Comparative Fatty Acid Compositional Profiles of Rhodotorula toruloides Haploid and Diploid Strains under Various Storage Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media and Culture Conditions
2.2. Cell Mass and Total Lipids
2.3. Fatty Acid Composition Analysis
2.4. Statistical Analyses
3. Results
3.1. Difference in Growth Patterns of Haploid and Diploid Strains
3.2. Lipid Accumulation Potential of Haploid and Diploid Strains
3.3. Analysis of Fatty Acid Compositional Difference of Haploid and Diploid Strains
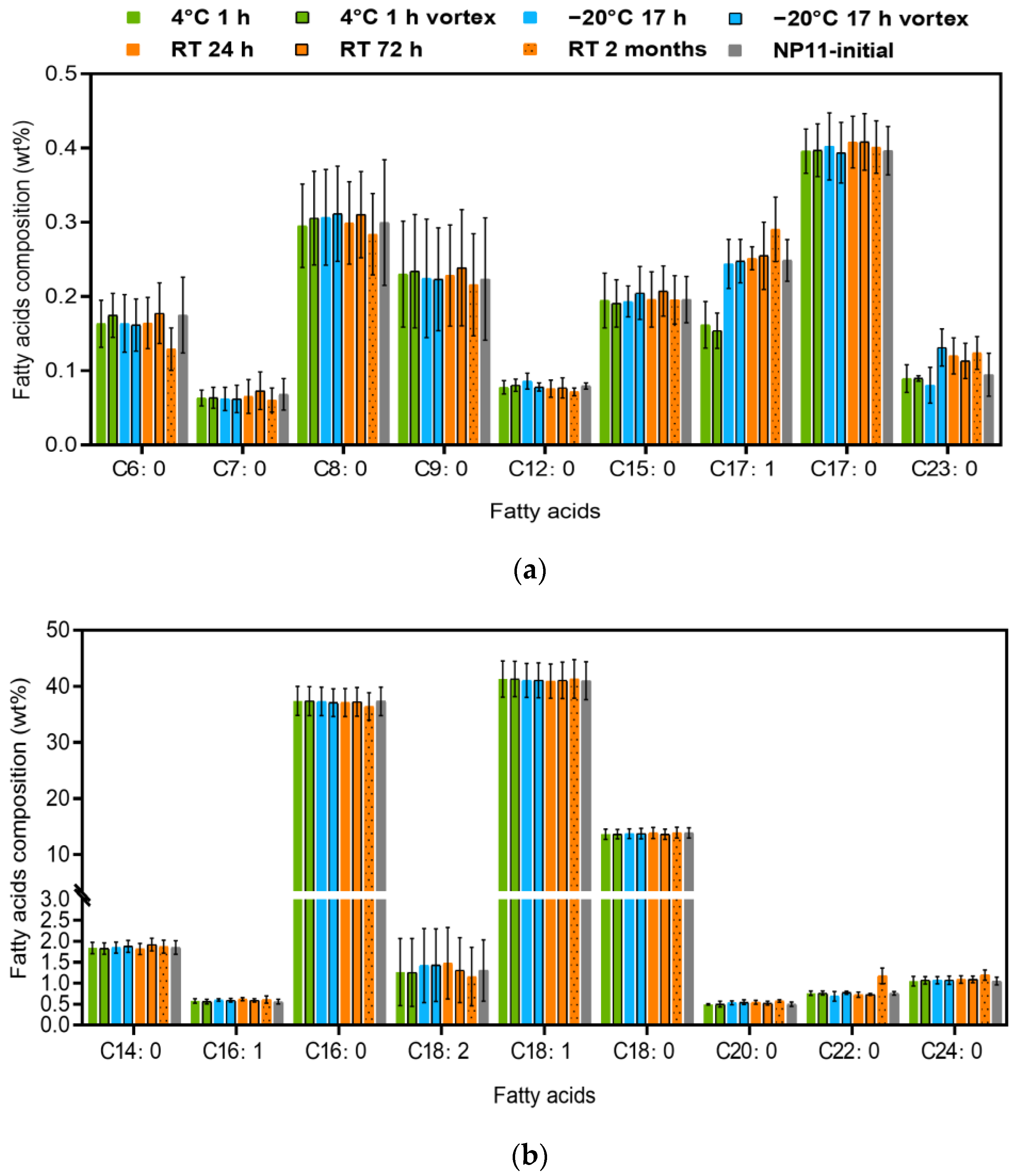
3.4. Stability of FAMEs under Different Preservation Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Nomenclature
| FAs | Fatty acids |
| SCFAs | Short-chain fatty acids |
| MCFAs | Medium-chain fatty acids |
| LCFAs | Long-chain fatty acids |
| VLCFAs | Very long-chain fatty acids |
| OCFAs | Odd-chain fatty acids |
| FAMEs | Fatty acid methyl esters |
References
- Chu, S.; Majumdar, A. Opportunities and challenges for a sustainable energy future. Nature 2012, 488, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Singer, S.D.; Souto, B.A.; Asomaning, J.; Ullah, A.; Bressler, D.C.; Chen, G.Q. Current progress in lipid-based biofuels: Feedstocks and production technologies. Bioresour. Technol. 2022, 351, 127020. [Google Scholar] [CrossRef] [PubMed]
- Carriquiry, M.A.; Du, X.; Timilsina, G.R. Second generation biofuels: Economics and policies. Energ. Policy 2011, 39, 4222–4234. [Google Scholar] [CrossRef]
- Madani, M.; Enshaeieh, M.; Abdoli, A. Single cell oil and its application for biodiesel production. Process Saf. Environ. Prot. 2017, 111, 747–756. [Google Scholar] [CrossRef]
- Chintagunta, A.D.; Zuccaro, G.; Kumar, M.; Kumar, S.P.J.; Garlapati, V.K.; Postemsky, P.D.; Kumar, N.S.S.; Chandel, A.K.; Simal-Gandara, J. Biodiesel production from lignocellulosic biomass using oleaginous microbes: Prospects for integrated biofuel production. Front. Microbiol. 2021, 12, 658284. [Google Scholar] [CrossRef]
- Nigam, P.S.; Singh, A. Production of liquid biofuels from renewable resources. Prog. Energ. Combust. 2011, 37, 52–68. [Google Scholar] [CrossRef]
- Szczepanska, P.; Hapeta, P.; Lazar, Z. Advances in production of high-value lipids by oleaginous yeasts. Crit. Rev. Biotechnol. 2022, 42, 1–22. [Google Scholar] [CrossRef]
- Wen, Z.; Zhang, S.; Odoh, C.K.; Jin, M.; Zhao, Z.K. Rhodosporidium toruloides—A potential red yeast chassis for lipids and beyond. FEMS Yeast Res. 2020, 20, foaa038. [Google Scholar] [CrossRef]
- Uprety, B.K.; Morrison, E.N.; Emery, R.J.N.; Farrow, S.C. Customizing lipids from oleaginous microbes: Leveraging exogenous and endogenous approaches. Trends Biotechnol. 2022, 40, 482–508. [Google Scholar] [CrossRef]
- Li, Y.; Liu, B.; Zhao, Z.K.; Bai, F. Optimization of culture conditions for lipid production by Rhodosporidium toruloides. Chin. J. Biotechnol. 2006, 22, 650–656. [Google Scholar] [CrossRef]
- Lamers, D.; van Biezen, N.; Martens, D.; Peters, L.; van de Zilver, E.; Jacobs-van Dreumel, N.; Wijffels, R.H.; Lokman, C. Selection of oleaginous yeasts for fatty acid production. BMC Biotechnol. 2016, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, S.; Liu, H.; Shen, H.; Lin, X.; Yang, F.; Zhou, Y.J.; Jin, G.; Ye, M.; Zou, H.; et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat. Commun. 2012, 3, 1112. [Google Scholar] [CrossRef] [PubMed]
- Coradetti, S.T.; Pinel, D.; Geiselman, G.M.; Ito, M.; Mondo, S.J.; Reilly, M.C.; Cheng, Y.F.; Bauer, S.; Grigoriev, I.V.; Gladden, J.M.; et al. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. eLife 2018, 7, e32110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Zhu, Z.; Shen, H.; Lin, X.; Jin, X.; Jiao, X.; Zhao, Z.K. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol. Biofuels 2018, 11, 148. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhao, X.; Shen, H.; Wang, Q.; Zhao, Z.K. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour. Technol. 2011, 102, 1803–1807. [Google Scholar] [CrossRef]
- Yang, X.B.; Sun, W.Y.; Shen, H.W.; Zhang, S.F.; Jiao, X.; Zhao, Z.K. Expression of phosphotransacetylase in Rhodosporidium toruloides leading to improved cell growth and lipid production. RSC Adv. 2018, 8, 24673–24678. [Google Scholar] [CrossRef]
- Fillet, S.; Ronchel, C.; Callejo, C.; Fajardo, M.-J.; Moralejo, H.; Adrio, J.L. Engineering Rhodosporidium toruloides for the production of very long-chain monounsaturated fatty acid-rich oils. Appl. Microbiol. Biotechnol. 2017, 101, 7271–7280. [Google Scholar] [CrossRef]
- Kamal, R.; Liu, Y.; Li, Q.; Huang, Q.; Wang, Q.; Yu, X.; Zhao, Z.K. Exogenous L-proline improved Rhodosporidium toruloides lipid production on crude glycerol. Biotechnol. Biofuels 2020, 13, 159. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.K.; Bai, F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzym. Microb. Technol. 2007, 41, 312–317. [Google Scholar] [CrossRef]
- Zhu, Z.; Hu, Y.; Teixeira, P.G.; Pereira, R.; Chen, Y.; Siewers, V.; Nielsen, J. Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nat. Catal. 2020, 3, 64–74. [Google Scholar] [CrossRef]
- Yu, T.; Zhou, Y.J.; Wenning, L.; Liu, Q.; Krivoruchko, A.; Siewers, V.; Nielsen, J.; David, F. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals. Nat. Commun. 2017, 8, 15587. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kamal, R.; Zhang, Y.; Zhou, R.H.; Lv, L.T.; Huang, Q.T.; Qian, S.R.G.L.; Zhang, S.F.; Zhao, Z.K. Expression of VHb improved lipid production in Rhodosporidium toruloides. Energies 2020, 13, 4446. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Liu, H.; Jiao, X.; Zhang, Q.; Zhang, S.; Zhao, Z.K. RNA interference in the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2019, 19, foz031. [Google Scholar] [CrossRef] [PubMed]
- de Godoy, L.M.; Olsen, J.V.; Cox, J.; Nielsen, M.L.; Hubner, N.C.; Frohlich, F.; Walther, T.C.; Mann, M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 2008, 455, 1251–1255. [Google Scholar] [CrossRef]
- Li, B.Z.; Cheng, J.S.; Ding, M.Z.; Yuan, Y.J. Transcriptome analysis of differential responses of diploid and haploid yeast to ethanol stress. J. Biotechnol. 2010, 148, 194–203. [Google Scholar] [CrossRef]
- Bao, R.Q.; Gao, N.; Lv, J.; Ji, C.F.; Liang, H.P.; Li, S.J.; Yu, C.X.; Wang, Z.Y.; Lin, X.P. Enhancement of torularhodin production in Rhodosporidium toruloides by agrobacterium tumefaciens-mediated transformation and culture condition optimization. J. Agric. Food Chem. 2019, 67, 1156–1164. [Google Scholar] [CrossRef]
- Erdbrugger, P.; Frohlich, F. The role of very long chain fatty acids in yeast physiology and human diseases. Biol. Chem. 2020, 402, 25–38. [Google Scholar] [CrossRef]
- Tiukova, I.A.; Prigent, S.; Nielsen, J.; Sandgren, M.; Kerkhoven, E.J. Genome-scale model of Rhodotorula toruloides metabolism. Biotechnol. Bioeng. 2019, 116, 3396–3408. [Google Scholar] [CrossRef]
- Liu, H.; Jiao, X.; Wang, Y.; Yang, X.; Sun, W.; Wang, J.; Zhang, S.; Zhao, Z.K. Fast and efficient genetic transformation of oleaginous yeast Rhodosporidium toruloides by using electroporation. FEMS Yeast Res. 2017, 17, fox017. [Google Scholar] [CrossRef]
- Lin, X.; Wang, Y.; Zhang, S.; Zhu, Z.; Zhou, Y.J.; Yang, F.; Sun, W.; Wang, X.; Zhao, Z.K. Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2014, 14, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Jiao, X.; Zhang, Y.; Liu, X.; Zhang, Q.; Zhang, S.; Zhao, Z.K. Developing a CRISPR/Cas9 system for genome editing in the basidiomycetous yeast Rhodosporidium toruloides. Biotechnol. J. 2019, 14, 1900036. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Ding, Y.; Gong, Z.; Yang, L.; Zhang, S.; Zhang, C.; Lin, X.; Shen, H.; Zou, H.; Xie, Z.; et al. Dynamics of the lipid droplet proteome of the oleaginous yeast Rhodosporidium toruloides. Eukaryot. Cell. 2015, 14, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhou, Y.J.; Krivoruchko, A.; Grininger, M.; Zhao, Z.K.; Nielsen, J. Expanding the product portfolio of fungal type I fatty acid synthases. Nat. Chem. Biol. 2017, 13, 360–362. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
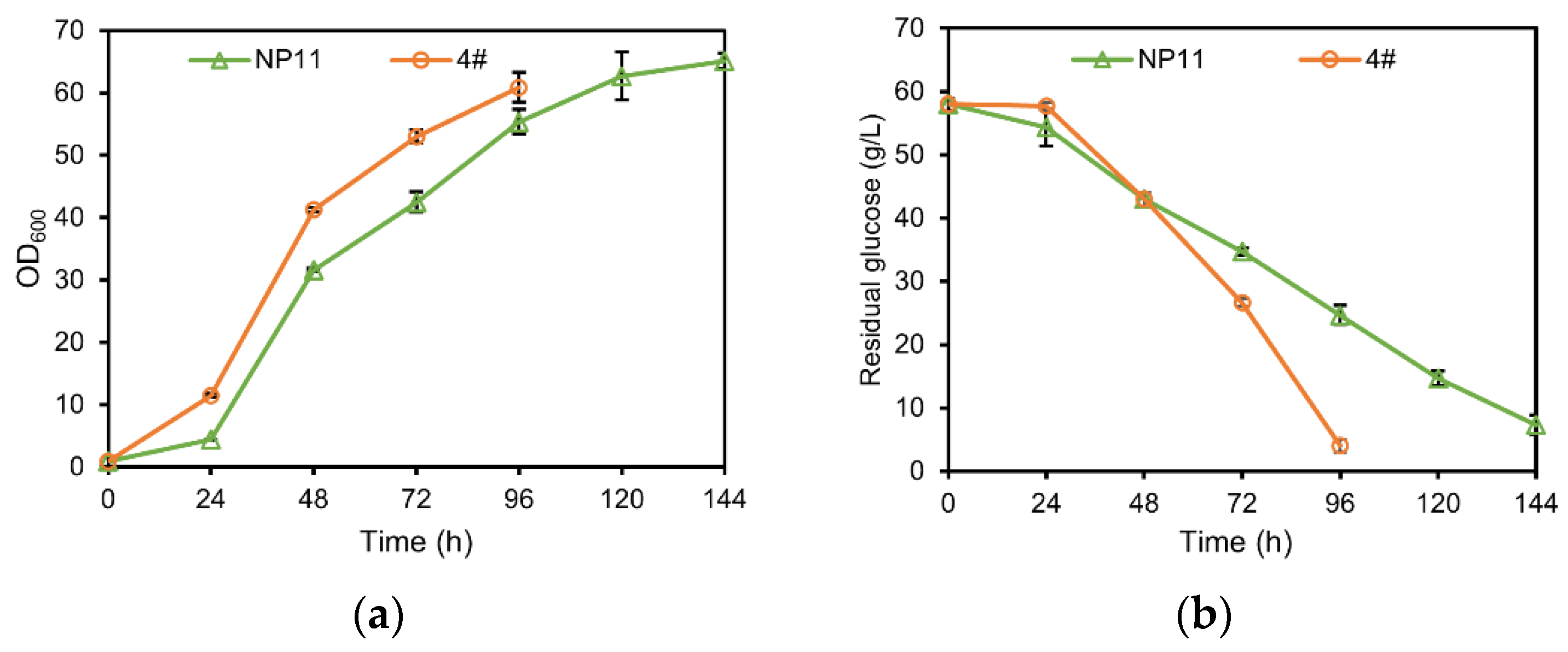
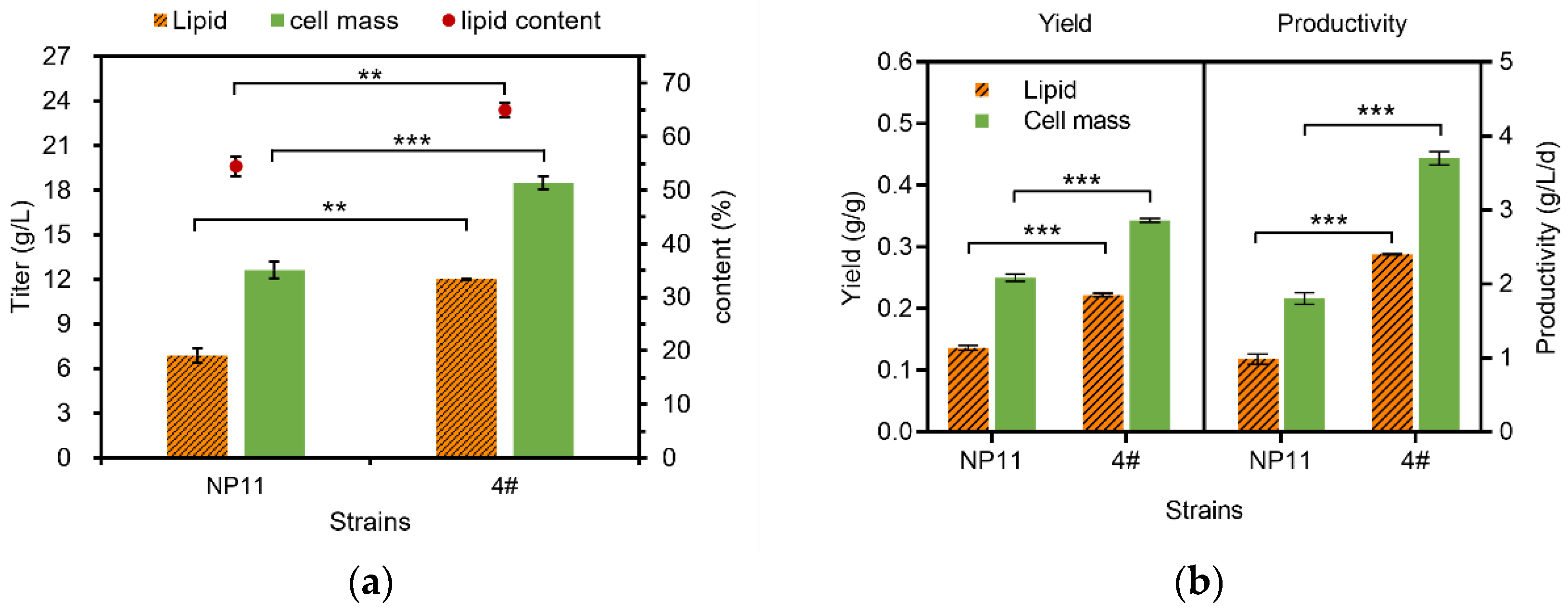
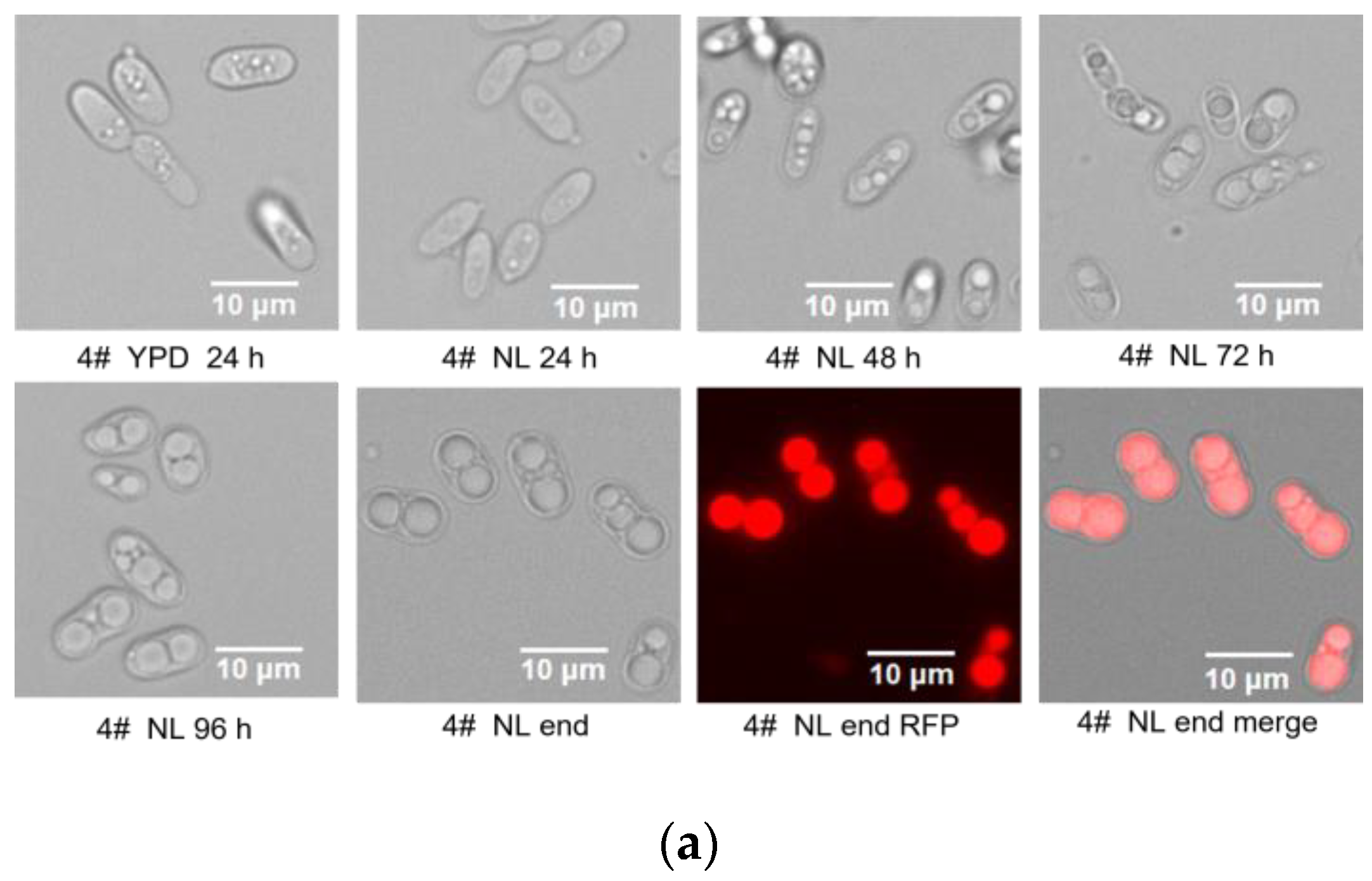
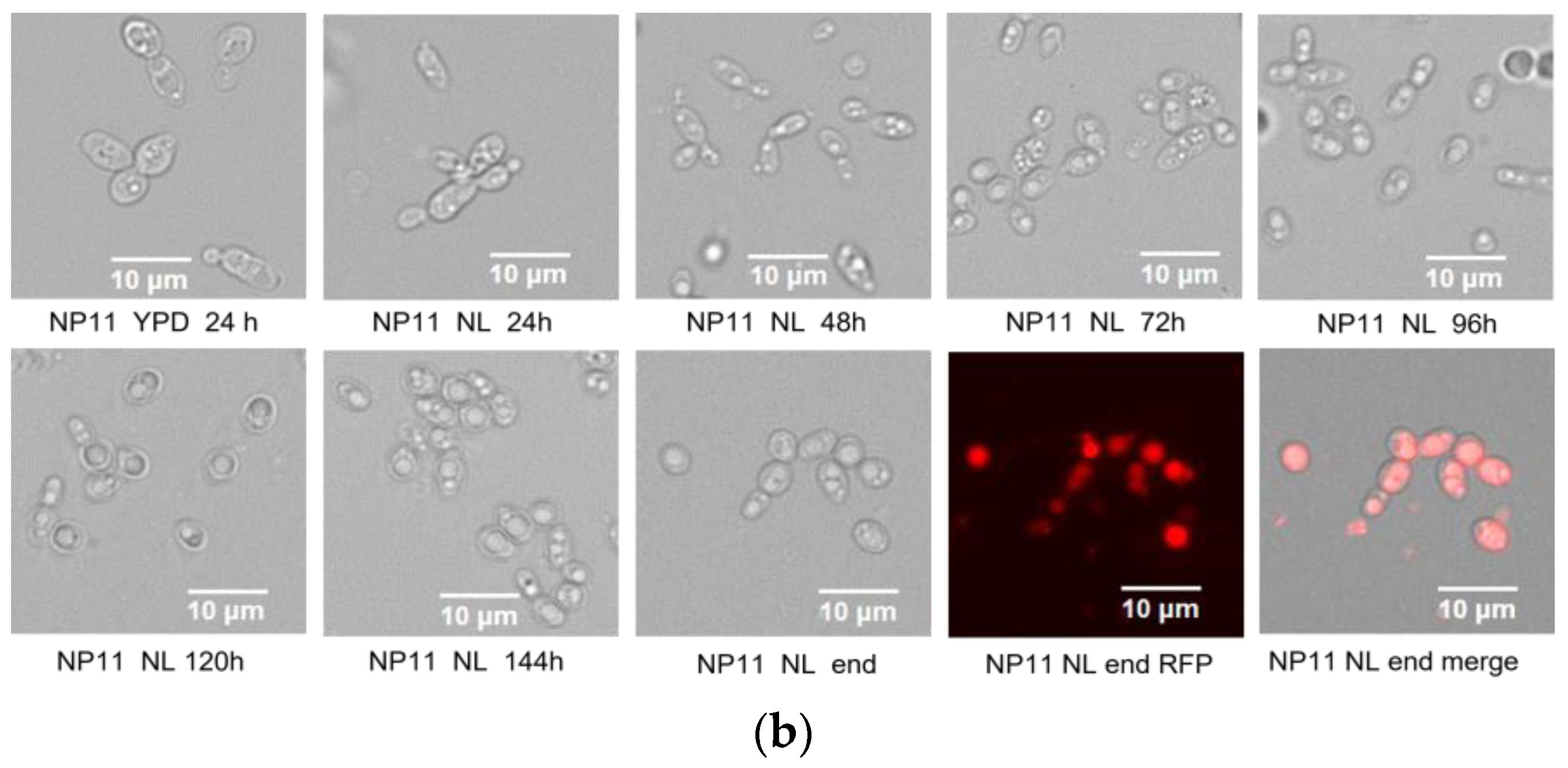
| Fatty Acid Profiles (wt%) | Haploid-NP11 | Diploid-4# | p-Value | FDR |
|---|---|---|---|---|
| C6:0 | 0.18 ± 0.05 | 0.06 ± 0.01 | 0.0609 | 0.1676 |
| C7:0 | 0.07 ± 0.02 | 0.04 ± 0.00 | 0.1276 | 0.2160 |
| C8:0 | 0.30 ± 0.08 | 0.16 ± 0.01 | 0.0963 | 0.1925 |
| Short-chain FAs (SCFAs) | 0.54 ± 0.16 | 0.26 ± 0.02 | 0.0861 | 0.1894 |
| C9:0 | 0.22 ± 0.08 | 0.17 ± 0.02 | 0.3161 | 0.4636 |
| C12:0 | 0.08 ± 0.00 | 0.13 ± 0.01 | 0.0001 *** | 0.0029 * |
| Medium-chain FAs (MCFAs) | 0.30 ± 0.09 | 0.30 ± 0.02 | 0.9984 | 0.9984 |
| C14:0 | 1.85 ± 0.16 | 2.32 ± 0.02 | 0.0359 * | 0.1315 |
| C15:0 | 0.20 ± 0.03 | 0.21 ± 0.01 | 0.5810 | 0.7102 |
| C16:1 | 0.56 ± 0.05 | 0.57 ± 0.01 | 0.8364 | 0.9200 |
| C16:0 | 37.32 ± 2.53 | 38.11 ± 0.42 | 0.6231 | 0.7215 |
| C17:1 | 0.25 ± 0.03 | 0.10 ± 0.01 | 0.0009 *** | 0.0094 * |
| C17:0 | 0.40 ± 0.03 | 0.35 ± 0.00 | 0.1211 | 0.2219 |
| C18:2 | 1.30 ± 0.73 | 0.42 ± 0.12 | 0.1679 | 0.2638 |
| C18:1 | 40.99 ± 3.38 | 41.43 ± 0.62 | 0.8365 | 0.8763 |
| C18:0 | 13.88 ± 0.93 | 14.32 ± 0.26 | 0.4770 | 0.6173 |
| Long-chain FAs (LCFAs) | 96.75 ± 7.87 | 97.81 ± 1.47 | 0.0526 | 0.1653 |
| C20:0 | 0.50 ± 0.05 | 0.53 ± 0.00 | 0.4068 | 0.5594 |
| C22:0 | 0.76 ± 0.04 | 0.54 ± 0.01 | 0.0010 ** | 0.0075 * |
| C23:0 | 0.09 ± 0.03 | 0.03 ± 0.00 | 0.0617 | 0.1509 |
| C24:0 | 1.05 ± 0.09 | 0.53 ± 0.01 | 0.0094 ** | 0.0517 |
| Verylong-chain FAs (VLCFAs) | 2.40 ± 0.21 | 1.63 ± 0.02 | 0.0224 * | 0.0987 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Kamal, R.; Li, Q.; Yu, X.; Wang, Q.; Zhao, Z.K. Comparative Fatty Acid Compositional Profiles of Rhodotorula toruloides Haploid and Diploid Strains under Various Storage Conditions. Fermentation 2022, 8, 467. https://doi.org/10.3390/fermentation8090467
Zhang Y, Kamal R, Li Q, Yu X, Wang Q, Zhao ZK. Comparative Fatty Acid Compositional Profiles of Rhodotorula toruloides Haploid and Diploid Strains under Various Storage Conditions. Fermentation. 2022; 8(9):467. https://doi.org/10.3390/fermentation8090467
Chicago/Turabian StyleZhang, Yue, Rasool Kamal, Qing Li, Xue Yu, Qian Wang, and Zongbao Kent Zhao. 2022. "Comparative Fatty Acid Compositional Profiles of Rhodotorula toruloides Haploid and Diploid Strains under Various Storage Conditions" Fermentation 8, no. 9: 467. https://doi.org/10.3390/fermentation8090467
APA StyleZhang, Y., Kamal, R., Li, Q., Yu, X., Wang, Q., & Zhao, Z. K. (2022). Comparative Fatty Acid Compositional Profiles of Rhodotorula toruloides Haploid and Diploid Strains under Various Storage Conditions. Fermentation, 8(9), 467. https://doi.org/10.3390/fermentation8090467






