitz, M.C.; Simister, R.; McQueen-Mason, S.J.; Gomez, L.D.; Rezende, C.A. Improved hydrolysis yields and silica recovery by design of experiments applied to acid-alkali pretreatment in rice husks. Ind. Crops Prod. 2021, 170, 113676. [Google Scholar] [CrossRef]
- Le, H.D.M.; Sørensen, R.; Knudsen, N.O.; Meyer, A.S. Implications of silica on biorefi neries – interactions with organic material and mineral elements in grasses. Biofuels Bioprod. Biorefining 2014, 9, 109–121. [Google Scholar] [CrossRef]
- Kaur, B.; Oberoi, H.S.; Chadha, B.S. Enhanced cellulase producing mutants developed from heterokaryotic Aspergillus strain. Bioresour. Technol. 2014, 156, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.B.; de Sousa Francisco Filho, Á.; Rêgo da Silva Rodrigues, J.; Rodrigues de Souza, R. Cellulase production by Aspergillus niger using urban lignocellulosic waste as substrate: Evaluation of different cultivation strategies. J. Environ. Manag. 2022, 305, 114431. [Google Scholar] [CrossRef]
- Kang, S.W.; Park, Y.S.; Lee, J.S.; Hong, S.I.; Kim, S.W. Production of cellulases and hemicellulases by Aspergillus niger KK2 from lignocellulosic biomass. Bioresour. Technol. 2004, 91, 153–156. [Google Scholar] [CrossRef]
- Sulyman, A.O.; Igunnu, A.; Malomo, S.O. Isolation, purification and characterization of cellulase produced by Aspergillus niger cultured on Arachis hypogaea shells. Heliyon 2020, 6, e05668. [Google Scholar] [CrossRef]
- Cunha, F.M.; Esperança, M.N.; Zangirolami, T.C.; Badino, A.C.; Farinas, C.S. Sequential solid-state and submerged cultivation of Aspergillus niger on sugarcane bagasse for the production of cellulase. Bioresour. Technol. 2012, 112, 270–274. [Google Scholar] [CrossRef] [Green Version]
- Wojtusik, M.; Villar, J.C.; Ladero, M.; Garcia-Ochoa, F. Physico-chemical kinetic modelling of hydrolysis of a steam-explosion pre-treated corn stover: A two-step approach. Bioresour. Technol. 2018, 268, 592–598. [Google Scholar] [CrossRef]
- Sakimoto, K.; Kanna, M.; Matsumura, Y. Kinetic model of cellulose degradation using simultaneous saccharification and fermentation. Biomass Bioenergy 2017, 99, 116–121. [Google Scholar] [CrossRef]
- Ryu, D.D.Y.; Lee, S.B. Enzymatic hydrolysis of cellulose: Determination of kinetic parameters. Chem. Eng. Commun. 1986, 45, 119–134. [Google Scholar] [CrossRef]
- Haldar, D.; Gayen, K.; Sen, D. Enumeration of monosugars’ inhibition characteristics on the kinetics of enzymatic hydrolysis of cellulose. Process. Biochem. 2018, 72, 130–136. [Google Scholar] [CrossRef]
- Paul, S.K.; Chakraborty, S. Mixing effects on the kinetics of enzymatic hydrolysis of lignocellulosic Sunn hemp fibres for bioethanol production. Chem. Eng. J. 2019, 377, 120103. [Google Scholar] [CrossRef]
- Jin, X.; Song, J.; Liu, G.Q. Bioethanol production from rice straw through an enzymatic route mediated by enzymes developed in-house from Aspergillus fumigatus. Energy 2020, 190, 116395. [Google Scholar] [CrossRef]
- Subsamran, K.; Mahakhan, P.; Vichitphan, K.; Vichitphan, S.; Sawaengkaew, J. Potential use of vetiver grass for cellulolytic enzyme production and bioethanol production. Biocatal. Agric. Biotechnol. 2019, 17, 261–268. [Google Scholar] [CrossRef]
- Chauhan, M.; Dutt, S.; Manjul, A.S.; Singh, B.; Garlapati, V.K. A sustainable approach of turning potato waste towards bioethanol production using indigenous microbes of Himachal Pradesh, India. Chemosphere 2022, 299, 134429. [Google Scholar] [CrossRef]
- Hosny, M.; El-Sheshtawy, H.S. Effect of biosurfactant on hydrolysis of municipal waste by cellulases producing bacteria for bioethanol production. Curr. Res. Green Sustain. Chem. 2022, 5, 100294. [Google Scholar] [CrossRef]
- Mihajlovski, K.; Pecarski, D.; Rajilić-Stojanović, M.; Dimitrijević-Branković, S. Valorization of corn stover and molasses for enzyme synthesis, lignocellulosic hydrolysis and bioethanol production by Hymenobacter sp. CKS3. Environ. Technol. Innov. 2021, 23, 101627. [Google Scholar] [CrossRef]
- Danso, B.; Ali, S.S.; Xie, R.; Sun, J. Valorisation of wheat straw and bioethanol production by a novel xylanase- and cellulase-producing Streptomyces strain isolated from the wood-feeding termite, Microcerotermes species. Fuel 2022, 310, 122333. [Google Scholar] [CrossRef]

| Components | Untreated Rice Hull (%) | H2O2 Pretreated Rice Hull (%) | Aqueous Ammonia Pretreated Rice Hull (%) |
|---|---|---|---|
| Cellulose | 29.26 | 38.77 | 43.80 |
| Hemicellulose | 20.79 | 17.71 | 15.64 |
| Lignin | 36.24 | 27.22 | 20.47 |
| Hot Water Solubility (HWS) | 4.49 | 4.08 | 9.03 |
| Ash | 9.22 | 12.22 | 11.06 |
| Element | Mass% (Untreated) | Mass% (H2O2 Pre-Treated Rice Husk) | Mass% (Aqueous Ammonia Pre-Treated Rice Husk) |
|---|---|---|---|
| C | 23.52 | 27.05 | 34.93 |
| O | 54.09 | 48.91 | 48.47 |
| Si | 22.40 | 24.05 | 16.60 |
| Microorganism | Substrate | FPase (Exoglucanase) | CMCase (Endoglucanase) | β-Glucosidase | Reference |
|---|---|---|---|---|---|
| Aspergillus niger KK2 | Rice Straw and Wheat Bran | 19.5 IU g−1 | 129 IU g−1 | 100 IU g−1 | [49] |
| Aspergillus niger NS-2 | Agricultural and Kitchen Waste Residues | 17 U g−1 | 310 U g−1 | 33 U g−1 | [50] |
| Aspergillus niger | Sugarcane Bagasse | - | 57 IU L−1 h−1 | - | [51] |
| Aspergillus nidulans and Aspergillus tubingensis | Sorghum Straw | 10.9 U mL−1 | 243.2 U mL−1 | 1114.3 U mL−1 | [47] |
| Aspergillus niger | Dry Leaves | 230.68 U L−1 | 413.49 U L−1 | - | [48] |
| Aspergillus niger | Rice Husk | 548.940 U mL−1 | 314.892 U mL−1 | - | This study |
| Ref. | Enzyme Source | Substrate | pH | T (°C) | KM | k2 | VM |
|---|---|---|---|---|---|---|---|
| This work | Aspergillus niger | Rice Husk | 5 | 50 °C | 1 × 10−3 Mol L−1 | 1.1 × 10−3 s−1 | 1.3 × 10−7 Mol L−1 s−1 |
| [53] | Trichoderma reesei | Cellulose | - | 40 °C | 4.70 × 10−3 Mol L−1 | 3.90 × 10−3 s−1 | 4.83 × 10−5 Mol L−1 s−1 |
| [32] | Aspergillus niger | Mixed Peels | 4.55 | 36 °C | 60.5 g dm3 | - | 28.2 g dm−3 d−1 |
| [54] | Trichoderma reesei | Sweco 270 | 4.80 | 50 °C | 36.2 mg mL−1 | - | 22.4 mg mL−1 hr−1 |
| [25] | Trichoderma reesei | Avicel | 4.80 | 50 °C | 3.8 mM | 5.6 × 10−4 s−1 | - |
| [28] | Trichoderma reesei | Sugarcane Bagasse | 4.80 | 50 °C | 8.78 g L−1 | - | 0.105 g L−1 min−1 |
| [55] | Trichoderma reesei | Cellulose | 4.80 | 50 °C | 45.66 g L−1 | - | 0.26 g L−1 h−1 |
| Substrate | Pre-Treatment Type | Microorganism | Total Reducing SUGARS (g L−1) | Ref. |
|---|---|---|---|---|
| Rice Straw | NaOH | Aspergillus fumigatus | 27.89 | [57] |
| Vetiver Grass | NaOH-H2SO4 | Aspergillus tubingensis HS1–5 | 21.10 | [58] |
| Potato Peel Waste | Untreated | B. amyloliquefaciens | 12.06 | [59] |
| Municipal Waste | Untreated | Bacillus subtilis | 9.08 | [60] |
| Rice Husk | 3% H2O2 solution (1:10) at 85 °C for 48 h; 20% aqueous ammonia at 100 °C for 5 h. | Aspergillus niger | 6.58 | [15] |
| Corn Stover | Untreated | Hymenobacter sp. CKS3 | 3.85 | [61] |
| Wheat Straw | Untreated | Streptomyces sp. MS-S2 | 3.6 | [62] |
| Rice Husk | 3% H2O2 solution (1:10) at 85 °C for 6 h; 20% aqueous ammonia at 100 °C for 5 h. | Aspergillus niger | 1.80 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efrinalia, W.; Novia, N.; Melwita, E. Kinetic Model for Enzymatic Hydrolysis of Cellulose from Pre-Treated Rice Husks. Fermentation 2022, 8, 417. https://doi.org/10.3390/fermentation8090417
Efrinalia W, Novia N, Melwita E. Kinetic Model for Enzymatic Hydrolysis of Cellulose from Pre-Treated Rice Husks. Fermentation. 2022; 8(9):417. https://doi.org/10.3390/fermentation8090417
Chicago/Turabian StyleEfrinalia, Winta, Novia Novia, and Elda Melwita. 2022. "Kinetic Model for Enzymatic Hydrolysis of Cellulose from Pre-Treated Rice Husks" Fermentation 8, no. 9: 417. https://doi.org/10.3390/fermentation8090417
APA StyleEfrinalia, W., Novia, N., & Melwita, E. (2022). Kinetic Model for Enzymatic Hydrolysis of Cellulose from Pre-Treated Rice Husks. Fermentation, 8(9), 417. https://doi.org/10.3390/fermentation8090417







