Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600
Abstract
1. Introduction
2. Materials and Methods
2.1. Media Composition and Culture Conditions
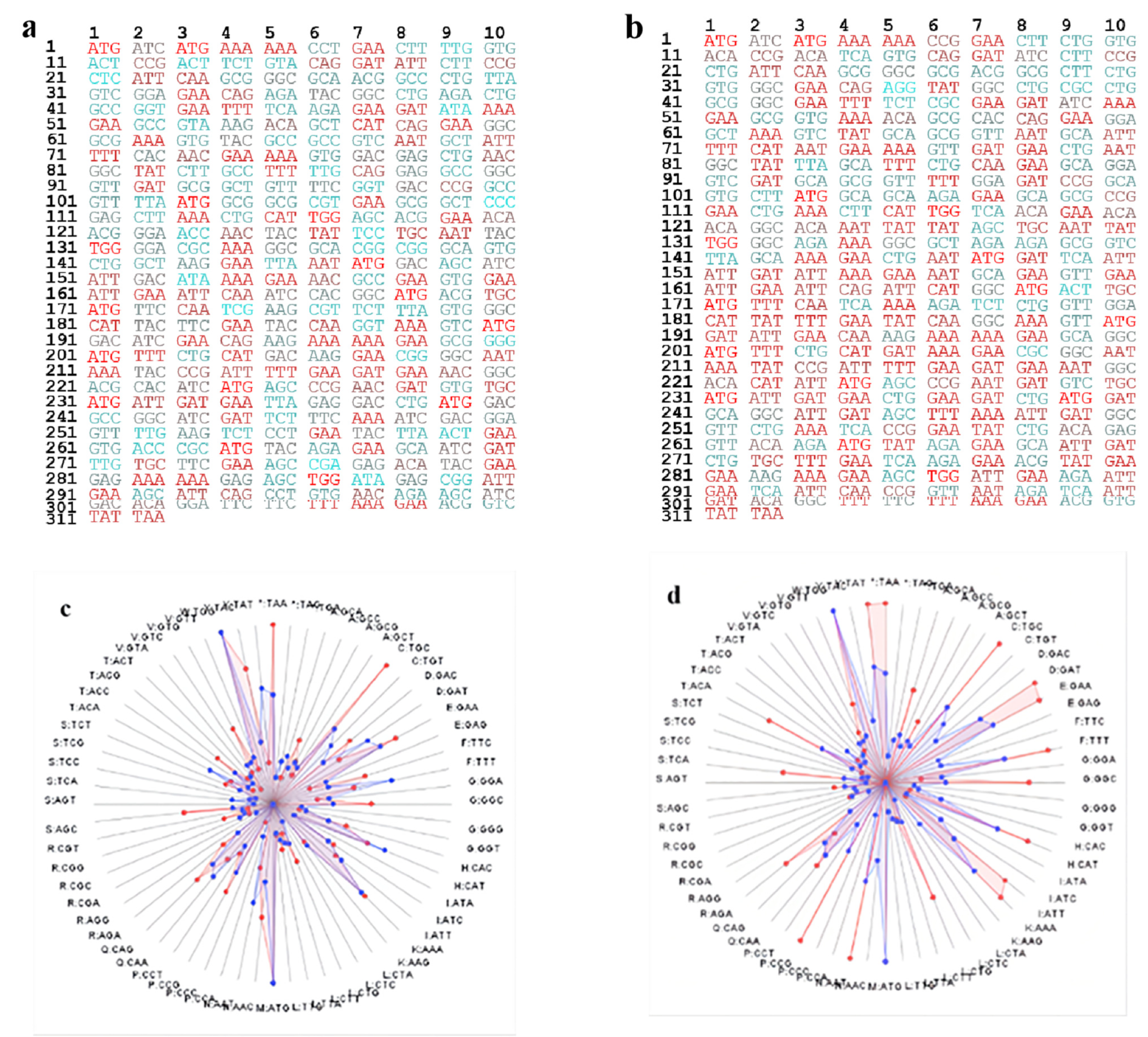
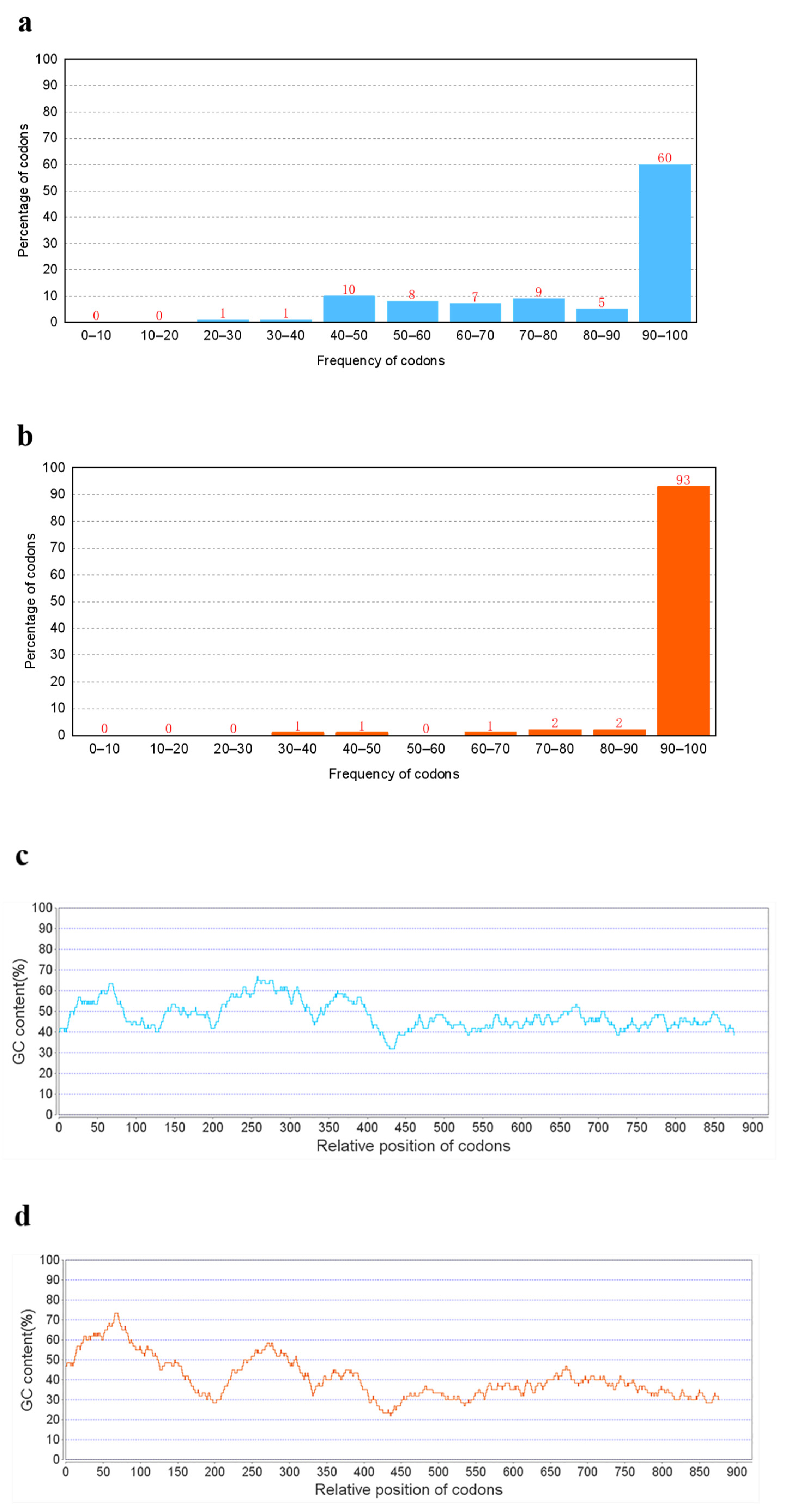
2.2. Gene Optimization Methodology
2.3. Plasmid Construction and Transformation for Collagenase Expression
2.4. Recombinant Expression and Purification of Collagenase
2.5. Sodium Dodecyl Sulphate–Polyacrylamide Gel Electrophoresis
2.6. Protein Quantification and Enzyme Assay of Collagenase
2.7. Effects of pH, Temperature, and Metal Ions on Collagenase Activity and Stability
2.8. Optimization of the Fermentation Conditions for the Recombinant Expression of Collagenase
3. Results and Discussion
3.1. Gene Optimization
3.2. Expression of Recombinant Collagenase
3.3. Purification of the Recombinantly Expressed Collagenase
3.4. Effect of pH, Temperature, and Metal Ions on the Stability of Collagenase
3.5. Optimization of Carbon Sources, Nitrogen Sources, Metal Ions for the Production of Collagenase by Recombinant B. subtilis WB600

3.6. Factors Affecting the Recombinant Production of Collagenase by B. subtilis WB600
3.6.1. Effect of Initial pH
3.6.2. Effect of Agitation Speed
3.6.3. Effect of Temperature on the Cell Growth and Expression of Collagenase
3.6.4. Effect of the Percentage of Inoculation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Taga, Y.; Tanaka, K.; Hattori, S.; Mizuno, K. In-depth correlation analysis demonstrates that 4-hydroxyproline at the Yaa position of Gly-Xaa-Yaa repeats dominantly stabilizes collagen triple helix. Matrix Biol. Plus. 2021, 10, 100067. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Fragai, M.; Luchinat, C.; Melikian, M.; Toccafondi, M.; Lauer, J.L.; Fields, G.B. Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis. J. Am. Chem. Soc. 2012, 134, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Okitsu, T.; Teramura, N.; Iijima, K.; Hayashida, O.; Teramae, H.; Hattori, S. Recombinant collagenase from Grimontia hollisae as a tissue dissociation enzyme for isolating primary cells. Sci. Rep. 2020, 10, 3927. [Google Scholar] [CrossRef]
- Varghese, A.; Chaturvedi, S.S.; Fields, G.B.; Karabencheva-Christova, T.G. A synergy between the catalytic and structural Zn(II) ions and the enzyme and substrate dynamics underlies the structure-function relationships of matrix metalloproteinase collagenolysis. J. Biol. Inorg. Chem. 2021, 26, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Laronha, H.; Caldeira, J. Structure and Function of Human Matrix Metalloproteinases. Cells 2020, 9, 1076. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Fu, Y.; Huang, S.; Liao, L.; Wu, Q.; Wang, Y.; Ge, F.; Fang, B. Identification and antioxidant activity of bovine bone collagen-derived novel peptides prepared by recombinant collagenase from Bacillus cereus. Food Chem. 2021, 349, 129143. [Google Scholar] [CrossRef]
- Cheng, X.; Abfalter, C.M.; Schönauer, E.; Ponnuraj, K.; Huemer, M.; Gadermaier, G.; Regl, C.; Briza, P.; Ferreira, F.; Huber, C.G.; et al. Cloning, Purification and Characterization of the Collagenase ColA Expressed by Bacillus cereus ATCC 14579. PLoS ONE 2016, 11, e0162433. [Google Scholar] [CrossRef]
- Ducka, P.; Eckhard, U.; Schonauer, E.; Kofler, S.; Gottschalk, G.; Brandstetter, H.; Nuss, D. A universal strategy for high-yield production of soluble and functional clostridial collagenases in E. coli . Appl. Microbiol. Biotechnol. 2009, 83, 1055–1065. [Google Scholar] [CrossRef][Green Version]
- Rochima, E.; Sekar, N.; Buwono, I.D.; Afrianto, E.; Pratama, R.I. Isolation and Characterization of Collagenase from Bacillus Subtilis (Ehrenberg, 1835); ATCC 6633 for Degrading Fish Skin Collagen Waste from Cirata Reservoir, Indonesia. Aquat. Procedia 2016, 7, 76–84. [Google Scholar] [CrossRef]
- Abood, A.; Salman, A.M.M.; Abdelfattah, A.M.; El-Hakim, A.E.; Abdel-Aty, A.M.; Hashem, A.M. Purification and characterization of a new thermophilic collagenase from Nocardiopsis dassonvillei NRC2aza and its application in wound healing. Int. J. Biol. Macromol. 2018, 116, 801–810. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, G.; Sehgal, A.; Bhardwaj, S.; Singh, S.; Buhas, C.; Judea-Pusta, C.; Uivarosan, D.; Munteanu, M.A.; Bungau, S. Multifaceted Role of Matrix Metalloproteinases in Neurodegenerative Diseases: Pathophysiological and Therapeutic Perspectives. Int. J. Mol. Sci. 2021, 22, 1413. [Google Scholar] [CrossRef] [PubMed]
- Folgueras, A.R.; Pendas, A.M.; Sanchez, L.M.; Lopez-Otin, C. Matrix metalloproteinases in cancer: From new functions to improved inhibition strategies. Int. J. Dev. Biol. 2004, 48, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Lauer-Fields, J.L.; Juska, D.; Fields, G.B. Matrix metalloproteinases and collagen catabolism. Biopolymers 2002, 66, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Spinale, F.G. Matrix metalloproteinases: Regulation and dysregulation in the failing heart. Circ. Res. 2002, 90, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, Z.; Zeng, Y.; Dong, Y.; Ma, L. Co-encapsulation of collagenase type I and silibinin in chondroitin sulfate coated multilayered nanoparticles for targeted treatment of liver fibrosis. Carbohydr. Polym. 2021, 263, 117964. [Google Scholar] [CrossRef]
- Yang, H.; Qu, J.; Zou, W.; Shen, W.; Chen, X. An overview and future prospects of recombinant protein production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2021, 105, 6607–6626. [Google Scholar] [CrossRef]
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 2018, 34, 145. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, X.; Wu, Y.; Niu, T.; Liu, Y.; Li, J.; Du, G.; Liu, L. Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab. Eng. 2018, 50, 109–121. [Google Scholar] [CrossRef]
- Wu, X.C.; Lee, W.; Tran, L.; Wong, S.L. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 1991, 173, 4952–4958. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.S.; Correia, A.; Esteves, A.C. Bacterial collagenases—A review. Crit. Rev. Microbiol. 2016, 42, 106–126. [Google Scholar] [CrossRef]
- Cao, S.; Li, D.; Ma, X.; Xin, Q.; Song, J.; Lu, F.; Li, Y. A novel unhairing enzyme produced by heterologous expression of keratinase gene (kerT) in Bacillus subtilis. World J. Microbiol. Biotechnol. 2019, 35, 122. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, O.; Yoshihara, K.; Katayama, S.; Minami, J.; Okabe, A. Purification and characterization of Clostridium perfringens 120-kilodalton collagenase and nucleotide sequence of the corresponding gene. J. Bacteriol. 1994, 176, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Bhattacharya, S.; Gachhui, R.; Hazra, S.; Mukherjee, J. U32 collagenase from Pseudoalteromonas agarivorans NW4327: Activity, structure, substrate interactions and molecular dynamics simulations. Int. J. Biol. Macromol. 2019, 124, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Alipour, H.; Raz, A.; Dinparast Djadid, N.; Zakeri, S. Expression of a New Recombinant Collagenase Protein of Lucilia Sericata in SF9 Insect Cell as a Potential Method for Wound Healing. Iran. J. Biotechnol. 2019, 17, e2429. [Google Scholar] [CrossRef]
- Jung, C.M.; Matsushita, O.; Katayama, S.; Minami, J.; Ohhira, I.; Okabe, A. Expression of the colH gene encoding Clostridium histolyticum collagenase in Bacillus subtilis and its application to enzyme purification. Microbiol. Immunol. 1996, 40, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Teramura, N.; Tanaka, K.; Iijima, K.; Hayashida, O.; Suzuki, K.; Hattori, S.; Irie, S. Cloning of a novel collagenase gene from the gram-negative bacterium Grimontia (Vibrio) hollisae 1706B and its efficient expression in Brevibacillus choshinensis. J. Bacteriol. 2011, 193, 3049–3056. [Google Scholar] [CrossRef]
- Li, J.; Cheng, J.H.; Teng, Z.J.; Sun, Z.Z.; He, X.Y.; Wang, P.; Shi, M.; Song, X.Y.; Chen, X.L.; Zhang, Y.Z.; et al. Taxonomic and Enzymatic Characterization of Flocculibacter collagenilyticus gen. nov., sp. nov., a Novel Gammaproteobacterium with High Collagenase Production. Front. Microbiol. 2021, 12, 621161. [Google Scholar] [CrossRef]
- Sakurai, Y.; Inoue, H.; Nishii, W.; Takahashi, T.; Iino, Y.; Yamamoto, M.; Takahashi, K. Purification and characterization of a major collagenase from Streptomyces parvulus. Biosci. Biotechnol. Biochem. 2009, 73, 21–28. [Google Scholar] [CrossRef]
- Pequeno, A.C.L.; Arruda, A.A.; Silva, D.F.; Duarte Neto, J.M.W.; Silveira Filho, V.M.; Converti, A.; Marques, D.A.V.; Porto, A.L.F.; Lima, C.A. Production and characterization of collagenase from a new Amazonian Bacillus cereus strain. Prep. Biochem. Biotechnol. 2019, 49, 501–509. [Google Scholar] [CrossRef]
- Wu, Q.; Li, C.; Li, C.; Chen, H.; Shuliang, L. Purification and characterization of a novel collagenase from Bacillus pumilus Col-J. Appl. Biochem. Biotechnol. 2010, 160, 129–139. [Google Scholar] [CrossRef]
- Klausmann, P.; Hennemann, K.; Hoffmann, M.; Treinen, C.; Aschern, M.; Lilge, L.; Morabbi Heravi, K.; Henkel, M.; Hausmann, R. Bacillus subtilis High Cell Density Fermentation Using a Sporulation-Deficient Strain for the Production of Surfactin. Appl. Microbiol. Biotechnol. 2021, 105, 4141–4151. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Kang, Z.; Zheng, E.; Liu, Y.; Gauld, J.W.; Wang, Q. Hydrolysis Mechanism of the Linkers by Matrix Metalloproteinase-9 Using QM/MM Calculations. J. Chem. Inf. Model. 2021, 61, 5203–5211. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Nakagawa, E.; Yoda, M.; Nakaichi, A.; Hibi, T.; Kimoto, H. Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7. Sci. Rep. 2019, 9, 14870. [Google Scholar] [CrossRef] [PubMed]
- Singh, W.; Fields, G.B.; Christov, C.Z.; Karabencheva-Christova, T.G. Effects of Mutations on Structure-Function Relationships of Matrix Metalloproteinase-1. Int. J. Mol. Sci. 2016, 17, 1727. [Google Scholar] [CrossRef]
- Sarchami, T.; Johnson, E.; Rehmann, L. Optimization of fermentation condition favoring butanol production from glycerol by Clostridium pasteurianum DSM 525. Bioresour. Technol. 2016, 208, 73–80. [Google Scholar] [CrossRef]
- Lima, C.A.; Marques, D.A.; Barros Neto, B.; Lima Filho, J.L.; Carneiro-da-Cunha, M.G.; Porto, A.L. Fermentation medium for collagenase production by Penicillium aurantiogriseum URM4622. Biotechnol Prog. 2011, 27, 1470–1477. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Zhao, X.; Zhang, Y.; Wang, X.; Nie, Y.; Xu, Y. Enhancement of the production of Bacillus naganoensis pullulanase in recombinant Bacillus subtilis by integrative expression. Protein Expr. Purif. 2019, 159, 42–48. [Google Scholar] [CrossRef]
- Yang, X.; Xiao, X.; Liu, D.; Wu, R.; Wu, C.; Zhang, J.; Huang, J.; Liao, B.; He, H. Optimization of Collagenase Production by Pseudoalteromonas sp. SJN2 and Application of Collagenases in the Preparation of Antioxidative Hydrolysates. Mar. Drugs 2017, 15, 377. [Google Scholar] [CrossRef]
- Amorim, C.; Silverio, S.C.; Goncalves, R.F.S.; Pinheiro, A.C.; Silva, S.; Coelho, E.; Coimbra, M.A.; Prather, K.L.J.; Rodrigues, L.R. Downscale fermentation for xylooligosaccharides production by recombinant Bacillus subtilis 3610. Carbohydr. Polym. 2019, 205, 176–183. [Google Scholar] [CrossRef]





| Ammonium Sulfate Concentration (%) | Specific Activity (U/mg) |
|---|---|
| 20 | 4498.19 |
| 30 | 2925.32 |
| 40 | 2165.25 |
| 50 | 2177.78 |
| 60 | 4921.74 |
| 70 | 6156.67 |
| 80 | 5509.75 |
| 90 | 4000.21 |
| Steps | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Purification (Fold) | Yield (%) |
|---|---|---|---|---|---|
| Cultivate supernatant | 114,516.78 | 57.29 | 1998.73 | 1 | 100 |
| Ammonium sulfate precipitation | 58,034.55 | 9.42 | 6156.67 | 3.08 | 50.67 |
| Ultrafiltration | 8329.20 | 0.94 | 8860.85 | 4.43 | 7.27 |
| Nickel column purification | 2445.44 | 0.26 | 9405.54 | 4.71 | 2.14 |
| Parameters | Enzyme Activity (U/mL) |
|---|---|
| Temperature (°C) | |
| 30 | 1449.39 ± 50 |
| 40 | 1600.96 ± 74 |
| 50 | 2055.65 ± 61 |
| 60 | 1202.49 ± 68 |
| 70 | 1035.44 ± 71 |
| 80 | 716.02 ± 53 |
| pH | |
| 5 | 1644.96 ± 56 |
| 6 | 850.47 ± 82 |
| 7 | 877.36 ± 53 |
| 8 | 1419.24 ± 76 |
| 9 | 2005.94 ± 69 |
| 10 | 1173.15 ± 64 |
| 11 | 680.16 ± 78 |
| Metal ions | |
| Control | 1599.33 ± 25 |
| Fe3+ | 1235.90 ± 34 |
| Zn2+ | 1816.08 ± 51 |
| Co2+ | 2343.30 ± 39 |
| Ca2+ | 2542.94 ± 43 |
| Mg2+ | 1908.97 ± 37 |
| Cu2+ | 174.95 ± 45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wang, L.; Zheng, K.; Liu, P.; Li, W.; Lin, J.; Liu, W.; Shan, S.; Sun, L.; Zhang, H. Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600. Fermentation 2022, 8, 449. https://doi.org/10.3390/fermentation8090449
Zhu Y, Wang L, Zheng K, Liu P, Li W, Lin J, Liu W, Shan S, Sun L, Zhang H. Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600. Fermentation. 2022; 8(9):449. https://doi.org/10.3390/fermentation8090449
Chicago/Turabian StyleZhu, Yaqing, Linlin Wang, Kaixuan Zheng, Ping Liu, Wenkang Li, Jian Lin, Wenjing Liu, Shoushui Shan, Liqin Sun, and Hailing Zhang. 2022. "Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600" Fermentation 8, no. 9: 449. https://doi.org/10.3390/fermentation8090449
APA StyleZhu, Y., Wang, L., Zheng, K., Liu, P., Li, W., Lin, J., Liu, W., Shan, S., Sun, L., & Zhang, H. (2022). Optimized Recombinant Expression and Characterization of Collagenase in Bacillus subtilis WB600. Fermentation, 8(9), 449. https://doi.org/10.3390/fermentation8090449





