Green Manufacturing of Steroids via Mycolicbacteria: Current Status and Development Trends
Abstract
:1. Introduction
2. Yesterday: Marker Degradation Technology
3. Today: Green Manufacturing of Steroids with Mycolicibacteria
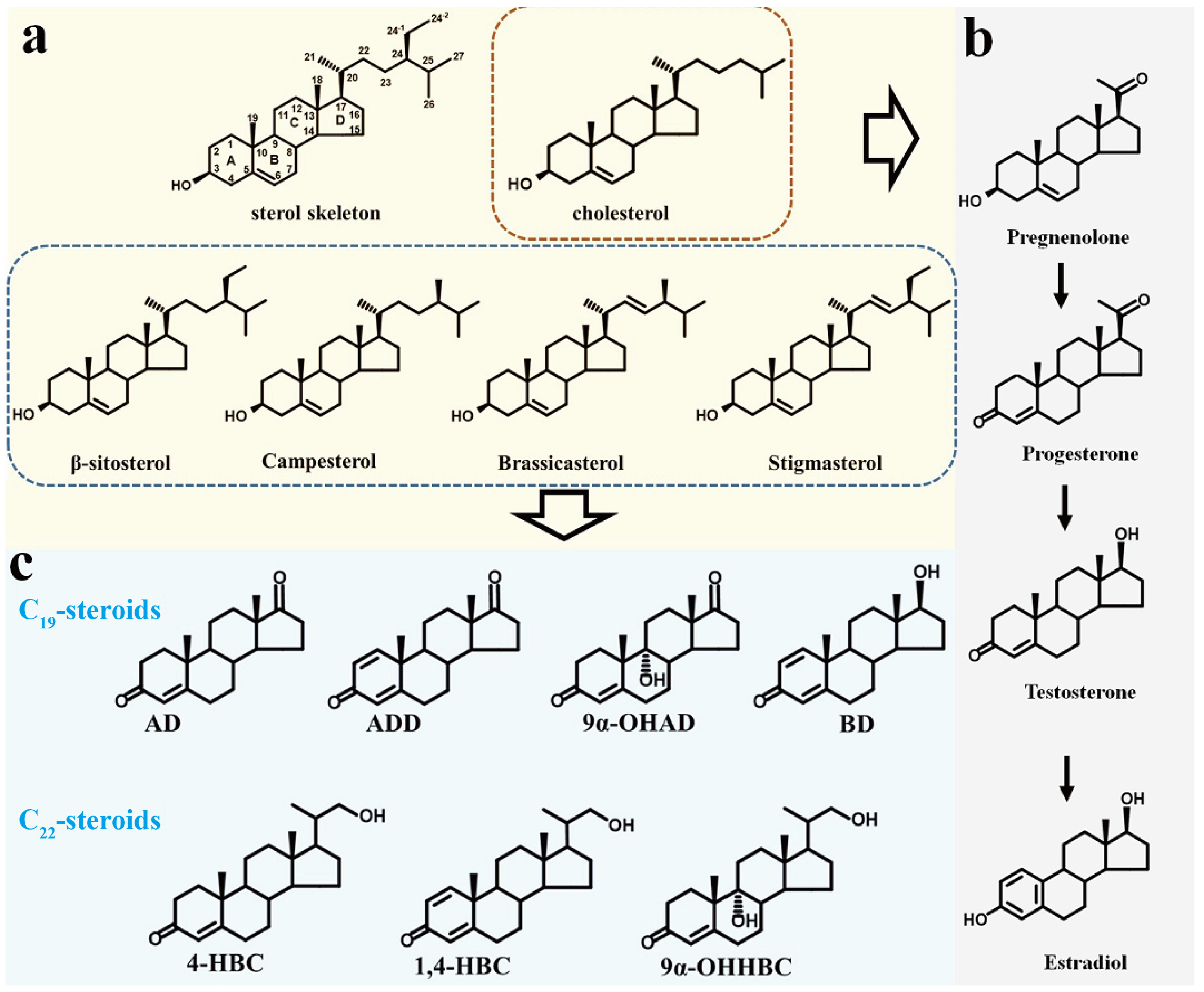
3.1. Analysis and Reconstruction of Steroids’ Metabolic Pathways
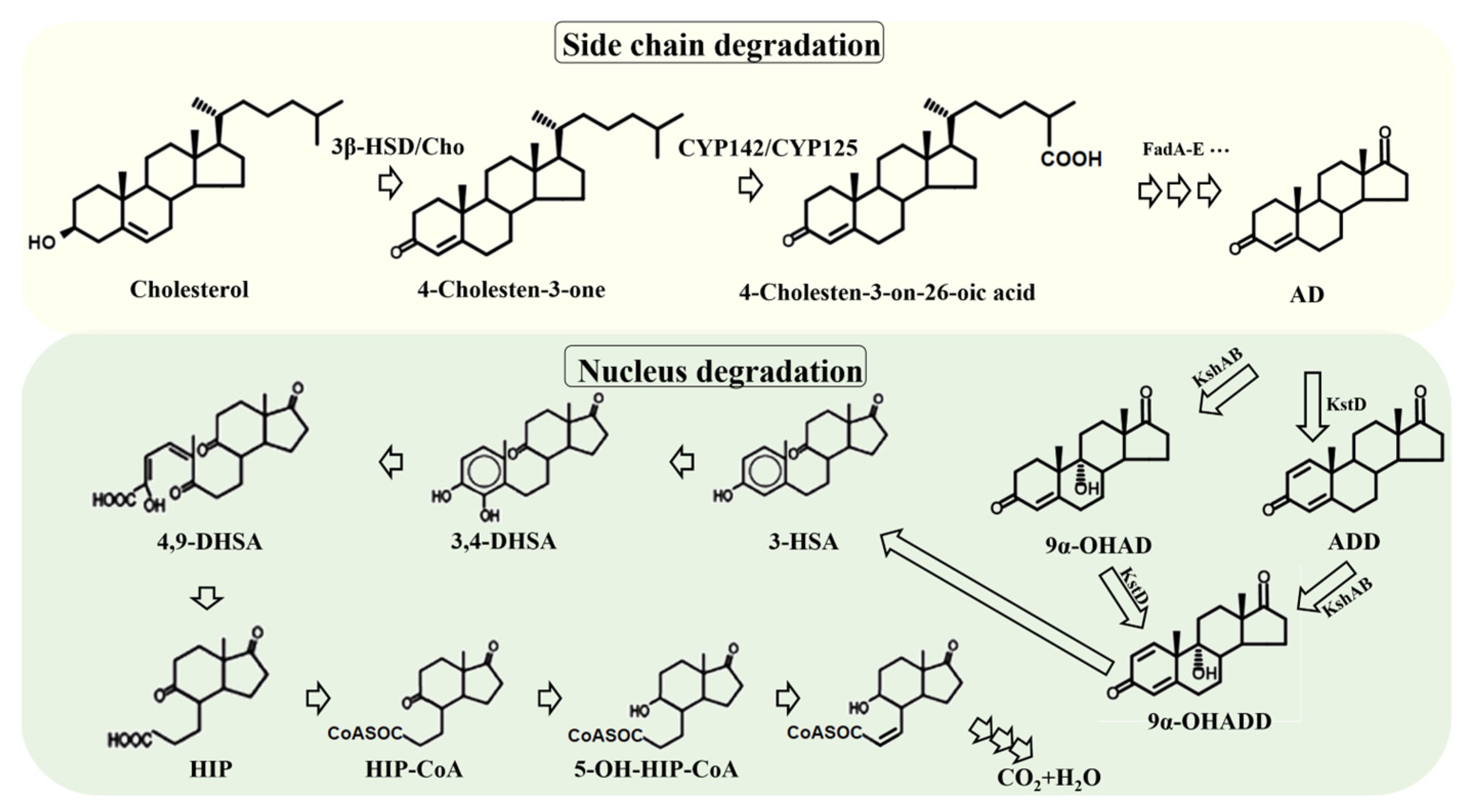
3.1.1. Reconstructing the Degradation Pathway of Steroid Nucleus
3.1.2. Reconstructing the Degradation Pathway of Steroid Side Chain
3.2. Optimization of Mycolicibacterial Cell Factories for Steroidal Industry
3.3. Production of Steroids via Mycolicbacteria Whole-Cell Biocatalysis
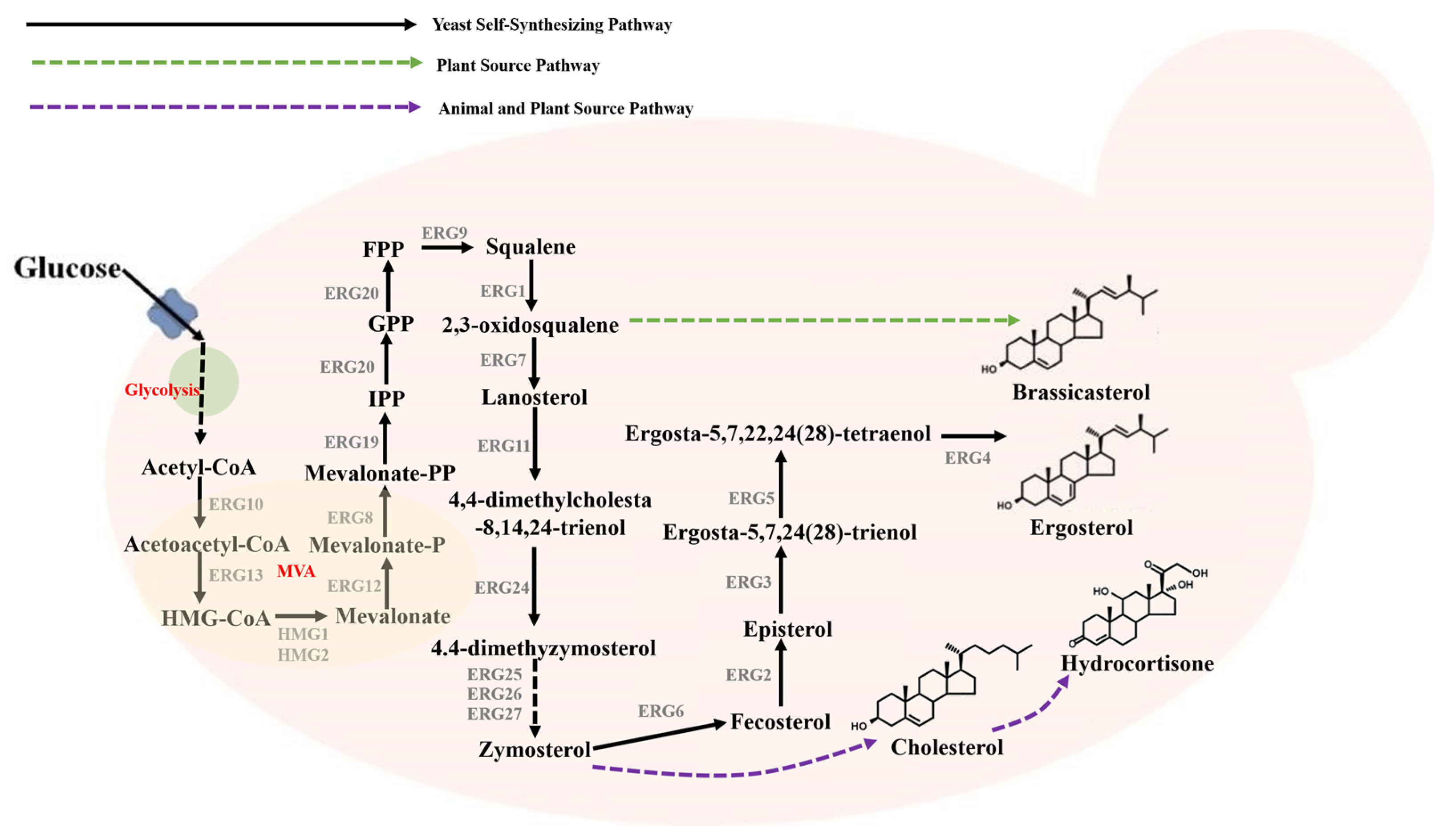
4. Tomorrow: De Novo Synthesis of Steroids
5. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Donova, M. Microbial steroid production technologies: Current trends and prospects. Microorganisms 2022, 10, 53. [Google Scholar] [CrossRef]
- Vladimir, K.; Vladimir, Z.; Aede, D.G. Twenty years of brassinosteroids: Steroidal plant hormones warrant better crops for the XXI century. Ann. Bot. 2000, 86, 441–447. [Google Scholar]
- Tuckey, R.C. Progesterone synthesis by the human placenta. Placenta 2005, 26, 273–281. [Google Scholar] [CrossRef]
- Dufourc, E.J. Sterols and membrane dynamics. J. Chem. Biol. 2008, 1, 63–77. [Google Scholar] [CrossRef]
- Weete, J.D.; Parish, E.J.; Nes, W.D. Chemistry, biochemistry, and function of sterols. Lipids 2000, 35, 241. [Google Scholar] [CrossRef]
- Wollam, J.; Antebi, A. Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 2011, 80, 885–916. [Google Scholar] [CrossRef]
- Vallée, M.; Vitiello, S.; Bellocchio, L.; Hebert-Chatelain, E.; Monlezun, S.; Martin-Garcia, E.; Kasanetz, F.; Baillie, G.; Panin, F.; Cathala, A.; et al. Pregnenolone can protect the brain from cannabis intoxication. Science 2014, 343, 94–98. [Google Scholar] [CrossRef]
- He, Y.Z.; Shi, J.J.; Nguyen, Q.T.; You, E.; Liu, H.B.; Ren, X.; Wu, Z.S.; Li, J.S.; Qiu, W.L.; Khoo, S.K. Development of highly potent glucocorticoids for steroid–resistant severe asthma. Proc. Natl. Acad. Sci. USA 2019, 116, 6932–6937. [Google Scholar] [CrossRef]
- Ahmed, M.H.; Hassan, A. Dexamethasone for the treatment of coronavirus disease (COVID-19): A review. SN Compr. Clin. Med. 2020, 2, 2637–2646. [Google Scholar] [CrossRef]
- Fernández-Cabezón, L.; Galán, B.; García, J.L. New insights on steroid biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Fragkaki, A.; Angelis, Y.; Koupparis, M.; Tsantili-Kakoulidou, A.; Kokotos, G.; Georgakopoulos, C. Structural characteristics of anabolic androgenic steroids contributing to binding to the androgen receptor and to their anabolic and androgenic activities: Applied modifications in the steroidal structure. Steroids 2009, 74, 172–197. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V.; Egorova, O.V. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012, 94, 1423–1447. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Ke, X.; Liu, Z.Q.; Zheng, Y.G. Rational development of mycobacteria cell factory for advancing the steroid biomanufacturing. World J. Microbiol. Biotechnol. 2022, 38, 191. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Corticosteroids: The drugs to beat. Eur. J. Pharmacol. 2006, 533, 2–14. [Google Scholar] [CrossRef]
- Ledford, H. Steroid is first drug shown to prevent deaths from COVID-19. Nature 2020, 582, 469. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Low dose steroid cuts death in ventilated patients by one third, trial finds. BMJ 2020, 369, 2422. [Google Scholar] [CrossRef]
- Lu, C.Y.; Liu, Y.; Chen, B.; Yang, H.; Hu, H.F.; Liu, Y.; Zhao, Y. Prognostic value of lymphocyte count in severe COVID-19 patients with corticosteroid treatment. Signal Transduct. Target. Ther. 2021, 6, 106. [Google Scholar] [CrossRef]
- Stong, R.A.; Kolodny, E.; Kelsey, R.G.; González-Hernández, M.P.; Vivanco, J.M.; Manter, D.K. Effect of plant sterols and tannins on Phytophthora ramorum growth and sporulation. J. Chem. Ecol. 2013, 39, 733–743. [Google Scholar] [CrossRef]
- Chiang, Y.R.; Wei, S.T.; Wang, P.H.; Wu, P.H.; Yu, C.P. Microbial degradation of steroid sex hormones: Implications for environmental and ecological studies. Microb. Biotechnol. 2020, 13, 926–949. [Google Scholar] [CrossRef]
- Rowland, S.J.; West, C.E.; Jones, D.; Scarlett, A.G.; Frank, R.A.; Hewitt, L.M. Steroidal aromatic “naphthenic acids” in oil sands process-affected water: Structural comparisons with environmental estrogens. Environ. Sci. Technol. 2011, 45, 9806–9815. [Google Scholar] [CrossRef]
- Marker, R.E.; Krueger, J. Sterols. CXII. sapogenins. XLI. the preparation of trillin and its conversion to progesterone. J. Am. Chem. Soc. 1940, 62, 3349–3350. [Google Scholar] [CrossRef]
- Johnson, A.C.; Williams, R.J.; Matthiessen, P. The potential steroid hormone contribution of farm animals to freshwaters, the United Kingdom as a case study. Sci. Total Environ. 2006, 362, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Mahajan, A. Sustainable approaches for steroid synthesis. Environ. Chem. Lett. 2018, 17, 879–895. [Google Scholar] [CrossRef]
- Mancino, V.; Cerra, B.; Piccinno, A.; Gioiello, A. Continuous Flow Synthesis of 16-Dehydropregnenolone Acetate, a Key Synthon for Natural Steroids and Drugs. Org. Process Res. Dev. 2018, 22, 600–607. [Google Scholar] [CrossRef]
- Chowdhury, P.; Borah, J.M.; Bordoloi, M.; Goswami, K.P. A Simple Efficient Process for the Synthesis of 16-Dehydropregnenolone Acetate(16-Dpa)–A Key Steroid Drug Intermediate from Diosgenin. J. Chem. Eng. Process Technol. 2011, 2, 1–7. [Google Scholar] [CrossRef]
- Baruah, D.; Das, R.N.; Konwar, D. Facile green synthesis of 16-dehydropregnenolone acetate (16-DPA) from diosgenin. Synth. Commun. 2016, 46, 79–84. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, P.; Pan, D.; Zhou, X. New Insights into the Modification of the Non-Core Metabolic Pathway of Steroids in Mycolicibacterium and the Application of Fermentation Biotechnology in C-19 Steroid Production. Int. J. Mol. Sci. 2023, 24, 5236. [Google Scholar] [CrossRef]
- Sedlaczek, L. Biotransformations of steroids. Crit. Rev. Biotechnol. 1988, 7, 187–236. [Google Scholar] [CrossRef]
- Ahmad, S.; Garg, S.K.; Johri, B.N. Biotransformation of sterols: Selective cleavage of the side chain. Biotechnol. Adv. 1992, 10, 1–67. [Google Scholar] [CrossRef]
- Sultana, N. Microbial biotransformation of bioactive and clinically useful steroids and some salient features of steroids and biotransformation. Steroids 2018, 136, 76–92. [Google Scholar] [CrossRef]
- Faramarzi, M.A.; Adrangi, S.; Yazdi, M.T. Microalgal biotransformation of steroids. J. Phycol. 2008, 44, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.C.; Fernandes, P.; Cabral, J.M.S.; Žnidaršič-Plazl, P.; Plazl, I. Continuous steroid biotransformations in microchannel reactors. New Biotechnol. 2012, 29, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.Y.; Dong, X. Microbial biotransformation: Recent developments on steroid drugs. Recent Pat. Biotechnol. 2009, 3, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Montagnon, T. Molecules that Changed the World; Wiley-VCH: Hoboken, NJ, USA, 2008; p. 286. [Google Scholar]
- Giorgi, V.; Menendez, P.; Garcia-Carnelli, C. Microbial transformation of cholesterol: Reactions and practical aspects-an update. World J. Microbiol. Biotechnol. 2019, 35, 131. [Google Scholar] [CrossRef] [PubMed]
- Mahato, S.B.; Garai, S. Advances in microbial steroid biotransformation. Steroids 1997, 62, 332–345. [Google Scholar] [CrossRef]
- Fernandes, P.; Cruz, A.; Angelova, B. Microbial conversion of steroid compounds: Recent developments. Enzym. Microb. Technol. 2003, 32, 688–705. [Google Scholar] [CrossRef]
- Feller, F.M.; Holert, J.; Yücel, O.; Philipp, B. Degradation of bile acids by soil and water bacteria. Microorganisms 2021, 9, 1759. [Google Scholar] [CrossRef]
- Dhar, A.; Samantha, T.B. Novel oxidative cleavage of C17-C20 bond in pregnane by a Pseudomonas sp. J. Steroid Biochem. Mol. Biol. 1993, 44, 101–104. [Google Scholar] [CrossRef]
- Tenneson, M.E.; Baty, J.D.; Bilton, R.F.; Mason, A.N. The degradation of cholic acid by Pseudomonas sp. N.C.I.B. 10590. Biochem. J. 1979, 184, 613–618. [Google Scholar] [CrossRef]
- Horvath, J.; Kramli, A. Microbiological oxidation of cholesterol with Azotobacter. Nature 1947, 160, 639. [Google Scholar] [CrossRef]
- Drzyzga, O.; Llorens, J.M.N.; Heras, L.F.D.L.; Fernández, E.G.; Perera, J. Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int. J. Syst. Evol. Microbiol. 2009, 59, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, N.P.; Tracey, R.P. Numerical taxonomy of cholesterol-degrading soil bacteria. J. Appl. Bacteriol. 1984, 57, 429–446. [Google Scholar] [CrossRef]
- Olivera, E.R.; Luengo, J.M. Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways. Genes 2019, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Galán, B.; Uhía, I.; García-Fernández, E.; Martínez, I.; Bahíllo, E.; Fuente, J.L.D.; Barredo, J.L.; Fernández-Cabezón, L.; García, J.L. Mycobacterium smegmatis is a suitable cell factory for the production of steroidic synthons. Microb. Biotechnol. 2017, 10, 138–150. [Google Scholar] [CrossRef]
- Li, M.; Zhu, Z.T.; Ta, X.Y.; Wang, F.Q.; Wei, D.Z. RNA-Seq analysis uncovers non-coding small RNA system of Mycobacterium neoaurum in the metabolism of sterols to accumulate steroid intermediates. Microb. Cell Fact. 2016, 15, 64. [Google Scholar] [CrossRef]
- Wang, H.W.; Song, S.K.; Peng, F.; Yang, F.; Chen, T.; Li, X.; Cheng, X.Y.; He, Y.J.; Huang, Y.Q.; Su, Z.D. Whole-genome and enzymatic analyses of an androstenedione-producing Mycobacterium strain with residual phytosterol-degrading pathways. Microb. Cell Fact. 2020, 19, 187. [Google Scholar] [CrossRef]
- Lin, C.W.; Wang, P.H.; Ismail, W.; Tsai, Y.W.; Nayal, A.E.; Yang, C.Y.; Yang, F.C.; Wang, C.H.; Chiang, Y.R. Substrate uptake and subcellular compartmentation of anoxic cholesterol catabolism in Sterolibacterium denitrificans. J. Biol. Chem. 2015, 290, 1155–1169. [Google Scholar] [CrossRef]
- Geize, R.V.D.; Yam, K.; Heuser, T.; Wilbrink, M.H.; Hara, H.; Anderton, M.C.; Sim, E.; Dijkhuizen, L.; Davies, J.E.; Mohn, W.W.; et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 2007, 104, 1947–1952. [Google Scholar] [CrossRef]
- Ouellet, H.; Johnston, J.B.; Ortiz de Montellano, P.R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef]
- Fernández-Cabezón, L.; García-Fernández, E.; Galán, B.; García, J.L. Molecular characterization of a new gene cluster for steroid degradation in Mycobacterium smegmatis. Environ. Microbiol. 2017, 19, 2546–2563. [Google Scholar] [CrossRef]
- Yao, K.; Wang, F.Q.; Zhang, H.C.; Wei, D.Z. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab. Eng. 2013, 15, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Xu, L.Q.; Wang, F.Q.; Wei, D.Z. Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab. Eng. 2014, 24, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Atrat, P.G.; Koch, B.; Szekalla, B.; Hörhold-Schubert, C. Application of newly synthesized detergents in the side chain degradation of plant sterols by Mycobacterium fortuitum. J. Basic Microbiol. 1992, 32, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Gao, X.Q.; Feng, J.X.; Wang, X.D.; Wei, D.Z. Influence of temperature on nucleus degradation of 4-androstene-3,17-dione in phytosterol biotransformation by Mycobacterium sp. Lett. Appl. Microbiol. 2015, 61, 63–68. [Google Scholar] [CrossRef]
- Gilbert, S.; Hood, L.; Seah, S.Y.K. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis. J. Bacteriol. 2017, 200, 512–517. [Google Scholar] [CrossRef]
- Wrońska, N.; Brzostek, A.; Szewczyk, R.; Soboń, A.; Dziadek, J.; Lisowska, K. The role of fadD19 and echA19 in sterol side chain degradation by Mycobacterium smegmatis. Molecules 2016, 21, 598. [Google Scholar] [CrossRef]
- Yang, X.X.; Dubnau, E.; Smith, I.; Sampson, N.S. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry 2007, 46, 9058–9067. [Google Scholar] [CrossRef]
- Uhía, I.; Galán, B.; Medrano, F.J.; García, J.L. Characterization of the KstR-dependent promoter of the gene for the first step of the cholesterol degradative pathway in Mycobacterium smegmatis. Microbiology 2011, 157, 2670–2680. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, J.; Cao, D.D.; Wang, X.D.; Wei, D.Z. Identification and in situ removal of an inhibitory intermediate to develop an efficient phytosterol bioconversion process using a cyclodextrin-resting cell system. RSC Adv. 2021, 11, 24787–24793. [Google Scholar] [CrossRef]
- Dresen, C.; Lin, L.Y.C.; Angelo, I.D.; Tocheva, E.I.; Strynadka, N.; Eltis, L.D. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 2010, 285, 22264–22275. [Google Scholar] [CrossRef]
- Liu, N.; Feng, J.; Zhang, R.; Chen, X.; Li, X.; Yao, P.; Wu, Q.; Ma, Y.; Zhu, D. Efficient microbial synthesis of key steroidal intermediates from bio-renewable phytosterols by genetically modified Mycobacterium fortuitum strains. Green Chem. 2019, 21, 4076–4085. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, K.; Liu, Y.; Xu, J.; Yang, Y.; Liu, J.; Hu, X. A three-step synthesis of estra-4,9-diene-3,17-dione. Can. J. Chem. 2018, 97, 267–269. [Google Scholar] [CrossRef]
- Szentirmai, A. Microbial physiology of sidechain degradation of sterols. J. Ind. Microbiol. 1990, 6, 101–115. [Google Scholar] [CrossRef]
- Lario, P.I.; Sampson, N.; Vrielink, A. Sub-atomic resolution crystal structure of cholesterol oxidase: What atomic resolution crystallography reveals about enzyme mechanism and the role of the FAD cofactor in redox activity. J. Mol. Biol. 2003, 326, 1635–1650. [Google Scholar] [CrossRef] [PubMed]
- Vrielink, A.; Lloyd, L.F.; Blow, D.M. Crystal structure of cholesterol oxidase from Brevibacterium sterolicum refined at 1.8 A resolution. J. Mol. Biol. 1991, 219, 533–554. [Google Scholar] [CrossRef]
- Zhang, R.J.; Liu, X.C.; Wang, Y.S.; Han, Y.C.; Sun, J.S.; Shi, J.P.; Zhang, B.G. Identification, function, and application of 3-ketosteroid Δ1-dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons. Microb. Cell Fact. 2018, 17, 77. [Google Scholar] [CrossRef]
- Wang, X.J.; Feng, J.H.; Zhang, D.L.; Wu, Q.Q.; Zhu, D.M.; Ma, Y.H. Characterization of new recombinant 3-ketosteroid-Δ1-dehydrogenases for the biotransformation of steroids. Appl. Microbiol. Biotechnol. 2017, 101, 6049–6060. [Google Scholar] [CrossRef]
- Guevara, G.; Fernández de Las, H.L.; Perera, J.; Llorens, J.M.N. Functional differentiation of 3-ketosteroidΔ1-dehydrogenase isozymes in Rhodococcus ruber strain Chol-4. Microb. Cell Fact. 2017, 16, 42. [Google Scholar] [CrossRef]
- Zhang, R.J.; Xu, X.X.; Cao, H.J.; Yuan, C.Y.; Yuminaga, Y.K.; Zhao, S.W.; Shi, J.P.; Zhang, B.G. Purification, characterization, and application of a high activity 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum DSM 1381. Appl. Microbiol. Biotechnol. 2019, 103, 6605–6616. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, M.C.; Han, R.M.; Zhao, Y.X.; Chen, K.W.; Qian, K.; Shao, M.L.; Yang, T.W.; Xu, M.J.; Xu, J.Z.; et al. A novel 3-phytosterone-9α-hydroxylase oxygenation component and its application in bioconversion of 4-androstene-3,17-dione to 9α-hydroxy-4-androstene-3,17-dione coupling with A NADH regeneration formate dehydrogenase. Molecules 2019, 24, 2534. [Google Scholar] [CrossRef]
- Bragin, E.Y.; Shtratnikova, V.Y.; Schelkunov, M.I.; Dovbnya, D.V.; Donova, M.V. Genome-wide response on phytosterol in 9-hydroxyandrostenedione-producing strain of Mycobacterium sp. VKM Ac-1817D. BMC Biotechnol. 2019, 19, 39. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Xu, L.Q.; Yao, K.; Xiong, L.B.; Tao, X.Y.; Liu, M.; Wang, F.Q.; Wei, D.Z. Engineered 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum: An efficient platform for production of steroid drugs. Appl. Environ. Microbiol. 2018, 84, e02777-17. [Google Scholar] [CrossRef] [PubMed]
- García, J.L.; Uhía, I.; Galán, B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb. Biotechnol. 2012, 5, 679–699. [Google Scholar] [CrossRef] [PubMed]
- Draper, P. The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 1998, 3, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Tahlan, K.; Wilson, R.; Kastrinsky, D.B.; Arora, K.; Nair, V.; Fischer, E.; Barnes, S.W.; Walker, J.R.; Alland, D.; Barry, C.E., III; et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2012, 56, 1797–1809. [Google Scholar] [CrossRef]
- Yuan, T.N.; Werman, J.M.; Yin, X.Y.; Yang, M.; Garcia-Diaz, M.; Sampson, N.S. Enzymatic β-oxidation of the cholesterol side chain in Mycobacterium tuberculosis bifurcates stereospecifically at hydration of 3-oxo-cholest-4,22-dien-24-oyl-CoA. ACS Infect. Dis. 2021, 7, 1739–1751. [Google Scholar] [CrossRef]
- Thomas, S.T.; Vanderven, B.C.; Sherman, D.R.; Russell, D.G.; Sampson, N.S. Pathway profiling in Mycobacterium tuberculosis: Elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 2011, 286, 43668–43678. [Google Scholar] [CrossRef]
- Thomas, S.T.; Sampson, N.S. Mycobacterium tuberculosis utilizes a unique heterotetrameric structure for dehydrogenation of the cholesterol side chain. Biochemistry 2013, 52, 2895–2904. [Google Scholar] [CrossRef]
- Ouellet, H.; Guan, S.H.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; Ortiz de Montellano, P.R. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef]
- García-Fernández, E.; Frank, D.J.; Galán, B.; Kells, P.M.; Podust, L.M.; García, J.L.; Ortiz de Montellano, P.R. A highly conserved mycobacterial cholesterol catabolic pathway. Environ. Microbiol. 2013, 15, 2342–2359. [Google Scholar] [CrossRef]
- Xiong, L.B.; Liu, H.H.; Xu, L.Q.; Sun, W.J.; Wang, F.Q.; Wei, D.Z. Improving the production of 22-hydroxy-23,24-bisnorchol-4-ene-3-one from sterols in Mycobacterium neoaurum by increasing cell permeability and modifying multiple genes. Microb. Cell Fact. 2017, 16, 89. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.Q.; Liu, Y.J.; Yao, K.; Liu, H.H.; Tao, X.Y.; Wang, F.Q.; Wei, D.Z. Unraveling and engineering the production of 23,24-bisnorcholenic steroids in sterol metabolism. Sci. Rep. 2016, 6, 21928. [Google Scholar] [CrossRef] [PubMed]
- Lisowska, K.; Korycka, M.; Hadław-Klimaszewska, O.; Ziółkowski, A.; Sedlaczek, L. Permeability of mycobacterial cell envelopes to sterols: Peptidoglycan as the diffusion barrier. J. Basic Microbiol. 1996, 36, 407–419. [Google Scholar] [CrossRef]
- Rumijowska, A.; Lisowska, K.; Ziótkowski, A.; Sedlaczek, L. Transformation of sterols by Mycobacterium vaccae: Effect of lecithin on the permeability of cell envelopes to sterols. World J. Microbiol. Biotechnol. 1997, 13, 89–95. [Google Scholar] [CrossRef]
- Sedlaczek, L.; Lisowska, K.; Korycka, M.; Rumijowska, A.; Ziółkowski, A.; Długoński, J. The effect of cell wall components on glycine-enhanced sterol side chain degradation to androstene derivatives by Mycobacteria. Appl. Microbiol. Biotechnol. 1999, 52, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.B.; Liu, H.H.; Song, X.W.; Meng, X.G.; Liu, X.Z.; Ji, Y.Q.; Wang, F.Q.; Wei, D.Z. Improving the biotransformation of phytosterols to 9α-hydroxy-4-androstene-3,17-dione by deleting embC associated with the assembly of cell envelope in Mycobacterium neoaurum. J. Biotechnol. 2020, 323, 341–346. [Google Scholar] [CrossRef]
- Xiong, L.B.; Liu, H.H.; Xu, L.Q.; Wei, D.Z.; Wang, F.Q. Role identification and application of SigD in the transformation of soybean phytosterol to 9α-hydroxy-4-androstene-3,17-dione in Mycobacterium neoaurum. J. Agric. Food Chem. 2017, 65, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.B.; Sun, W.J.; Liu, Y.J.; Wang, F.Q.; Wei, D.Z. Enhancement of 9α-hydroxy-4-androstene-3,17-dione production from soybean phytosterols by deficiency of a regulated intramembrane proteolysis metalloprotease in Mycobacterium neoaurum. J. Agric. Food Chem. 2017, 65, 10520–10525. [Google Scholar] [CrossRef]
- Su, L.; Shen, Y.; Zhang, W.; Gao, T.; Shang, Z.; Wang, M. Cofactor engineering to regulate NAD+/NADH ratio with its application to phytosterols biotransformation. Microb. Cell Fact. 2017, 16, 182. [Google Scholar] [CrossRef]
- Sun, W.J.; Wang, L.; Liu, H.H.; Liu, Y.J.; Ren, Y.H.; Wang, F.Q.; Wei, D.Z. Characterization and engineering control of the effects of reactive oxygen species on the conversion of sterols to steroid synthons in Mycobacterium neoaurum. Metab. Eng. 2019, 56, 97–110. [Google Scholar] [CrossRef]
- Zhao, A.Q.; Zhan, X.Q.; Li, Y.M.; Wang, Z.; Lv, Y.K.; Liu, J.L.; Alam, M.A.; Xiong, W.L.; Xu, J.L. Mycolicibacterium cell factory for the production of steroid-based drug intermediates. Biotechnol. Adv. 2021, 53, 107860. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Peng, H.; Zhang, W.; Zheng, M.; Tian, W.; Zhang, Y.; Liu, H.; Lin, Z.; Deng, Z.; Qu, X. Production of heterodimeric diketopiperazines employing a Mycobacterium-based whole-cell biocatalysis system. J. Org. Chem. 2021, 86, 11189–11197. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.A.A.; Sultan, A.; Adnan, H.S. A whole-cell biocatalysis application of steroidal drugs. Orient. J. Chem. 2013, 29, 389–403. [Google Scholar]
- Zhao, Y.Q.; Shen, Y.B.; Ma, S.; Luo, J.M.; Ouyang, W.; Zhou, H.J.; Tang, R.; Wang, M. Production of 5α-androstene-3,17-dione from phytosterols by co-expression of 5α-reductase and glucose-6-phosphate dehydrogenase in engineered Mycobacterium neoaurum. Green Chem. 2019, 21, 1809–1815. [Google Scholar] [CrossRef]
- Zhu, Z.L.; Gao, X.; Song, Z.; Li, C.; Lu, F.P.; Tanokura, M.; Qin, H.M. Development of engineered ferredoxin reductase systems for the efficient hydroxylation of steroidal substrates. ACS Sustain. Chem. Eng. 2020, 8, 16720–16730. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Liu, Y.J.; Ji, W.T.; Liu, K.; Gao, B.; Tao, X.Y.; Zhao, M.; Wang, F.Q.; Wei, D.Z. One-pot biosynthesis of 7β-hydroxyandrost-4-ene-3,17-dione from phytosterols by cofactor regeneration system in engineered Mycolicibacterium neoaurum. Microb. Cell Fact. 2022, 21, 59. [Google Scholar] [CrossRef]
- Gu, Y.H.; Jiao, X.; Ye, L.D.; Yu, H.W. Metabolic engineering strategies for de novo biosynthesis of sterols and steroids in yeast. Bioresour. Bioprocess. 2021, 8, 1. [Google Scholar] [CrossRef]
- Prasad, B.D.; Sahni, S.; Krishna, P.; Kumari, D.; Mahato, A.K.; Jambhulkar, S.J.; Kumar, P.; Ranjan, T.; Kumar, A. De novo transcriptome assembly and identification of brassinosteroid biosynthetic pathway in safflower. J. Plant Growth Regul. 2022, 41, 1854–1870. [Google Scholar] [CrossRef]
- Szczebara, F.M.; Chandelier, C.; Villeret, C.; Masurel, A.; Bourot, S.; Duport, C.; Blanchard, S.; Groisillier, A.; Testet, E.; Costaglioli, P.; et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003, 21, 143–149. [Google Scholar] [CrossRef]
- Russell, D.W. Cholesterol biosynthesis and metabolism. Cardiovasc. Drugs Ther. 1992, 6, 103–110. [Google Scholar] [CrossRef]
- Sonawane, P.D.; Pollier, J.; Panda, S.; Szymanski, J.; Massalha, H.; Yona, M.; Unger, T.; Malitsky, S.; Arendt, P.; Pauwels, L.; et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 2016, 3, 16205. [Google Scholar] [CrossRef] [PubMed]
- Duport, C.; Spagnoli, R.; Degryse, E.; Pompon, D. Self-sufficient biosynthesis of pregnenolone and progesterone in engineered yeast. Nat. Biotechnol. 1998, 16, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.A.V.; Cassuriaga, A.P.A.; Moraes, L.; Greque de Morais, M. Biosynthesis and potential applications of terpenes produced from microalgae. Bioresour. Technol. Rep. 2022, 19, 101166. [Google Scholar] [CrossRef]
- Eichenberger, M.; Hansson, A.; Fischer, D.; Dürr, L.; Naesby, M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, 46. [Google Scholar] [CrossRef]
- Sun, J.C.; Sun, W.T.; Zhang, G.L.; Lv, B.; Li, C. High efficient production of plant flavonoids by microbial cell factories: Challenges and opportunities. Metab. Eng. 2022, 70, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.G.; Zhao, H.M. Engineering robust microorganisms for organic acid production. J. Ind. Microbiol. Biotechnol. 2022, 49, 67. [Google Scholar] [CrossRef] [PubMed]
- Du, H.X.; Xiao, W.H.; Wang, Y.; Zhou, X.; Zhang, Y.; Liu, D.; Yuan, Y.J. Engineering Yarrowia lipolytica for campesterol overproduction. PLoS ONE 2016, 11, e0146773. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Yao, M.D.; Liu, H.; Zhou, X.; Xiao, W.H.; Yuan, Y.J. Improved campesterol production in engineered Yarrowia lipolytica strains. Biotechnol. Lett. 2017, 39, 1033–1039. [Google Scholar] [CrossRef]
- Brocard-Masson, C.; Bonnin, I.; Dumas, B. Process for Preparing Genetically Transformed Yeasts Capable of Producing a Molecule of Interest at a High Titre. U.S. Patent WO2012175453, 27 December 2012. [Google Scholar]


| Aspect of Research | Key Findings |
|---|---|
| Engineering Mycolicibacterium Strains | Engineered Mycolicibacterium strains have been used to accumulate steroids and intermediates by disrupting specific steroid degradation pathways. |
| Heterologous Enzymes | Efficient heterologous enzymes like P450s have been cloned and expressed in engineered Mycolicbacteria to enhance sterol molecule modification, improving biomanufacturing efficiency for steroids. |
| Sterol Degradation Pathways | Sterol degradation pathways in micro-organisms include aerobic and anaerobic metabolism, with aerobic metabolism being predominant in industrial applications. |
| Rate-Limiting Steps | Rate-limiting steps in cholesterol degradation include the action of cholesterol oxidase (ChO), KstD, and Ksh enzymes. |
| Energy Balance | The NAD+/NADH ratio and control of reactive oxygen species (ROS) are crucial for efficient steroid production in Mycolicbacteria. |
| Metabolic Regulation | Omics analyses have been used to optimize Mycolicbacteria strains for steroid production. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Li, X.; Xiong, L.; Liu, K.; Liu, Y.; Xue, Z.; Han, R. Green Manufacturing of Steroids via Mycolicbacteria: Current Status and Development Trends. Fermentation 2023, 9, 890. https://doi.org/10.3390/fermentation9100890
Zhao M, Li X, Xiong L, Liu K, Liu Y, Xue Z, Han R. Green Manufacturing of Steroids via Mycolicbacteria: Current Status and Development Trends. Fermentation. 2023; 9(10):890. https://doi.org/10.3390/fermentation9100890
Chicago/Turabian StyleZhao, Ming, Xiangfei Li, Liangbin Xiong, Kun Liu, Yan Liu, Zhenglian Xue, and Rumeng Han. 2023. "Green Manufacturing of Steroids via Mycolicbacteria: Current Status and Development Trends" Fermentation 9, no. 10: 890. https://doi.org/10.3390/fermentation9100890
APA StyleZhao, M., Li, X., Xiong, L., Liu, K., Liu, Y., Xue, Z., & Han, R. (2023). Green Manufacturing of Steroids via Mycolicbacteria: Current Status and Development Trends. Fermentation, 9(10), 890. https://doi.org/10.3390/fermentation9100890









