Abstract
Pyrimidine biosynthesis and ribonucleoside metabolism in species of Pseudomonas was the focus of this review, in relation to their current taxonomic assignments in different homology groups. It was of interest to learn whether pyrimidine biosynthesis in taxonomically related species of Pseudomonas was regulated in a similar fashion by pyrimidine base supplementation or by pyrimidine limitation of pyrimidine auxotrophic strains. It was concluded that the regulation of pyrimidine biosynthesis in Pseudomonas species could not be correlated with their taxonomic assignment into a specific homology group. Pyrimidine ribonucleoside metabolism in Pseudomonas species primarily involved the pyrimidine ribonucleoside salvage enzymes nucleoside hydrolase and cytosine deaminase, independently of the Pseudomonas homology group to which the species was assigned. Similarly, pyrimidine base catabolism was shown to be active in different taxonomic homology groups of Pseudomonas. Although the number of studies exploring the catabolism of the pyrimidine bases uracil and thymine was limited in scope, it did appear that the presence of the pyrimidine base reductive pathway of pyrimidine catabolism was a commonality observed for the species of Pseudomonas investigated. There also appeared to be a connection between pyrimidine ribonucleoside degradation and the catabolism of pyrimidine bases in providing a cellular source of carbon or nitrogen independently of which homology group the species of Pseudomonas were assigned to.
Keywords:
pyrimidine; biosynthesis; ribonucleoside; catabolism; uracil; Pseudomonas; regulation; limitation 1. Introduction
Although a number of studies have investigated pyrimidine biosynthesis in Gram-negative [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26] and Gram-positive bacteria [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41] as well as in eukaryotes [42,43,44,45,46,47,48,49], relatively few studies have examined pyrimidine biosynthesis in species of Pseudomonas. Similarly, several investigations have explored pyrimidine ribonucleoside metabolism [50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69] and pyrimidine base catabolism [70,71,72,73,74,75,76,77,78,79,80] in a variety of prokaryotic and eukaryotic species, but few reviews have provided an in-depth analysis of pyrimidine ribonucleoside metabolism and pyrimidine base catabolism metabolism in species of Pseudomonas. Therefore, this review is focused on exploring the regulation of pyrimidine biosynthesis, pyrimidine ribonucleoside metabolism and pyrimidine base catabolism in species of Pseudomonas. A number of studies have been performed investigating the regulation of pyrimidine biosynthesis in species of Pseudomonas, but prior studies have not examined whether this regulation varies according to which homology group the species has been assigned to. Similarly, no prior studies have performed a comparison of the regulation of pyrimidine ribonucleoside or pyrimidine base metabolism in species of Pseudomonas based on their taxonomic assignment. The species of Pseudomonas have been taxonomically assigned to five homology groups based on RNA-DNA hybridization [81,82,83]. These homology groups include the Pseudomonas aeruginosa group, the Pseudomonas chlororaphis group, the Pseudomonas fluorescens group, the Pseudomonas putida group and the Pseudomonas syringae group. The regulation of pyrimidine biosynthesis, as well as pyrimidine ribonucleoside metabolism and pyrimidine base catabolism, has been investigated in some of the Pseudomonas homology groups, which allows a comparative biochemical analysis to be performed relative to Pseudomonas species assigned to the same homology group. Although the majority of pyrimidine biosynthesis studies being compared have used the same cell processing techniques, there does exist some variation in cell preparation (such as pH, temperature and substrate conditions) that needs to be considered when viewing the data in the tables. The majority of the pyrimidine biosynthesis studies conducted have involved investigations where the species of Pseudomonas were grown in medium containing succinate or glucose as a carbon source. These carbon sources were chosen to support pseudomonad growth, since succinate has been reported to cause catabolite repression of enzyme synthesis, while glucose does not. The carbon source glucose is often chosen since it supports a high level of pseudomonad growth compared to alternative carbon sources. Overall, there exists a need to study the regulation of pyrimidine metabolism from the perspective of the taxonomic assignment of pseudomonads into homology groups.
2. Pyrimidine Biosynthesis
2.1. Pseudomonas aeruginosa Homology Group
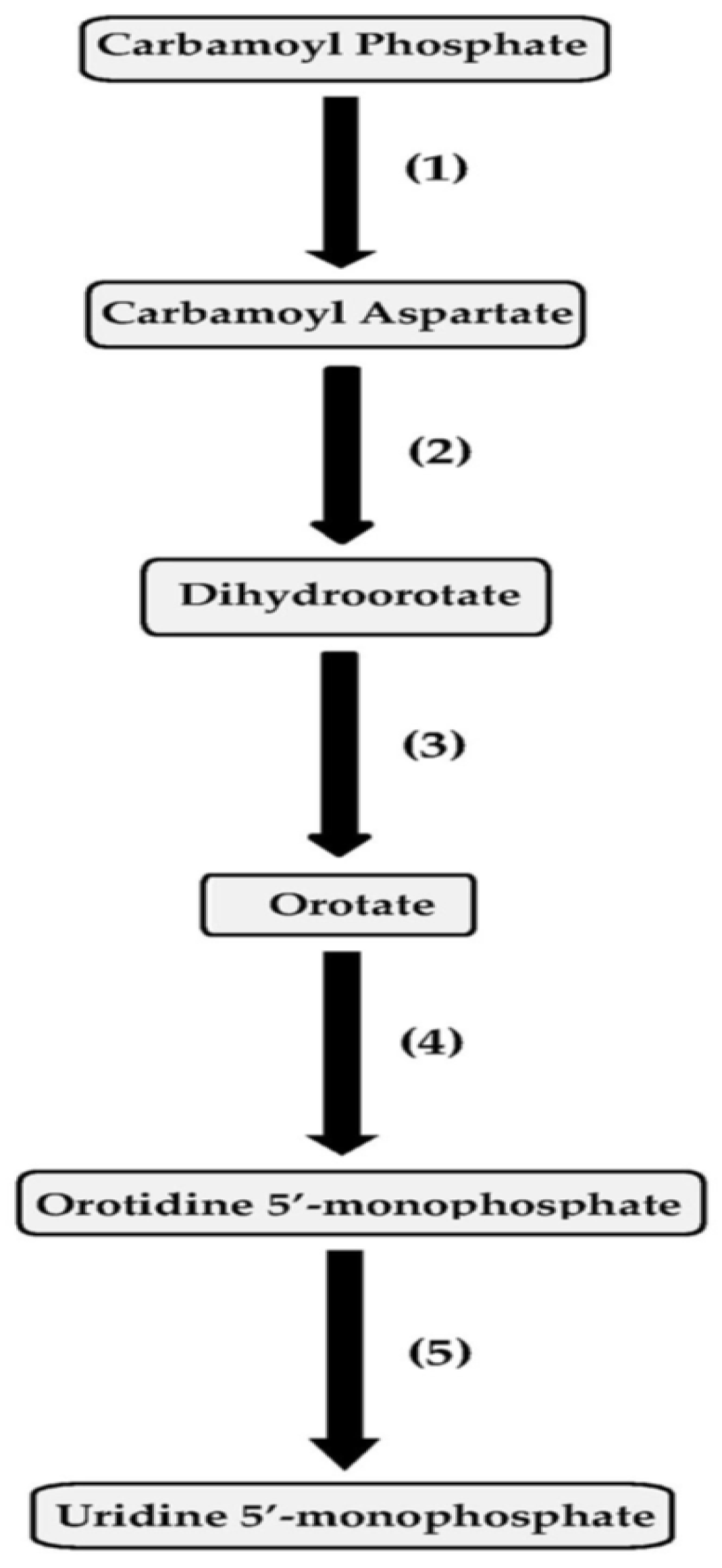
The pyrimidine biosynthetic pathway enzymes in pseudomonads include aspartate transcarbamoylase, dihydroorotase, dihydroorotate dehydrogenase, orotate phosphoribosyltransferase and orotidine 5′-monophosphate (OMP) decarboxylase (Figure 1). The species taxonomically assigned to the Pseudomonas aeruginosa group where the regulation of pyrimidine biosynthesis has been investigated previously include P. aeruginosa, Pseudomonas alcaligenes, Pseudomonas citronellolis, Pseudomonas mendocina, Pseudomonas nitroreducens, Pseudomonas oleovorans, Pseudomonas pseudoalcaligenes and Pseudomonas resinovorans [84,85,86,87,88,89,90,91,92,93,94,95,96].
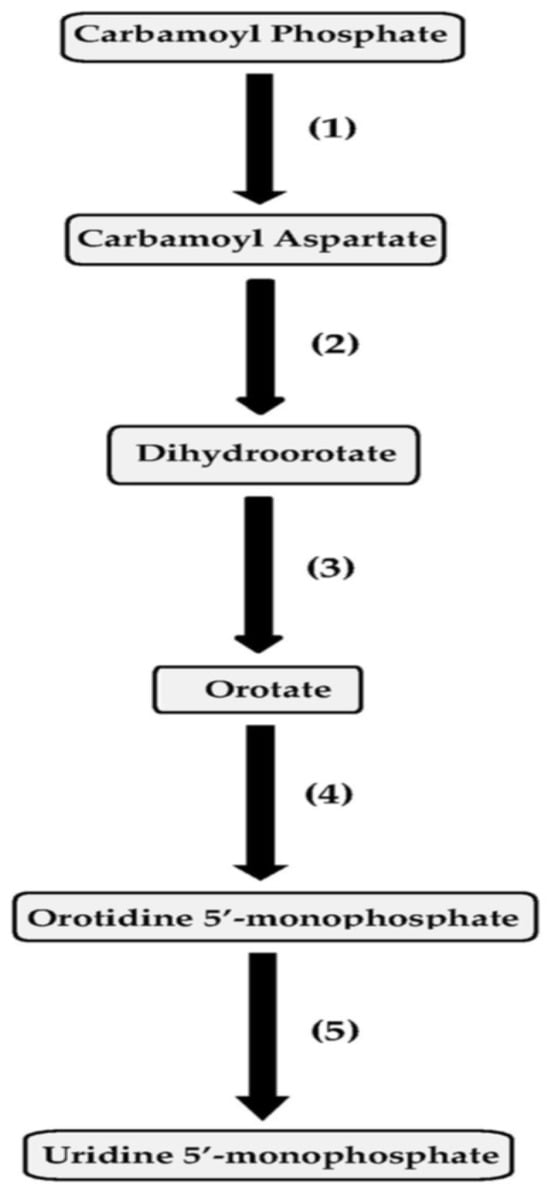
Figure 1.
Pyrimidine biosynthetic pathway and its enzymes. The enzymes include (1) aspartate transcarbamoylase; (2) dihydroorotase; (3) dihydroorotate dehydrogenase; (4) orotate phosphoribosyltransferase; and (5) orotidine 5′-monophosphate (OMP) decarboxylase.
In Table 1, the regulation of pyrimidine biosynthesis in species assigned to the P. aeruginosa homology group is compared relative to enzyme repression by a uracil-related metabolite and the level of enzyme synthesis following pyrimidine limitation of a pyrimidine auxotroph. No clear pattern is apparent in Table 1 other than that pyrimidine-depleted glucose-grown mutant cells produced greater elevations in enzyme activity than did the succinate-grown cells. In an early investigation of the control of pyrimidine biosynthesis in P. aeruginosa PAO1 cells [84], no repression of transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase synthesis by a uracil-related metabolite was noted when the 0.2% glucose-containing cells were supplemented with 112 µg/mL uracil [84]. Using isolated mutant strains of P. aeruginosa PAO1 glucose-grown cells lacking either dehydrogenase, phosphoribosyltransferase or decarboxylase activity, pyrimidine limitation of these cells for up to 120 min resulted in very little derepression of transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase synthesis. In a later study [85], the regulation of pyrimidine biosynthesis in P. aeruginosa was again investigated in 0.2% glucose-containing ATCC 15692 cells. When the medium was supplemented with 40 µg/mL orotate, the activity of transcarbamoylase, dihydroorotase, dehydrogenase, phosphoribosyltransferase or decarboxylase increased by up to four-fold [85]. If the ATCC 15692 glucose-grown cells were supplemented with 40 µg/mL uracil, repression of transcarbamoylase, phosphoribosyltransferase or decarboxylase activity was observed. This indicated that the repression of pyrimidine biosynthetic enzyme synthesis was occurring. A mutant was isolated that was deficient in dehydrogenase activity. When the glucose-grown mutant strain cells were subjected to pyrimidine-limited growth conditions for 2 h, it was found that transcarbamoylase, phosphoribosyltransferase or decarboxylase activity increased by 3.1-fold, 1.7-fold or 1.5-fold, respectively [85]. In contrast to the initial investigation exploring the regulation of pyrimidine biosynthesis in P. aeruginosa PAO1, the later investigation was able to demonstrate that the repression of pyrimidine biosynthetic enzyme synthesis by a uracil-related compound was occurring in ATCC 15692 cells.

Table 1.
Comparison of pyrimidine biosynthetic activity profiles in the Pseudomonas aeruginosa DNA homology group. Abbreviations: AT, aspartate transcarbamoylase; DO, dihydroorotase; DD, dihydroorotate dehydrogenase; OP, orotate phosphoribosyltransferase; and OD, OMP decarboxylase. Number in parentheses following each enzyme listed under effect of uracil addition on carbon source glucose or succinate indicates relative activity in the cells compared to activity in unsupplemented cells. Number in parentheses following each enzyme listed under pyrimidine limitation (2 h) on carbon source glucose or succinate indicates relative activity in the cells compared to activity in uracil-grown cells. ND, not determined. Pseudomonas pseudoalcaligenes is unable to utilize glucose as a carbon source.
In P. alcaligenes ATCC 14909, the presence of the pyrimidine base orotic acid in the growth medium appeared to repress the level of pyrimidine biosynthetic enzyme activity independently of whether glucose or succinate was used as a carbon source [86]. Interestingly, the addition of uracil to the growth medium of ATCC 14909 had little effect on the pyrimidine biosynthetic enzyme activity, regardless of the carbon source present [86]. It appeared that a metabolite of orotic acid was capable of repressing the synthesis of the pathway enzymes. Using a mutant strain deficient in orotate phosphoribosyltransferase, pyrimidine limitation studies were conducted using glucose or succinate as a carbon source [86]. It was found that the derepression of pyrimidine biosynthetic enzyme synthesis was evident. The activity of transcarbamoylase, dihydroorotase and decarboxylase was found to increase by nearly double in the pyrimidine-limited phosphoribosyltransferase mutant strain cells.
In P. citronellolis ATCC 13674, the regulation of pyrimidine biosynthetic enzyme synthesis was shown to be controlled by a uracil-related metabolite depending upon the carbon source utilized [87]. In succinate-grown ATCC 13674 cells, it was demonstrated that uracil was converted to a metabolite that repressed the activity of the five pathway enzymes. When glucose was utilized as the carbon source, it was also shown that the majority of the pyrimidine biosynthetic enzymes’ activity was depressed when uracil was supplemented into the medium [87]. Similarly, ATCC 13674 cells grown on either glucose or succinate as a carbon source supplemented with orotic acid showed a depression in transcarbamoylase, dihydroorotase or decarboxylase activity to an analogous degree observed when uracil was supplemented [87]. Pyrimidine limitation experiments were conducted for 4 h using a mutant strain deficient in decarboxylase activity, grown on either glucose or succinate as a carbon source. It was shown that transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase activity was elevated approximately double following pyrimidine limitation, in succinate-grown compared to mutant cells grown with saturating levels of uracil [87]. When the succinate-grown mutant cells were limited in pyrimidines for 4 h, a 6.4-fold, 14.4-fold or 5.8-fold increase, respectively, in dihydroorotase, dehydrogenase or phosphoribosyltransferase activity was observed [87]. It was concluded that a uracil-related metabolite was highly involved in the repression of pyrimidine biosynthetic enzyme synthesis in P. citronellolis [87].
In P. mendocina ATCC 25411, regulation of the pyrimidine biosynthetic enzymes in the bacterium P. mendocina was examined when its cells were grown on succinate as a carbon source. When succinate-grown P. mendocina cells were grown in the presence of orotic acid, the pyrimidine pathway enzymes transcarbamoylase, dehydrogenase and phosphoribosyltransferase’s activity was significantly depressed compared to their activity in the wild-type strain cells [88]. When the succinate-grown ATCC 25411 cells were supplemented with uracil, dihydroorotase or decarboxylase activity was observed to be greatly diminished relative to the unsupplemented cells [88]. If the ATCC 25411 cells were grown on glucose as a carbon source and supplemented with orotic acid or uracil, the pyrimidine biosynthetic enzyme activity appeared to be repressed by their metabolites, compared to their enzyme activity in the unsupplemented cells [89]. A mutant strain deficient in orotate phosphoribosyltransferase activity was utilized to determine the effect of pyrimidine limitation upon the pyrimidine biosynthetic pathway enzymes’ activity. When the succinate-grown mutant cells were limited in pyrimidine for 4 h, it was seen that transcarbamoylase, dehydrogenase or decarboxylase activity increased by 1.8-fold, 3.1-fold or 2.1-fold, respectively [88]. Interestingly, the pyrimidine limitation of glucose-grown cells of the mutant strain caused a depression in pyrimidine biosynthetic enzyme activity following 4 h of limitation, in comparison to the mutant cells grown in excess uracil [89]. It appeared that the regulation of pyrimidine biosynthesis in P. mendocina was not only controlled by pyrimidine metabolites but also by the carbon source being utilized [89].
In P. nitroreducens ATCC 33634, it was observed that the orotic acid or uracil supplementation of glucose-grown cells depressed the levels of pyrimidine biosynthetic enzyme activity [90]. This indicated that a pyrimidine-related compound was important in the regulation of pathway synthesis. In succinate-grown wild-type cells, the addition of uracil to the growth medium was shown to depress the transcarbamylase, dihydroorotase, dehydrogenase and decarboxylase activity [90]. This demonstrated the regulatory role of a uracil-related metabolite in controlling pyrimidine biosynthetic pathway enzyme synthesis. Pyrimidine limitation studies were performed using an orotate phosphoribosyltransferase mutant strain isolated from P. nitroreducens ATCC 33634 [90]. It was noted that pyrimidine limitation of the glucose-grown mutant cells for 4 h resulted in transcarbamoylase or dihydroorotase activity being elevated by approximately four-fold, while the dehydrogenase and decarboxylase activity increased by approximately double compared to the activity in the mutant cells grown in saturating uracil concentrations [90]. When succinate was utilized as the carbon source, pyrimidine limitation of the phosphoribosyltransferase mutant cells caused transcarbamoylase, dihydroorotase or dehydrogenase activity to increase by at least three-fold relative to the mutant cells grown with excess uracil [90]. A mutant strain lacking OMP decarboxylase activity was also isolated from P. nitroreducens [90]. Pyrimidine limitation of this mutant strain grown on glucose as a carbon source elevated transcarbamoylase, dihydroorotase, dehydrogenase and phosphoribosyltransferase activity by up to 10-fold compared to the mutant cells supplemented with uracil [90]. When succinate was the carbon source, pyrimidine limitation of the decarboxylase mutant strain cells produced at least a doubling of the transcarbamoylase, dihydroorotase, dehydrogenase and phosphoribosyltransferase activity relative to the mutant cells grown in excess uracil [90]. Based on the findings in P. nitroreducens, pyrimidine biosynthesis is highly controlled by metabolites of pyrimidines relative to enzyme synthesis.
When succinate-grown P. pseudoalcaligenes ATCC 17440 cells were grown in the presence of uracil, the activity of the five biosynthetic enzymes was depressed, with dihydroorotase activity being reduced by more than half relative to the activity in unsupplemented cells [91]. It was found that the wild-type strain was unable to transport either uridine or cytidine. A mutant was isolated that exhibited reduced orotate phosphoribosyltransferase activity compared to its activity in the wild-type strains supplemented with uracil. An increase in aspartate transcarbamoylase and dihydroorotate dehydrogenase activity by more than three-fold, and dihydroorotase and OMP decarboxylase activity approximately 1.5-fold, was found when the mutant cells were subjected to pyrimidine limitation, relative to their activity in the uracil-grown mutant cells [91]. It appeared that the regulation of the pyrimidine biosynthetic enzyme synthesis by a uracil-related metabolite was evident.
In P. oleovorans ATCC 8062, it was observed that the carbon source had an effect on the pyrimidine biosynthetic enzyme activity relative to pyrimidine supplementation [92]. When ATCC 8062 cells were grown on succinate as a carbon source, the supplementation of orotic acid or uracil generally resulted in the five pyrimidine biosynthetic activity being elevated. It was demonstrated that a metabolite of uracil was capable of inducing the synthesis of transcarbamoylase and dihydroorotase. In contrast, the activity of dehydrogenase, phosphoribosyltransferase and decarboxylase was depressed in the glucose-grown ATCC 8062 cells when orotic acid and uracil were supplemented into the medium. Mutants deficient in either aspartate transcarbamoylase or dihydroorotase activity were isolated from P. oleovorans [92]. When the mutant deficient in dihydroorotase activity was subjected to pyrimidine limitation, the carbon source had an effect on the level of pyrimidine biosynthetic enzyme synthesis. It was witnessed that the derepression of dihydroorotase, phosphoribosyltransferase and decarboxylase activity occurred following pyrimidine limitation in the glucose-grown mutant cells but not in the succinate-grown mutant cells. In the mutant strain deficient in transcarbamoylase activity, derepression of the pyrimidine biosynthetic enzyme activity was observed independently of the carbon source utilized, compared to the activity in mutants grown in excess uracil [92]. It has been suggested that P. pseudoalcaligenes ATCC 17440 should be reclassified as P. oleovorans [93]. If the regulation of pyrimidine biosynthesis in P. pseudoalcaligenes and P. oleovorans is compared [92,93], they appear to be dissimilar, despite their DNA homology indicating their taxonomic relatedness.
In P. resinovorans ATCC 14235, transcarbamoylase or dihydroorotase activity appeared to be repressed in succinate-grown cells supplemented with uracil [94]. Orotic acid inclusion in the succinate-grown wild-type cells generally increased the pyrimidine biosynthetic enzyme activity. Orotic acid supplementation of the glucose-grown ATCC 14235 cells also caused an increase in the pyrimidine biosynthetic enzyme activity [94]. Repression by a uracil-related metabolite of pyrimidine biosynthetic enzyme synthesis was indicated when the glucose-grown wild-type strain was supplemented with uracil. An orotate phosphoribosyltransferase was isolated from P. resinovorans and used for pyrimidine limitation experiments. When the succinate-grown mutant strain cells were limited in pyrimidines for 2 h, the pyrimidine biosynthetic enzyme activity was at least doubled compared to the activity in excess-uracil-grown mutant cells [94]. The pyrimidine limitation of glucose-grown mutant cells for 4 h increased the pyrimidine biosynthetic enzyme activity by more than three-fold relative to the activity in mutant cells grown under saturating uracil conditions. The findings showed that the carbon source was a factor in the level of derepression observed for pyrimidine biosynthetic enzyme synthesis in P. resinovorans [94].
2.2. Pseudomonas chlororaphis Homology Group
The species taxonomically assigned to the Pseudomonas chlororaphis group where the regulation of pyrimidine biosynthesis has been investigated previously include Pseudomonas aurantiaca, P. chlororaphis, Pseudomonas fragi, Pseudomonas lundensis and Pseudomonas taetrolens [97,98,99,100,101]. Pyrimidine biosynthesis regulation in the P. chlororaphis homology group species was compared relative to biosynthetic enzyme repression by a uracil-related metabolite or the effect of pyrimidine mutant limitation on enzyme activity (Table 2). Table 2 does not reveal a specific pattern demonstrating relatedness between the species classified in the P. chlororaphis homology group.

Table 2.
Comparison of pyrimidine biosynthetic activity profiles in the Pseudomonas chlororaphis DNA homology group. Abbreviations: AT, aspartate transcarbamoylase; DO, dihydroorotase; DD, dihydroorotate dehydrogenase; OP, orotate phosphoribosyltransferase; and OD, OMP decarboxylase. Number in parentheses following each enzyme listed under effect of uracil addition on carbon source glucose or succinate indicates relative activity in the cells compared to activity in unsupplemented cells. Number in parentheses following each enzyme listed under pyrimidine limitation (2 h) on carbon source glucose or succinate indicates relative activity in the cells compared to activity in uracil-grown cells. ND, not determined. Pseudomonas fragi is unable to utilize succinate as a carbon source.
In P. aurantiaca ATCC 33663 cells, regulation by a pyrimidine-related compound in the repression of pyrimidine biosynthetic enzyme synthesis appeared to be influenced by the carbon source present [97]. When the wild-type strain cells were grown on succinate as a carbon source, orotic acid or uracil supplementation had a repressive effect on the biosynthetic enzyme activity relative to the unsupplemented wild-type cells. Using glucose-grown ATCC 33663 cells, it was observed that only orotic acid supplementation produced a depression in the pathway enzyme activity. Using an orotate phosphoribosyltransferase mutant isolated from P. aurantiaca, pyrimidine limitation experiments conducted using succinate-grown cells produced an elevation in dehydrogenase or decarboxylase activity, with each enzyme’s activity increasing by at least three-fold [97]. Pyrimidine depletion experiments performed with glucose-grown cells of the phosphoribosyltransferase mutant strain caused dihydroorotase and decarboxylase activity to increase by at least double compared to the activity in uracil-grown mutant cells. The regulation of pyrimidine biosynthetic enzyme synthesis by a pyrimidine-related compound did appear to occur in P. aurantiaca [97].
In P. chlororaphis ATCC 17414, the effect of pyrimidine base supplementation on the levels of pyrimidine biosynthetic enzyme synthesis was studied relative to either glucose or succinate as a carbon source [98]. It was demonstrated that transcarbamoylase and decarboxylase activity was repressed by orotic acid or uracil supplementation to the culture medium. A mutant strain was isolated that was deficient in OMP decarboxylase activity and it was subjected to pyrimidine limitation. Pyrimidine limitation of the decarboxylase mutant cells for 1 h caused the derepression of the synthesis of transcarbamoylase, dihydroorotase, dehydrogenase and phosphoribosyltransferase, compared to the activity in the uracil-grown mutant cells [98]. Little derepression of pyrimidine biosynthetic enzyme synthesis in the glucose-grown mutant cells was observed after 2 h of pyrimidine limitation, compared to the activity in the mutant strain grown in saturating uracil [98]. It was concluded that the regulation of pyrimidine biosynthetic enzyme synthesis was dependent on the length of pyrimidine limitation, with longer periods of limitation causing activity to decrease.
In P. fragi ATCC 4973 glucose-grown cells, the effect of the supplementation of dihydroorotate, orotate or uracil on the level of the pyrimidine biosynthetic enzyme activity was investigated [99]. It was noted that dihydroorotate or orotate supplementation caused the depression of phosphoribosyltransferase activity, while uracil supplementation produced a decrease in transcarbamoylase or dihydroorotase activity [99]. When a dihydroorotase mutant strain isolated from ATCC 4973 was limited in pyrimidines for 2 or 4 h, the derepression of transcarbamoylase, dehydrogenase, phosphoribosyltransferase or decarboxylase activity was seen, compared to each enzyme’s activity in uracil-grown mutant cells [99]. Similarly, when an OMP decarboxylase mutant strain was depleted of pyrimidines for 2 or 4 h, transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase activity was also elevated by at least double compared to the uracil-grown mutant cells [99].
In P. lundensis ATCC 49968, pyrimidine supplementation was shown to be dependent on the carbon source utilized [100]. Uracil supplementation of ATCC 49968 cells grown on glucose resulted in the depression of transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase, while orotate supplementation of the glucose-grown cells only elevated the pyrimidine biosynthetic enzyme activity relative to the activity in unsupplemented cells. In succinate-grown ATCC 49968 cells, the supplementation of either orotic acid or uracil caused the depression of only phosphoribosyltransferase activity, while the remaining pyrimidine biosynthetic enzymes’ activity was found to have increased compared to the activity in the uracil-grown cells. It was of interest to learn whether the presence of orotic acid in the culture medium was inducing the synthesis of pyrimidine biosynthetic enzymes. It was confirmed that the synthesis of transcarbamoylase, dihydroorotase or decarboxylase was induced by orotic acid in the glucose-grown ATCC 49968 cells [100]. A transcarbamoylase and a phosphoribosyltransferase mutant strain were isolated from ATCC 49968. Using the transcarbamoylase mutant strain, pyrimidine limitation of the succinate-grown cells for 2 h produced a 3.6-fold increase in transcarbamoylase activity and a 1.9-fold increase in decarboxylase activity compared their activity in the uracil-grown mutant cells [100]. A 1.5-fold or 2.3-fold increase in phosphoribosyltransferase or decarboxylase activity, respectively, was seen in glucose-grown transcarbamoylase mutant cells relative to the activity in uracil-grown mutant cells [100]. It was also demonstrated that dihydroorotase or decarboxylase activity approximately doubled when glucose-grown cells of the mutant cells were supplemented with orotic acid. In the phosphoribosyltransferase mutant strain grown on succinate as a carbon source, pyrimidine limitation for 2 h caused a 2-fold or 3.2-fold increase in dehydrogenase or decarboxylase activity, respectively [100]. When the phosphoribosyltransferase mutant strain was grown on glucose as a carbon source, dihydroorotase, dehydrogenase or decarboxylase activity increased by 3.1-fold, 3.1-fold or 4.4-fold, respectively, after 2 h of pyrimidine limitation, compared to each enzyme’s activity in the uracil-grown mutant cells. For P. lundensis, it was clear that the induction of pyrimidine biosynthetic enzyme synthesis by orotic acid was an important factor in how the pathway was regulated in ATCC 49968 [100].
In succinate-grown P. taetrolens ATCC 4683, orotic acid or uracil supplementation was shown to have differing effects. Orotic acid supplementation produced an increase in the pyrimidine biosynthetic enzyme activity, while uracil supplementation repressed the activity of dihydroorotase, dehydrogenase or decarboxylase compared to their activity in unsupplemented cells [101]. An OMP decarboxylase mutant isolated from P. taetrolens ATCC 4683 was limited in pyrimidines for 2 or 4 h. It was observed that after 2 h of pyrimidine-limiting conditions, dihydroorotase or phosphoribosyltransferase increased by 2.3-fold or 1.6-fold, respectively, relative to each one’s activity in uracil-grown mutant cells [101]. When the mutant cells were starved of pyrimidines for 4 h, dihydroorotase, dehydrogenase or phosphoribosyltransferase activity increased by 1.9-fold, 1.7-fold or 1.4-fold, respectively, compared to each enzyme’s activity in uracil-grown mutant cells [101]. It appeared that a uracil-related metabolite was important in the regulation of pyrimidine biosynthesis in P. taetrolens.
2.3. Pseudomonas fluorescens Homology Group
The regulation of pyrimidine biosynthesis has been investigated in certain species taxonomically assigned to the Pseudomonas fluorescens chlororaphis group, including P. fluorescens, Pseudomonas mucidolens, Pseudomonas reptilivora, Pseudomonas synxantha and Pseudomonas veronii [102,103,104,105,106,107,108,109]. The regulation of pyrimidine biosynthesis in the P. fluorescens homology group species was compared by examining uracil-related metabolite repression or the influence of pyrimidine auxotroph limitation on the biosynthetic enzyme levels (Table 3). Although no pattern is apparent from the data in Table 1, it does appear that the pyrimidine-limited glucose-grown mutant cells produced greater elevations in enzyme activity than did the succinate-grown cells.

Table 3.
Comparison of pyrimidine biosynthetic activity profiles in the Pseudomonas fluorescens DNA homology group. Abbreviations: AT, aspartate transcarbamoylase; DO, dihydroorotase; DD, dihydroorotate dehydrogenase; OP, orotate phosphoribosyltransferase; and OD, OMP decarboxylase. Number in parentheses following each enzyme listed under effect of uracil addition on carbon source glucose or succinate indicates relative activity in the cells compared to activity in unsupplemented cells. Number in parentheses following each enzyme listed under pyrimidine limitation (2 h) on carbon source glucose or succinate indicates relative activity in the cells compared to activity in uracil-grown cells.
The regulation of pyrimidine biosynthesis by pyrimidines in P. fluorescens has been explored in prior studies [102,103]. In P. fluorescens biotype A126, uracil addition to the culture medium containing glucose as a carbon source repressed the pyrimidine biosynthetic enzyme activity compared to their activity in unsupplemented cells [102]. The supplementation of orotic acid to the glucose-grown biotype A126 cell repressed the transcarbamoylase, dehydrogenase, phosphoribosyltransferase and decarboxylase activity, while increasing the dihydroorotase activity by more than double, when compared to the activity in unsupplemented cells [102]. It was shown that dihydroorotase was induced by the presence of orotic acid in the glucose-containing medium [102]. Cells of a dihydroorotase mutant strain isolated from biotype A126 were subjected to pyrimidine limitation for 1 h, which resulted in transcarbamoylase, dehydrogenase and decarboxylase activity at least doubling compared to each one’s activity in uracil-grown mutant cells [102]. The type strain of P. fluorescens biotype A, namely ATCC 13525, was also investigated [103]. Independent of the carbon source used, it was found that uracil supplementation repressed transcarbamoylase, phosphoribosyltransferase or decarboxylase activity in ATCC 13525 cells, while only phosphoribosyltransferase or decarboxylase activity was repressed by orotic acid addition in ATCC 13525 cells, compared to their activity in uracil-grown cells [103]. Similar to the previous study [102], the supplementation of orotic acid to the medium used to grow the ATCC 13525 cells elevated dihydroorotase activity independent of the carbon source [103]. Using a phosphoribosyltransferase mutant strain isolated from ATCC 13525, it was observed that the derepression of pyrimidine biosynthetic enzyme activity was greater if the cells were grown on succinate instead of glucose [103]. After 4 h of pyrimidine limitation in succinate-grown mutant cells, the activity of transcarbamoylase, dihydroorotase and decarboxylase was elevated by 1.6-fold, 5-fold or 3.4-fold, respectively, compared to their activity in uracil-grown mutant cells [103]. The findings from these studies [102,103] seemed to confirm that dihydroorotase synthesis was regulated by a pyrimidine-related compound.
In the taxonomically related species P. jessenii ATCC 700870, the regulation of pyrimidine biosynthesis by a pyrimidine-related compound was examined. The supplementation of orotic acid to succinate-grown ATCC 700870 cells had little effect on the pyrimidine biosynthetic enzyme activity, while a decrease in the levels of transcarbamoylase, dihydroorotase and dehydrogenase was observed when glucose-grown ATCC 700870 cells were supplemented with orotic acid, relative to each one’s activity in unsupplemented cells [105]. In contrast, the uracil supplementation of ATCC 700870 cells reduced the pyrimidine biosynthetic enzyme activity independently of the carbon source, compared to their activity in unsupplemented cells [105]. Pyrimidine limitation of a P. jessenii orotate phosphoribosyltransferase mutant strain for 1 or 2 h produced approximately a two-fold increase in transcarbamoylase, dihydroorotase or decarboxylase activity compared to each one’s activity in uracil-grown mutant cells [105]. This indicated that pyrimidine biosynthesis in ATCC 700870 cells was subject to the regulation by a pyrimidine-related compound.
In a previous work, the regulation of pyrimidine biosynthesis in P. mucidolens ATCC 4685 was investigated [106]. In succinate-grown ATCC 4685 cells, orotic acid or uracil supplementation had a minimal effect upon the levels of the pyrimidine biosynthetic enzyme activity compared to the activity in unsupplemented cells [106]. In the glucose-grown ATCC 4685 cells, uracil supplementation depressed the activity of transcarbamoylase, dehydrogenase, phosphoribosyltransferase or decarboxylase, while orotic acid addition affected the pyrimidine biosynthetic enzyme activity slightly [106]. Pyrimidine limitation of a glucose-grown P. mucidolens orotate phosphoribosyltransferase mutant strain for 2 or 4 h produced more than a two-fold increase in the pyrimidine biosynthetic enzyme activity relative to the activity in uracil-grown mutant cells [106]. When the P. mucidolens orotate phosphoribosyltransferase mutant strain was grown on succinate, the reduced derepression of the pyrimidine biosynthetic enzyme activity was seen [106]. The regulation of pyrimidine biosynthesis in P. mucidolens by a pyrimidine-related compound and a carbon source was indicated by the pyrimidine supplementation and pyrimidine limitation findings.
Pyrimidine biosynthesis was shown to be regulated by pyrimidines in P. reptilivora ATCC 14836 [107]. In succinate-grown ATCC 14836 cells, uracil supplementation lowered transcarbamoylase, dehydrogenase, phosphoribosyltransferase or decarboxylase activity, while orotic acid supplementation was less effective in repressing the pathway enzymes’ activity relative to each one’s activity in unsupplemented cells [107]. In glucose-grown ATCC 14836 cells, orotic acid supplementation demonstrated a greater ability to repress the pyrimidine biosynthetic enzyme activity than if the cells were supplemented with uracil [107]. Independently of the carbon source, the pyrimidine limitation of P. mucidolens aspartate transcarbamoylase mutant cells for 1 or 2 h produced up to a 7-fold, 2.5-fold or 2.4-fold increase in dihydroorotase, phosphoribosyltransferase or decarboxylase activity [107]. Similar to other species assigned within the P. fluorescens group, it appeared that dihydroorotase synthesis was regulated by a pyrimidine-related compound.
In P. synxantha ATCC 9890, a previous investigation explored whether pyrimidine biosynthesis was controlled by a pyrimidine-related compound [108]. Glucose-grown ATCC 9890 cells were impacted by orotic acid or uracil supplementation, with decarboxylase activity being depressed more than the other pyrimidine biosynthetic enzymes’ activity, compared to their activity in unsupplemented cells [108]. In succinate-grown ATCC 9890 cells, orotic acid supplementation was more effective in repressing the pyrimidine biosynthetic enzyme activity than uracil supplementation. Pyrimidine limitation of a succinate-grown P. synxantha orotate phosphoribosyltransferase mutant strain for 4 h produced an increase in the pyrimidine biosynthetic enzyme activity relative to their activity in uracil-grown mutant cells [108]. When the glucose-grown mutant cells were limited in pyrimidines for 4 h, only dihydroorotase, dehydrogenase and decarboxylase activity was elevated compared to their activity in uracil-grown mutant cells [108]. This showed that the carbon source affected the level of pyrimidine biosynthetic enzyme synthesis derepression. The regulation of pyrimidine pathway enzyme synthesis in P. synxantha was derepressed [108] in a similar fashion to P. jessenii [105].
In P. veronii ATCC 700474, the regulation of pyrimidine biosynthesis was influenced by pyrimidine-related metabolites [109]. Independently of the carbon source, orotic acid supplementation appeared to elevate the pyrimidine biosynthetic enzyme activity in ATCC 700474. Similarly, the uracil supplementation of succinate-grown ATCC 700474 cells caused the pyrimidine biosynthetic enzyme activity to increase [109]. In general, in glucose-grown ATCC 700474 cells, uracil supplementation was observed to repress the pyrimidine biosynthetic enzyme activity relative to their activity in unsupplemented cells [109]. Pyrimidine depletion experiments were conducted using a mutant strain isolated from ATCC 700474 that was deficient in decarboxylase activity [109]. When the P. veronii mutant strain was grown on succinate or glucose as a carbon source and subjected to pyrimidine limitation for 1 or 2 h, the pyrimidine biosynthetic enzyme activity was found to be roughly double the pathway enzyme activity detected in the mutant strain grown in excess uracil [109]. It appeared that pyrimidine biosynthesis in P. veronii was transcriptionally regulated by pyrimidines but to a lesser degree than what was observed for other species assigned to the P. fluorescens group [109].
2.4. Pseudomonas putida Homology Group
In the species taxonomically assigned to the Pseudomonas putida homology group, the regulation of pyrimidine biosynthesis has been studied previously in P. putida, Pseudomonas fulva, Pseudomonas monteilii and Pseudomonas oryzihabitans [110,111,112,113,114,115,116]. In Table 4, the regulation of pyrimidine biosynthesis in the P. putida homology group species relative to enzyme repression by a uracil-related metabolite or the effect of pyrimidine limitation of pyrimidine-requiring mutants on the pyrimidine biosynthetic enzyme activity is shown. No conclusive pattern emerged when comparing the species assigned to the P. putida homology group (Table 4). The regulation of pyrimidine biosynthesis by pyrimidines has been investigated in P. putida biotypes A and B [110,111,112]. In P. putida biotype A90 cells, it was reported that the addition of uracil to an asparagine-containing medium exhibited no repressive effect upon the pyrimidine biosynthetic enzyme activity [110]. The possible derepression of the pyrimidine biosynthetic enzyme activity was studied using the pyrimidine limitation of an OMP decarboxylase mutant strain isolated from biotype A90 cells. It was found that the activity of transcarbamoylase, dihydroorotase, dehydrogenase and phosphoribosyltransferase was elevated by at least 1.6-fold after pyrimidine limitation of the mutant cells grown in the asparagine-containing medium [110]. In P. putida ATCC 17536, characterized as a biotype B strain, the control of pyrimidine biosynthesis by pyrimidines was also studied [111]. It appeared that the regulation of the pyrimidine biosynthetic activity was significantly affected by the carbon source present. When the ATCC 17536 cells were grown on the carbon source succinate, uracil supplementation only repressed transcarbamoylase activity, while orotic acid supplementation repressed the levels of transcarbamoylase, dehydrogenase, phosphoribosyltransferase and decarboxylase activity [111]. In glucose-grown ATCC 17536 cells, orotic acid or uracil supplementation repressed all the pyrimidine biosynthetic enzymes’ activity, compared to their activity in the unsupplemented cells. When the glucose-grown dihydroorotase mutant cells were subjected to pyrimidine-limiting conditions for 4 h, it was shown that the pyrimidine biosynthetic enzyme activity was elevated up to eight-fold compared to their activity in the uracil-grown mutant cells [111]. It was demonstrated that orotic acid induced dehydrogenase, phosphoribosyltransferase or decarboxylase activity in glucose-grown dihydroorotase mutant cells [111]. In succinate-grown cells of the dihydroorotase mutant, pyrimidine limitation for 4 h resulted in more than a three-fold elevation in phosphoribosyltransferase and decarboxylase activity, but little derepression of transcarbamoylase and dehydrogenase activity was seen [111]. When a mutant strain deficient in phosphoribosyltransferase activity was starved of pyrimidine for 4 h, transcarbamoylase, dihydroorotase and dehydrogenase activity was not affected by pyrimidine depletion, while decarboxylase activity roughly doubled, independent of the carbon source used [111]. Regardless of the biotype of P. putida, the regulation of pyrimidine biosynthesis by the level of pyrimidine ribonucleotides was indicated.

Table 4.
Comparison of pyrimidine biosynthetic activity profiles in the Pseudomonas putida DNA homology group. Abbreviations: AT, aspartate transcarbamoylase; DO, dihydroorotase; DD, dihydroorotate dehydrogenase; OP, orotate phosphoribosyltransferase; and OD, OMP decarboxylase. Number in parentheses following each enzyme listed under effect of uracil addition on carbon source glucose or succinate indicates relative activity in the cells compared to activity in unsupplemented cells. Number in parentheses following each enzyme listed under pyrimidine limitation (2 h) on carbon source glucose or succinate indicates relative activity in the cells compared to activity in uracil-grown cells.
Pyrimidine biosynthesis was examined in P. fulva ATCC 31418 for regulation by a pyrimidine-related compound [113]. The supplementation of orotic acid to the ATCC 31418 cells exhibited little repression of the pyrimidine biosynthetic enzyme activity, independent of the carbon source utilized, relative to their activity in unsupplemented cells. The addition of uracil to glucose-grown ATCC 31418 cells depressed the activity of dihydroorotase, phosphoribosyltransferase and decarboxylase, while the uracil supplementation of succinate-grown ATCC 31418 cells diminished the activity of dihydroorotase and dehydrogenase [113]. Pyrimidine limitation studies of a decarboxylase mutant strain isolated from ATCC 31418 were conducted. When glucose-grown cells of the mutant strain were limited in pyrimidines for 4 h, it was noted that transcarbamoylase, dihydroorotase, dehydrogenase or phosphoribosyltransferase activity was elevated by 5.5-fold, 13.1-fold, 2.5-fold or 10.6-fold, respectively, compared to their enzyme activity in the mutant cells grown in excess uracil [113]. In contrast, no derepression of pyrimidine biosynthetic enzyme activity in succinate-grown mutant cells limited for 4 h was observed relative to the mutant cells grown with saturating uracil conditions [113]. The carbon source appeared to be an important factor in the regulation of pyrimidine biosynthesis by pyrimidines in P. fulva cells.
Another species investigated for the regulation of the pyrimidine biosynthetic pathway by pyrimidines was P. monteilii ATCC 700476 [114]. In succinate-grown ATCC 700476 cells, orotic acid or uracil supplementation produced a depression in the pyrimidine biosynthetic enzyme activity compared to the wild-type strain grown in unsupplemented succinate minimal medium [114]. In glucose-grown ATCC 700476 cells, orotic acid addition caused transcarbamoylase, dihydroorotase, dehydrogenase or decarboxylase activity to decrease relative to each one’s activity in unsupplemented cells [114]. Similarly, uracil addition to succinate-grown ATCC 700476 cells decreased transcarbamoylase, dehydrogenase, phosphoribosyltransferase or decarboxylase activity compared to each one’s activity in unsupplemented cells. A strain deficient in dihydroorotase or orotate phosphoribosyltransferase activity was isolated from ATCC 700476 [114]. In the glucose-grown cells of the dihydroorotase mutant strain, pyrimidine limitation for 2 or 4 h only produced a slight increase in dehydrogenase activity. The succinate-grown cells of the dihydroorotase mutant cells limited in pyrimidines for 4 h showed the 4.8-fold derepression of transcarbamoylase activity relative to its activity in uracil-grown mutant cells [114]. Interestingly, pyrimidine limitation of the glucose-grown phosphoribosyltransferase mutant cells for 4 h caused all the pyrimidine biosynthetic enzymes’ activity to increase, with the decarboxylase activity being elevated by eight-fold [114]. Similarly, pyrimidine limitation of the succinate-grown phosphoribosyltransferase mutant cells for 4 h caused all the pyrimidine biosynthetic enzymes’ activity to increase by at least double compared to their activity in uracil-grown mutant cells [114]. It appeared that a deficiency in phosphoribosyltransferase activity led to greater pyrimidine biosynthetic enzyme derepression following pyrimidine nucleotide depletion than if the mutant strain was deficient in dihydroorotase activity.
In P. oryzihabitans ATCC 43272, pyrimidine biosynthesis was investigated to determine whether the carbon source and pyrimidines were factors in controlling the pathway enzymes [115,116]. It was witnessed that orotic acid supplementation of the ATCC 43272 cells affected the pyrimidine biosynthetic enzyme activity very little, independent of the carbon source utilized, but generally increased their activity [115,116]. Similarly, uracil supplementation of the succinate-grown ATCC 43272 cells produced a slight elevation in the pyrimidine biosynthetic enzyme activity compared to the activity in unsupplemented cells. It appeared that uracil-grown ATCC 43272 cells did repress the dehydrogenase and phosphoribosyltransferase activity, while the other pathway enzymes’ activity was unchanged when glucose served as the carbon source, relative to the activity in unsupplemented cells. Pyrimidine limitation experiments were performed using an OMP decarboxylase mutant strain isolated from ATCC 43272 cells [115,116]. Pyrimidine limitation of the glucose-grown mutant cells for 1 h resulted in approximately a two-fold increase in dihydroorotase and dehydrogenase activity, compared to each one’s activity in uracil-grown mutant cells [116]. On the other hand, pyrimidine limitation of the succinate-grown decarboxylase mutant cells for 1 or 2 h produced approximately a three-fold elevation in transcarbamoylase or dihydroorotase activity relative to each one’s activity in uracil-grown mutant cells. The carbon source did seem to influence the degree of derepression of pyrimidine biosynthetic enzyme synthesis observed. Similar to P. monteilii [114], the greater derepression of pyrimidine biosynthetic enzyme synthesis was seen in the succinate-grown cells compared to the glucose-grown cells.
3. Pyrimidine Ribonucleoside Metabolism
3.1. Pseudomonas aeruginosa Homology Group
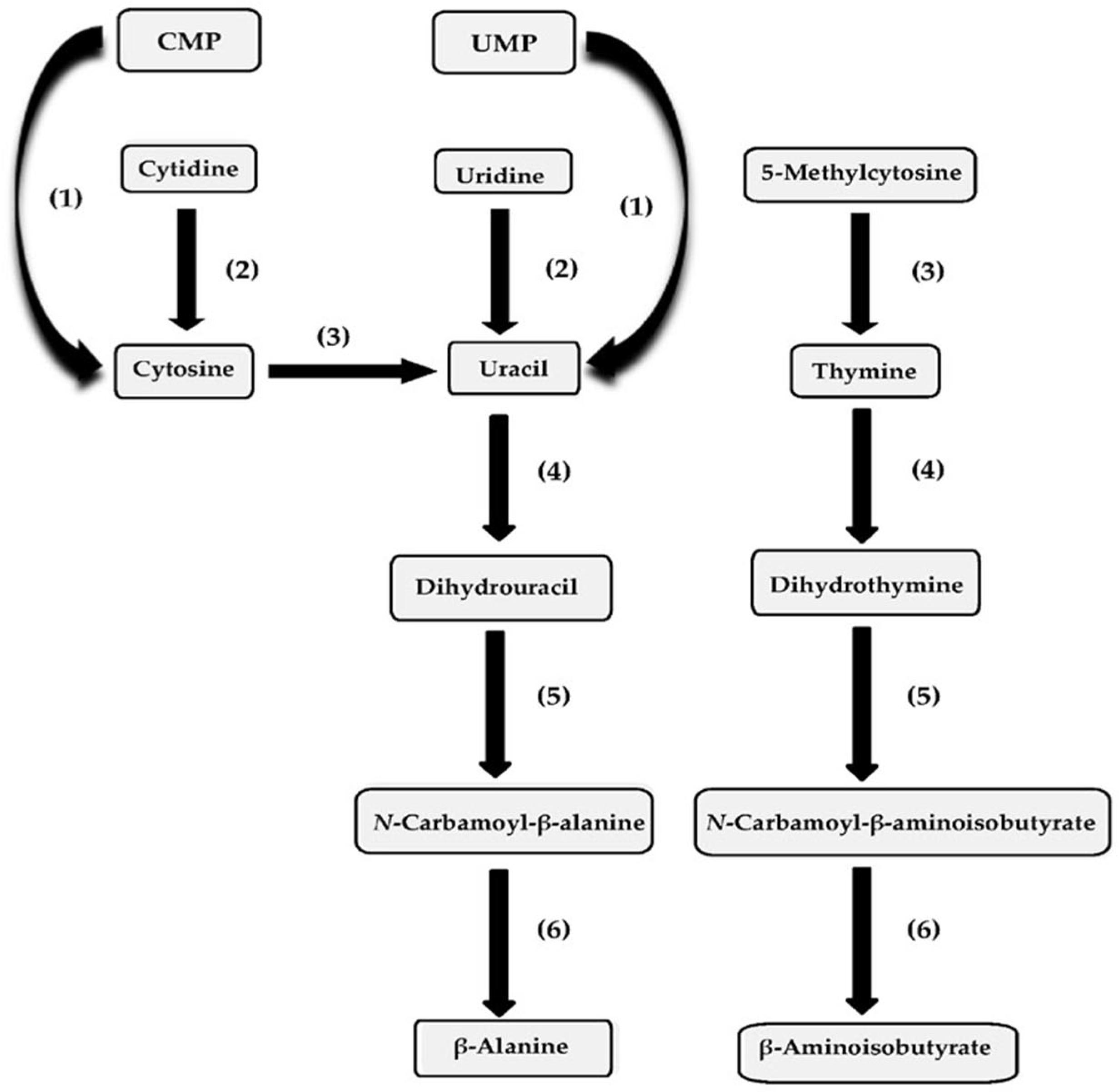
It has been determined that a number of pyrimidine salvage enzymes can be involved in pyrimidine ribonucleoside metabolism in species of Pseudomonas and taxonomically related species [117,118,119]. In Figure 2, the relationship between pyrimidine ribonucleoside metabolism and subsequent pyrimidine base catabolism is shown. With respect to comparing the kinetic parameters of the pyrimidine biosynthetic enzymes’ specific activity with those of the pyrimidine ribonucleoside catabolic-specific activity, the levels of the catabolic-specific activity tended to be higher, with their enzyme synthesis usually being inducible. In the P. aeruginosa homology group, pyrimidine ribonucleoside metabolism in the species P. aeruginosa, P. mendocina, P. alcaligenes, P. pseudoalcaligenes and P. oleovorans has been investigated. A number of studies have been conducted investigating pyrimidine ribonucleoside metabolism in Pseudomonas species assigned to this homology group. In P. aeruginosa ATCC 15692, the enzymes nucleoside hydrolase and cytosine deaminase were found to be active [118,120]. In ATCC 15692 cells, catabolite repression of nucleoside hydrolase synthesis by succinate as a carbon source was evident, since its activity with glucose or ribose as a carbon source was approximately doubled [120]. When ammonium sulfate served as the nitrogen source, it was shown that nucleoside hydrolase activity in ATCC 15692 cells increased by 1.4-fold, 2-fold, 1.7-fold or 1.7-fold if cytosine, 5-methylcytosine, uridine or cytidine, respectively, was substituted for glucose as the carbon source [120]. When ammonium sulfate served as the nitrogen source, the cytosine deaminase activity in ATCC 15692 cells was higher when succinate or ribose was the carbon source, compared to glucose as the carbon source [120]. If uracil, cytosine, 5-methylcytosine, thymine, uridine or cytidine was substituted for ammonium sulfate as a nitrogen source in glucose-grown ATCC 15692 cells, it was observed that the cytosine deaminase activity was elevated by 19.3-fold, 5.3-fold, 20.8-fold, 15.9-fold, 2.6-fold or 8.2-fold, respectively, relative to the deaminase activity in the ammonium sulfate-grown cells [120].
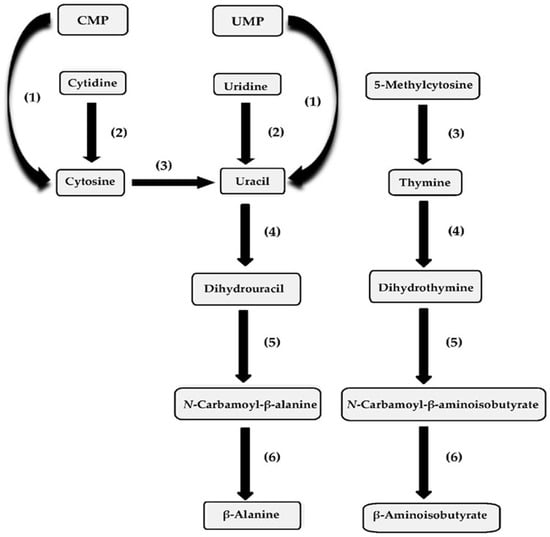
Figure 2.
Pyrimidine base, ribonucleoside and mononucleotide metabolism in Pseudomonas species and the catabolic enzymes involved in their metabolism. The enzymes include (1) pyrimidine nucleotide N-ribosidase; (2) nucleoside hydrolase; (3) cytosine deaminase; (4) dihydropyrimidine dehydrogenase; (5) dihydropyrimidinase; and (6) β-ureidopropionase.
Both nucleoside hydrolase and cytosine deaminase activity was detected in P. mendocina ATCC 25411 cells grown in a medium containing glucose as a carbon source and ammonium sulfate as a nitrogen source [121]. If asparagine was substituted for ammonium sulfate as a nitrogen source in glucose-grown ATCC 25411 cells, both nucleoside hydrolase and cytosine deaminase activity increased slightly [121]. When glycerol was substituted for glucose as a carbon source in ammonium sulfate-grown ATCC 25411 cells, nucleoside hydrolase activity was elevated by more than five-fold, while cytosine deaminase activity remained unaffected, compared to their activity in the cells grown on glucose as a carbon source and ammonium sulfate as a nitrogen source [121].
In P. alcaligenes ATCC 14909, nucleoside hydrolase and cytosine deaminase activity was measurable in cells grown in a medium containing succinate as a carbon source and ammonium sulfate as a nitrogen source [122]. Interestingly, the substitution of asparagine or ammonium sulfate as a nitrogen source in the succinate-grown cells depressed both the hydrolase and deaminase activity by more than four-fold compared to their activity in the ammonium sulfate-grown cells [122].
The presence of nucleoside hydrolase and cytosine deaminase in P. pseudoalcaligenes ATCC 17440 cells grown on succinate as a carbon source and ammonium sulfate as a nitrogen source was studied [122]. It was observed that nucleoside hydrolase activity dropped if asparagine was substituted for ammonium sulfate as a nitrogen source in succinate-grown ATCC 17440 [122]. Nucleoside hydrolase was elevated by at least 1.6-fold if cytosine or uracil was substituted for ammonium sulfate in the succinate-grown cells [122]. The substitution of asparagine, cytosine or uracil for ammonium sulfate as a nitrogen source caused an elevation of 1.6-fold, 5.0-fold or 2.1-fold, respectively, in the cells grown on succinate as a carbon source [122]. Clearly, cytosine deaminase synthesis was more influenced by the nitrogen source than was nucleoside hydrolase synthesis.
Considering that P. oleovorans cells contain pyrimidine nucleotide N-ribosidase activity instead of nucleoside hydrolase, pyrimidine ribonucleoside metabolism in P. oleovorans differed from the metabolism seen in the other species of the P. aeruginosa homology group [123,124,125,126]. It was found that the nitrogen source influenced the levels of pyrimidine ribonucleotide N-ribosidase and cytosine deaminase in succinate-grown P. oleovorans ATCC 8062 cells [126]. When ATCC 8062 cells were grown on dihydrouracil as a nitrogen source and succinate as a carbon source, the nucleoside hydrolase activity was increased by 1.9-fold compared to its activity in cells grown on succinate as a carbon source and ammonium sulfate as a nitrogen source [126]. The cytosine deaminase activity in ATCC 8062 cells was shown to be more strongly influenced by the nitrogen source than was pyrimidine ribonucleotide N-ribosidase [125]. The cytosine deaminase activity increased by 2.5-fold, 4.8-fold or 2.3-fold, respectively, when cytosine, dihydrouracil or dihydrothymine served as a nitrogen source and succinate served as the carbon source [126]. It should be noted that the synthesis of both pyrimidine ribonucleotide N-ribosidase and cytosine deaminase was increased when dihydrouracil was the nitrogen source and succinate was the carbon source [126]. A prior study indicated that P. pseudoalcaligenes should be classified taxonomically as a strain of P. oleovorans [93]. Although both species are taxonomically related, it is clear that pyrimidine ribonucleoside metabolism in P. pseudoalcaligenes and P. oleovorans is distinctly different.
3.2. Pseudomonas chlororaphis Homology Group
Pyrimidine ribonucleoside metabolism has been investigated to a limited extent in P. chloroaphis, P. fragi and P. taetrolens. In a single study [117], it has been reported that P. chlororaphis, P. fragi and P. taetrolens contains the enzymes nucleoside hydrolase and cytosine deaminase, which, in combination, degrade cytidine to cytosine and ribose.
3.3. Pseudomonas fluorescens Homology Group
It was found that nucleoside hydrolase and cytosine deaminase were active in P. fluorescens [118,127,128]. Nucleoside hydrolase and cytosine deaminase activity was detected in P. fluorescens biotype A126 cells grown on a medium containing glucose, succinate, glycerol or ribose as a carbon source and ammonium sulfate as a nitrogen source [127,128]. It was observed that the nucleoside hydrolase activity in A126 cells was elevated by nearly nine-fold on a medium containing ribose as a carbon source, compared to the activity in succinate-grown cells [128]. When uridine or cytidine served as a carbon source in an ammonium sulfate-containing medium, the nucleoside hydrolase activity in P. fluorescens was observed to have increased by 16-fold or nearly 21-fold, respectively, relative to its activity in succinate-grown cells [128]. The cytosine deaminase activity in the P. fluorescens A126 cells was influenced by the type of nitrogen source included in a glucose-containing medium. It was noted that the substitution of the amino acid asparagine for ammonium sulfate as a nitrogen source produced a 2.5-fold increase in cytosine deaminase activity in A126 cells. When cytosine was substituted for ammonium sulfate in a glucose-containing medium, the cytosine deaminase activity in A126 cells was increased by 3.4-fold [128]. It was concluded that the nucleoside hydrolase synthesis in P. fluorescens was influenced by the carbon source present, while the nitrogen source was a greater factor in regulating the synthesis of cytosine deaminase. In biotype F cells of P. fluorescens, it was seen in ATCC 12983 cells that nucleoside hydrolase activity was not affected by the carbon source [129]. In contrast, the substitution of asparagine for ammonium sulfate as a nitrogen source in a glucose-containing medium resulted in a 1.9-fold increase in cytosine deaminase activity [129]. It may be that the nitrogen source regulation of cytosine deaminase synthesis may be what the two biotypes of P. fluorescens have in common.
3.4. Pseudomonas putida Homology Group
In the P. putida homology group, the only species in which pyrimidine ribonucleoside metabolism has been explored is P. putida. It was determined that cytidine was degraded by the enzymes nucleoside hydrolase, uridine phosphorylase and cytosine deaminase [118]. Another study found that the pyrimidine base cytosine served as a chemoattractant for P. putida, with cytosine deaminase degrading cytosine to uracil, which provides a nitrogen source to the cells [130,131].
4. Pyrimidine Base Catabolism
4.1. Pseudomonas aeruginosa Homology Group
It has been determined that the reductive pathway of pyrimidine catabolism is active in many pseudomonads and taxonomically related species [132]. The pyrimidine reductive pathway enzymes include dihydropyrimidine dehydrogenase, dihydropyrimidinase and β-ureidopropionase (Table 2), which were found to be active in P. aeruginosa PAO1 cell extracts [133]. It was also noted that the carbon source present affected the levels of the reductive catabolic enzymes very little [133]. The induction of reductive pathway enzyme synthesis in PAO1 following growth on uracil as a nitrogen source was demonstrated [133]. Growth on thymine, dihydrouracil or dihydrothymine as a nitrogen source generally had little impact on the pyrimidine reductive pathway enzyme activity in PAO1 [133]. The products of the reductive pathway are β-alanine and β-aminoisobutyric acid, which are subject to transamination to release nitrogen for growth.
The pyrimidine base reductive catabolic enzymes dihydropyrimidine dehydrogenase and dihydropyrimidinase were found to be active in P. pseudoalcaligenes ATCC 17440 and P. alcaligenes ATCC 14909 grown on succinate as a carbon source and ammonium sulfate as a nitrogen source [122]. When asparagine was substituted for ammonium sulfate as a nitrogen source for the ATCC 17440 cells, the dehydrogenase and dihydropyrimidinase activity was elevated [122]. If cytosine, uracil, thymine or dihydrothymine served as a nitrogen source, dehydrogenase activity rose in ATCC 17440 cells by at least three-fold compared to its activity in ammonium sulfate-grown cells [122]. Relative to ammonium sulfate-grown ATCC 17440 cells, the dihydropyrimidinase activity increased by at least 27-fold when cytosine, uracil, thymine, dihydrouracil or dihydrothymine was considered [122]. In P. alcaligenes ATCC 14909, dehydrogenase and dihydropyrimidinase activity was measurable in asparagine-grown cells [122].
4.2. Pseudomonas chlororaphis Homology Group
Only a single study has explored pyrimidine base catabolism in P. chlororaphis ATCC 17414 [134], and it was shown that the reductive catabolic enzymes dihydropyrimidine dehydrogenase and dihydropyrimidinase were active in cells grown on glucose as a carbon source and ammonium sulfate as a nitrogen source [134]. It was found that the growth of ATCC 17414 on uracil, thymine, dihydrouracil or dihydrothymine as a nitrogen source and glucose as a carbon source increased the dihydropyrimidine dehydrogenase activity by at least 14-fold compared to its activity in ammonium sulfate-grown cells [134]. The growth of ATCC 17414 cells on uracil or dihydrothymine as a nitrogen source and glucose as a carbon source caused a 16.1-fold or 122-fold change, respectively, in dihydropyrimidinase activity relative to its activity in ammonium sulfate-grown cells [134]. In ATCC 17414 cells, uracil or dihydrothymine as a nitrogen source appeared responsible for stimulating pyrimidine degradation in P. chlororaphis [134].
4.3. Pseudomonas fluorescens Homology Group
A single study has examined pyrimidine base catabolism in P. fluorescens biotype A126 cells, and the three pyrimidine reductive pathway enzymes were active in cells grown on succinate as a carbon source and ammonium sulfate as a nitrogen source [135]. When uracil, dihydrouracil or β-aminoisobutyric acid served as a nitrogen source, succinate-grown cells showed at least a two-fold increase in dehydrogenase activity, compared to its activity in succinate-grown cells utilizing ammonium sulfate as a nitrogen source [135]. When succinate-grown biotype A126 cells utilized uracil, cytosine, thymine, dihydrouracil, dihydrothymine, β-alanine or β-aminoisobutyric acid as a nitrogen source, dihydropyrimidinase or β-ureidopropionase activity was elevated by more than two-fold or five-fold, respectively, relative to its activity in succinate-grown cells utilizing ammonium sulfate as a nitrogen source [135].
4.4. Pseudomonas putida Homology Group
It was determined that the reductive pathway enzymes of pyrimidine catabolism were active in P. putida ATCC 17536 [136]. When uracil or thymine served as a nitrogen source for succinate-grown ATCC 17536 cells, it was observed that dihydropyrimidine dehydrogenase activity increased by 4.6-fold or 2.6-fold, respectively, compared to its activity in succinate-grown cells using ammonium sulfate as a nitrogen source [136]. It has been determined that a pydR protein regulates the expression of dihydropyrimidine dehydrogenase synthesis in P. putida [137]. The dihydropyrimidinase activity in succinate-grown cells utilizing uracil, thymine, dihydrouracil or dihydrothymine as a nitrogen source increased by more than 12-fold relative to its activity in cells grown on ammonium sulfate as a nitrogen source [136]. It was seen that β-ureidopropionase activity more than doubled when uracil or β-alanine served as the nitrogen source for succinate-grown ATCC 17536 cells, compared to the amidohydrolase activity in ammonium sulfate-grown cells [136]. The induction of pyrimidine reductive pathway enzyme synthesis by growth on uracil as a nitrogen source in succinate-grown cells was confirmed [136]. It did appear that the activity of dihydropyrimidinase was elevated to a greater degree than the dehydrogenase or β-ureidopropionase activity when grown on pyrimidine bases or dihydropyrimidine bases as a source of nitrogen. In P. putida strain RU-KM3S, it was shown that dihydropyrimidinase and β-ureidopropionase were subject to transcriptional regulation by hydantoin induction, which was subject to carbon catabolite repression [138].
5. Conclusions
It can be concluded from a comparison of the regulation of pyrimidine biosynthesis, pyrimidine ribonucleoside metabolism or pyrimidine base catabolism in species of Pseudomonas, based on their taxonomic assignments involving RNA-DNA hybridization studies, that there is no correlation between metabolic regulation and taxonomic homology group assignment. Instead, there may be some similarities in the regulation of pyrimidine metabolism relative to the individual species within the homology group to which a species has been assigned. It may be that pyrimidine metabolism cannot be used for the taxonomic differentiation of Pseudomonas species. It should also be noted that far more studies have been conducted into the regulation of pyrimidine biosynthesis in Pseudomonas species than those that have explored pyrimidine ribonucleoside and base catabolism. There is a need for additional studies to be conducted into pyrimidine ribonucleoside and base catabolism in species of Pseudomonas if a more thorough analysis of any correlation between the regulation of pyrimidine ribonucleoside metabolism and its taxonomic assignment within a homology group can be made.
Funding
This research was funded by the Welch Foundation Grant T-0014.
Data Availability Statement
Data sharing not applicable.
Acknowledgments
The author wishes to acknowledge the mentorship provided by the late Gerry O’Donovan in encouraging the study of pyrimidine metabolism in microorganisms.
Conflicts of Interest
The author declares no conflict of interest.
References
- Yates, R.A.; Pardee, A.B. Pyrimidine biosynthesis in Escherichia coli. J. Biol. Chem. 1956, 221, 743–756. [Google Scholar] [CrossRef]
- Beckwith, J.R.; Pardee, A.B.; Austrian, R.; Jacob, F. Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J. Mol. Biol. 1962, 5, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Hayward, W.S.; Belser, W.L. Regulation of pyrimidine biosynthesis in Serratia marcescens. Proc. Natl. Acad. Sci. USA 1965, 53, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Neuhard, J. Pyrimidine nucleotide metabolism and pathways of thymidine triphosphate biosynthesis in Salmonella typhimurium. J. Bacteriol. 1968, 96, 1519–1527. [Google Scholar] [CrossRef]
- O’Donovan, G.A.; Neuhard, J. Pyrimidine metabolism in microorganisms. Bacteriol. Rev. 1970, 34, 278–343. [Google Scholar] [CrossRef]
- Beck, C.F.; Ingraham, J.L.; Neuhard, J.; Thomassen, E. Metabolism of pyrimidines and pyrimidine nucleosides by Salmonella typhimurium. J. Bacteriol. 1972, 110, 219–228. [Google Scholar] [CrossRef]
- O’Donovan, G.A.; Gerhart, J.C. Isolation and partial characterization of regulatory mutants of the pyrimidine pathway in Salmonella typhimurium. J. Bacteriol. 1972, 109, 1085–1096. [Google Scholar] [CrossRef]
- Schwartz, M.; Neuhard, J. Control of expression of the pyr genes in Salmonella typhimurium: Effects of variations in uridine and cytidine nucleotide pools. J. Bacteriol. 1975, 121, 814–822. [Google Scholar] [CrossRef]
- Kelln, R.A.; Kinahan, J.J.; Foltermann, K.F.; O’Donovan, G.A. Pyrimidine biosynthetic enzymes of Salmonella typhimurium, repressed specifically by growth in the presence of cytidine. J. Bacteriol. 1975, 124, 764–774. [Google Scholar] [CrossRef]
- Jyssum, S. Pyrimidine biosynthesis in Neisseria meningitidis: 2. Regulation of enzyme synthesis. Acta Pathol. Microbiol. Scand. B 1983, 91, 257–260. [Google Scholar] [CrossRef]
- Nowlan, S.F.; Kantrowitz, E.R. Identification of a trans-acting regulatory factor involved in the control of the pyrimidine pathway in E. coli. Mol. Genet. Genom. 1983, 192, 264–271. [Google Scholar] [CrossRef]
- Turnbough, C.L., Jr. Regulation of Escherichia coli aspartate transcarbamylase synthesis by guanosine tetraphosphate and pyrimidine ribonucleoside triphosphates. J. Bacteriol. 1983, 153, 998–1007. [Google Scholar] [CrossRef]
- Bouvier, J.; Patte, J.C.; Stragier, P. Multiple regulatory signals in the control region of the Escherichia coli carAB operon. Proc. Natl. Acad. Sci. USA 1984, 81, 4139–4143. [Google Scholar] [CrossRef]
- Levin, H.L.; Schachman, H.K. Regulation of aspartate transcarbamoylase synthesis in Escherichia coli: Analysis of deletion mutations in the promoter region of the pyrBI operon. Proc. Natl. Acad. Sci. USA 1985, 82, 4643–4647. [Google Scholar] [CrossRef]
- Wilson, H.R.; Chan, P.T.; Turnbough, C.L., Jr. Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J. Bacteriol. 1987, 169, 3051–3058. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.F. Regulation of Salmonella typhimurium pyr gene expression: Effect of changing both purine and pyrimidine nucleotide pools. Microbiology 1989, 135, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Turnbough, C.L., Jr. Multiple control mechanisms for pyrimidine-mediated regulation of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 1989, 171, 3337–3342. [Google Scholar] [CrossRef]
- O’Donovan, G.A.; Herlick, S.; Beck, D.E.; Dutta, P.K. UTP/CTP ratio, an important regulatory parameter for ATCase expression. Arch. Microbiol. 1989, 153, 19–25. [Google Scholar] [CrossRef]
- Choi, K.Y.; Zalkin, H.O. Regulation of Escherichia coli pyrC by the purine regulon repressor protein. J. Bacteriol. 1990, 172, 3201–3207. [Google Scholar] [CrossRef]
- Wilson, H.R.; Turnbough, C.L., Jr. Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J. Bacteriol. 1990, 172, 3208–3213. [Google Scholar] [CrossRef]
- Baker, K.E.; Ditullio, K.P.; Neuhard, J.; Kelln, R.A. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli requires the dicarboxylate transport protein encoded by dctA. J. Bacteriol. 1996, 178, 7099–7105. [Google Scholar] [CrossRef] [PubMed]
- Kholti, A.; Charlier, D.; Gigot, D.; Huysveld, N.; Roovers, M.; Glansdorff, N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J. Mol. Biol. 1998, 280, 571–582. [Google Scholar] [CrossRef]
- Rodríguez, M.; Good, T.A.; Wales, M.E.; Hua, J.P.; Wild, J.R. Modeling allosteric regulation of de novo pyrimidine biosynthesis in Escherichia coli. J. Theor. Biol. 2005, 234, 299–310. [Google Scholar] [CrossRef]
- Turnbough, C.L., Jr.; Switzer, R.L. Regulation of pyrimidine biosynthetic gene expression in bacteria: Repression without repressors. Microbiol. Mol. Biol. 2008, 72, 266–300. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Terol, G.; Gallego-Jara, J.; Sola-Martínez, R.A.; Ortega, Á.; Martinez Vivancos, A.; Canovas Diaz, M.; de Diego Puente, T. Regulation of the pyrimidine biosynthetic pathway by lysine acetylation of E. coli OPRTase. FEBS J. 2023, 290, 442–464. [Google Scholar] [CrossRef] [PubMed]
- Akberdin, I.R.; Kozlov, K.N.; Kazantsev, F.V.; Fadeev, S.I.; Likhoshvai, V.A.; Khlebodarova, T.M. Impact of negative feedbacks on de novo pyrimidines biosynthesis in Escherichia coli. Int. J. Mol. Sci. 2023, 24, 4806. [Google Scholar] [CrossRef]
- Waleh, N.S.; Ingraham, J.L. Pyrimidine ribonucleoside monophosphokinase and the mode of RNA turnover in Bacillus subtilis. Arch. Microbiol. 1976, 110, 49–54. [Google Scholar] [CrossRef]
- Lerner, C.G.; Stephenson, B.T.; Switzer, R.L. Structure of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster. J. Bacteriol. 1987, 169, 2202–2206. [Google Scholar] [CrossRef]
- Switzer, R.L.; Turner, R.J.; Lu, Y. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: Control of gene expression by an mRNA-binding protein. Prog. Nucleic Acid Res. Mol. Biol. 1988, 62, 329–367. [Google Scholar]
- Asahi, S.; Doi, M.; Tsunemi, Y.; Akiyama, S.I. Regulation of pyrimidine nucleotide biosynthesis in cytidine deaminase-negative mutants of Bacillus subtilis. Agric. Biol. Chem. 1989, 53, 97–102. [Google Scholar] [CrossRef]
- Quinn, C.L.; Stephenson, B.T.; Switzer, R.L. Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J. Biol. Chem. 1991, 266, 9113–9127. [Google Scholar] [CrossRef] [PubMed]
- Ghim, S.Y.; Neuhard, J. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J. Bacteriol. 1994, 176, 3698–3707. [Google Scholar] [CrossRef]
- Ghim, S.Y.; Nielsen, P.; Neuhard, J. Molecular characterization of pyrimidine biosynthesis genes from the thermophile Bacillus caldolyticus. Microbiology 1994, 140, 479–491. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Turner, R.J.; Lu, Y.; Switzer, R.L. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 1994, 176, 3708–3722. [Google Scholar] [CrossRef]
- Lu, Y.; Turner, R.J.; Switzer, R.L. Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol. 1995, 177, 1315–1325. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Andersen, P.S.; Martinussen, J.; Hammer, K. Sequence analysis and identification of the pyrKDbF operon from Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J. Bacteriol. 1996, 178, 5005–5012. [Google Scholar] [CrossRef][Green Version]
- Elagöz, A.; Abdi, A.; Hubert, J.C.; Kammerer, B. Structure and organisation of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: A PCR strategy for sequencing without cloning. Gene 1996, 182, 37–43. [Google Scholar] [CrossRef]
- Martinussen, J.; Schallert, J.; Andersen, B.; Hammer, K. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 2001, 183, 2785–2794. [Google Scholar] [CrossRef]
- Thia-Toong, T.L.; Roovers, M.; Durbecq, V.; Gigot, D.; Glansdorff, N.; Charlier, D. Genes of de novo pyrimidine biosynthesis from the hyperthermoacidophilic crenarchaeote Sulfolobus acidocaldarius: Novel organization in a bipolar operon. J. Bacteriol. 2002, 184, 4430–4441. [Google Scholar] [CrossRef]
- Buvelot, H.; Roth, M.; Jaquet, V.; Lozkhin, A.; Renzoni, A.; Bonetti, E.J.; Krause, K.H. Hydrogen peroxide affects growth of S. aureus through downregulation of genes involved in pyrimidine biosynthesis. Front. Immunol. 2021, 12, 673985. [Google Scholar] [CrossRef]
- Chen, S.; He, X.; Qin, Z.; Li, G.; Wang, W.; Nai, Z.; Tian, Y.; Liu, D.; Jiang, X. Loss in the antibacterial ability of a PyrR gene regulating pyrimidine biosynthesis after using CRISPR/Cas9-mediated knockout for metabolic engineering in Lactobacillus casei. Microorganisms 2023, 11, 2371. [Google Scholar] [CrossRef] [PubMed]
- Lovatt, C.J.; Albert, L.S.; Tremblay, G.C. Regulation of pyrimidine biosynthesis in intact cells of Cucurbita pepo. Plant Physiol. 1979, 64, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Jacques, S.; Sung, Z.R. Regulation of pyrimidine and arginine biosynthesis investigated by the use of phaseolotoxin and 5-fluorouracil. Plant Physiol. 1981, 67, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Giermann, N.; Schröder, M.; Ritter, T.; Zrenner, R. Molecular analysis of de novo pyrimidine synthesis in solanaceous species. Plant Mol. Biol. 2002, 50, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Boldt, R.; Zrenner, R. Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant. 2003, 117, 297–304. [Google Scholar] [CrossRef]
- Huang, M.; Graves, L.M. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci. 2003, 60, 321–336. [Google Scholar] [CrossRef]
- Sigoillot, F.D.; Berkowski, J.A.; Sigoillot, S.M.; Kotsis, D.H.; Guy, H.I. Cell cycle-dependent regulation of pyrimidine biosynthesis. J. Biol. Chem. 2003, 278, 3403–3409. [Google Scholar] [CrossRef]
- Evans, D.R.; Guy, H.I. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. J. Biol. Chem. 2004, 279, 33035–33038. [Google Scholar] [CrossRef]
- Kafer, C.; Zhou, L.; Santoso, D.; Guirgis, A.; Weers, B.; Park, S.; Thornburg, R. Regulation of pyrimidine metabolism in plants. Front. Biosci. Landmark 2004, 9, 1611–1625. [Google Scholar]
- Robertson, B.C.; Jargiello, P.; Blank, J.; Hoffee, P.A. Genetic regulation of ribonucleoside and deoxyribonucleoside catabolism in Salmonella typhimurium. J. Bacteriol. 1970, 102, 628–635. [Google Scholar] [CrossRef]
- Nygaard, P. Nucleoside-catabolizing enzymes in Salmonella typhimurium induction by ribonucleosides. Eur. J. Biochem. 1973, 36, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Mygind, B.; Munch-Petersen, A. Transport of pyrimidine nucleosides in cells of Escherichia coli K 12. Eur. J. Biochem. 1975, 59, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Leer, J.C.; Hammer-Jespersen, K.; Schwartz, M. Uridine phosphorylase from Escherichia coli: Physical and chemical characterization. Eur. J. Biochem. 1977, 75, 217–224. [Google Scholar] [CrossRef]
- Mitchell, A.; Finch, L.R. Enzymes of pyrimidine metabolism in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 1979, 137, 1073–1080. [Google Scholar] [CrossRef]
- Williams, J.C.; Lee, C.E.; Wild, J.R. Genetic and biochemical characterization of distinct transport systems for uracil, uridine and cytidine in Salmonella typhimurium. Mol. Genet. Genom. 1980, 178, 121–130. [Google Scholar] [CrossRef]
- Andersen, L.; Kilstrup, M.; Neuhard, J. Pyrimidine, purine and nitrogen control of cytosine deaminase synthesis in Escherichia coli K12. Involvement of the glnLG and purR genes in the regulation of codA expression. Arch. Microbiol. 1989, 152, 115–118. [Google Scholar] [CrossRef]
- Kilstrup, M.; Meng, L.M.; Neuhard, J.; Nygaard, P. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J. Bacteriol. 1989, 171, 2124–2127. [Google Scholar] [CrossRef]
- Danielsen, S.; Kilstrup, M.; Barilla, K.; Jochimsen, B.; Neuhard, J. Characterization of the Escherichia coli codBA operon encoding cytosine permease and cytosine deaminase. Mol. Microbiol. 1992, 6, 1335–1344. [Google Scholar] [CrossRef]
- Tu, A.H.; Turnbough, C.L., Jr. Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription. J. Bacteriol. 1997, 179, 6665–6673. [Google Scholar] [CrossRef]
- Hayden, M.S.; Linsley, P.S.; Wallace, A.R.; Marquardt, H.; Kerr, D.E. Cloning, overexpression, and purification of cytosine deaminase from Saccharomyces cerevisiae. Protein Expr. Purif. 1998, 12, 173–184. [Google Scholar] [CrossRef]
- Tiraby, M.; Cazaux, C.; Baron, M.; Drocourt, D.; Reynes, J.-P.; Tiraby, G. Concomitant expression of E. coli cytosine deaminase and uracil phosphoribosyltransferase improves the cytotoxicity of 5-fluorocytosine. FEMS Microbiol. Lett. 1998, 167, 41–49. [Google Scholar] [CrossRef]
- Muse, W.B.; Rosario, C.J.; Bender, R.A. Nitrogen regulation of the codBA (cytosine deaminase) operon from Escherichia coli by the nitrogen assimilation control protein, NAC. J. Bacteriol. 2003, 185, 2920–2926. [Google Scholar] [CrossRef]
- Loh, K.D.; Gyaneshwar, P.; Markenscoff Papadimitriou, E.; Fong, R.; Kim, K.S.; Parales, R.; Zhou, Z.; Inwood, W.; Kustu, S. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 5114–5119. [Google Scholar] [CrossRef] [PubMed]
- Mihara, H.; Hidese, R.; Yamane, M.; Kurihara, T.; Esaki, N. The iscS gene deficiency affects the expression of pyrimidine metabolism genes. Biochem. Biophys. Res. Commun. 2008, 372, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.; Flörchinger, M.; Kunz, H.-H.; Traub, M.; Wartenberg, R.; Wolfgang Jeblick, W.; Neuhaus, H.E.; Möhlmann, T. Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 2009, 21, 876–891. [Google Scholar] [CrossRef]
- Mach, J. Uridine ribohydrolase and the balance between nucleotide degradation and salvage. Plant Cell 2009, 21, 699. [Google Scholar] [CrossRef][Green Version]
- Fang, H.; Zhang, C.; Xie, X.; Xu, Q.; Zhou, Y.; Chen, N. Enhanced cytidine production by a recombinant Escherichia coli strain using genetic manipulation strategies. Ann. Microbiol. 2014, 64, 1203–1210. [Google Scholar] [CrossRef]
- García-Bayona, L.; Garavito, M.F.; Lozano, G.L.; Vasquez, J.J.; Myers, K.; Fry, W.E.; Bernal, A.; Zimmerman, B.H.; Restrepo, S. De Novo pyrimidine biosynthesis in the oomycete plant pathogen Phytophthora infestans. Gene 2014, 537, 312–321. [Google Scholar] [CrossRef]
- Garavito, M.F.; Narváez-Ortiz, H.Y.; Zimmermann, B.H. Pyrimidine metabolism: Dynamic and versatile pathways in pathogens and cellular development. J. Genet. Genom. 2015, 42, 195–205. [Google Scholar] [CrossRef]
- Ban, J.; Ljubinka Vitale, L.; Kos, E. Thymine and uracil catabolism in Escherichia coli. Microbiology 1972, 73, 267–272. [Google Scholar] [CrossRef][Green Version]
- Šimaga, Š.; Kos, E. Properties and regulation of pyrimidine catabolism in Escherichia coli. Int. J. Biochem. 1981, 13, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Matthews, M.M.; Traut, T.W. Regulation of N-carbamoyl-beta-alanine amidohydrolase, the terminal enzyme in pyrimidine catabolism, by ligand-induced change in polymerization. J. Biol. Chem. 1987, 262, 7232–7237. [Google Scholar] [CrossRef] [PubMed]
- Kern, L.; De Montigny, J.; Lacroute, F.; Jund, R. Regulation of the pyrimidine salvage pathway by the FUR1 gene product of Saccharomyces cerevisiae. Curr. Genet. 1991, 19, 333–337. [Google Scholar] [CrossRef] [PubMed]
- West, T.P. Isolation and characterization of an Escherichia coli B mutant strain defective in uracil catabolism. Can. J. Microbiol. 1998, 44, 1106–1109. [Google Scholar] [CrossRef]
- Soong, C.-L.; Ogawa, J.; Sakuradani, E.; Shimizu, S. Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 2002, 277, 7051–7058. [Google Scholar] [CrossRef]
- Van Kuilenburg, A.B.P.; Meinsma, R.; Beke, E.; Assmann, B.; Ribes, A.; Lorente, I.; Busch, R.; Mayatepek, E.; Nico, G.G.M.; Abeling, N.G.G.M.; et al. β-Ureidopropionase deficiency: An inborn error of pyrimidine degradation associated with neurological abnormalities. Hum. Mol. Genet. 2004, 13, 2793–2801. [Google Scholar] [CrossRef]
- Shimada, T.; Hirao, K.; Kori, A.; Yamamoto, K.; Ishihama, A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 2007, 66, 744–757. [Google Scholar] [CrossRef]
- Shimada, T.; Ishihama, A.; Busby, S.J.; Grainger, D.C. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008, 36, 3950–3955. [Google Scholar] [CrossRef]
- Hidese, R.; Mihara, H.; Kurihara, T.; Nobuyoshi Esaki, N. Escherichia coli dihydropyrimidine dehydrogenase is a novel NAD-dependent heterotetramer essential for the production of 5,6-dihydrouracil. J. Bacteriol. 2011, 193, 989–993. [Google Scholar] [CrossRef]
- Andersson Rasmussen, A.; Kandasamy, D.; Beck, H.; Crosby, S.D.; Björnberg, O.; Schnackerz, K.D.; Piškur, J. Global expression analysis of the yeast Lachancea (Saccharomyces) kluyveri reveals new URC genes involved in pyrimidine catabolism. Eukaryot. Cell 2014, 13, 31–42. [Google Scholar] [CrossRef]
- De Vos, P.; De Ley, J. Intra- and intergeneric similarities of Pseudomonas and Xanthomonas ribosomal ribonucleic acid cistrons. Int. J. Syst. Bacteriol. 1983, 33, 487–509. [Google Scholar] [CrossRef][Green Version]
- Anzai, Y.; Kim, H.; Park, J.-Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Lalucat, J.; Garcia-Valdes, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Isaac, J.H.; Holloway, B.W. Control of pyrimidine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 1968, 96, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Ralli, P.; Srivastava, A.C.; O’Donovan, G. Regulation of the pyrimidine biosynthetic pathway in a pyrD knockout mutant of Pseudomonas aeruginosa. J. Basic Microbiol. 2007, 47, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Santiago, M.F.; West, T.P. Effect of carbon source on pyrimidine biosynthesis in Pseudomonas alcaligenes ATCC 14909. Microbiol. Res. 2003, 158, 195–199. [Google Scholar] [CrossRef]
- West, T.P. Pyrimidine nucleotide synthesis in Pseudomonas citronellolis. Can. J. Microbiol. 2004, 50, 455–459. [Google Scholar] [CrossRef]
- Santiago, M.F.; West, T.P. Regulation of pyrimidine synthesis in Pseudomonas mendocina. J. Basic Microbiol. 2002, 42, 75–79. [Google Scholar] [CrossRef]
- Santiago, M.F.; West, T.P. Influence of carbon source on pyrimidine synthesis in Pseudomonas mendocina. J. Basic Microbiol. 2003, 43, 534–538. [Google Scholar] [CrossRef]
- West, T.P. Pyrimidine nucleotide synthesis in Pseudomonas nitroreducens and the regulatory role of pyrimidines. Microbiol. Res. 2014, 169, 954–958. [Google Scholar] [CrossRef]
- West, T.P. Control of the pyrimidine biosynthetic pathway in Pseudomonas pseudoalcaligenes. Arch. Microbiol. 1994, 162, 75–79. [Google Scholar] [CrossRef]
- Haugaard, L.E.; West, T.P. Pyrimidine biosynthesis in Pseudomonas oleovorans. J. Appl. Microbiol. 2002, 92, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Saha, R.; Sproer, C.; Beck, B.; Bagley, S. Pseudomonas oleovorans subsp. lubricantis subsp. nov., and reclassification of Pseudomonas pseudoalcaligenes ATCC 17440T as later synonym of Pseudomonas oleovorans ATCC 8062T. Curr. Microbiol. 2010, 60, 294–300. [Google Scholar] [CrossRef]
- West, T.P. Regulation of pyrimidine synthesis in Pseudomonas resinovorans. Lett. Appl. Microbiol. 2005, 40, 473–478. [Google Scholar] [CrossRef]
- Abdurahman, N.; Hughes, L.E. Accumulation of pyrimidine intermediate orotate decreases virulence factor production in Pseudomonas aeruginosa. Curr. Microbiol. 2015, 71, 229–234. [Google Scholar]
- Al Ahmar, R.; Kirby, B.D.; Yua, H.D. Pyrimidine biosynthesis regulates the small-colony variant and mucoidy in Pseudomonas aeruginosa through sigma factor competition. J. Bacteriol. 2019, 201, e00575-18. [Google Scholar] [CrossRef]
- Domakonda, A.; West, T.P. Control of pyrimidine nucleotide formation in Pseudomonas aurantiaca. Arch. Microbiol. 2020, 202, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Bani Ahmad, A.; West, T.P. Regulation of the pyrimidine biosynthetic pathway in the bacterium Pseudomonas chlororaphis. In Pyrimidines and Their Importance; Ward, R.G., Ed.; Nova Press Inc.: Hauppauge, NY, USA, 2023; pp. 191–230. [Google Scholar]
- West, T.P. Control of pyrimidine synthesis in Pseudomonas fragi. Lett. Appl. Microbiol. 2002, 35, 380–384. [Google Scholar] [CrossRef]
- West, T.P. Regulation of pyrimidine formation in Pseudomonas lundensis. Can. J. Microbiol. 2009, 55, 261–268. [Google Scholar] [CrossRef]
- West, T.P. Regulation of pyrimidine nucleotide formation in Pseudomonas taetrolens ATCC 4683. Microbiol. Res. 2004, 159, 29–33. [Google Scholar] [CrossRef]
- Chu, C.P.; West, T.P. Pyrimidine biosynthetic pathway of Pseudomonas fluorescens. J. Gen. Microbiol. 1990, 136, 875–880. [Google Scholar] [CrossRef] [PubMed]
- West, T.P. Effect of carbon source on pyrimidine formation in Pseudomonas fluorescens ATCC 13525. Microbiol. Res. 2005, 160, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Gallie, J.; Libby, E.; Bertels, F.; Remigi, P.; Jendresen, C.B.; Desprat, N.; Buffing, M.F.; Sauer, U.; Beaumont, H.J.E.; Martinussen, J.; et al. Bistability in a metabolic network underpins the de novo evolution of colony switching in Pseudomonas fluorescens. PLoS Biol. 2015, 13, e1002109. [Google Scholar] [CrossRef] [PubMed]
- Murahari, E.C.; West, T.P. The pyrimidine biosynthetic pathway and its regulation in Pseudomonas jessenii. Antonie Leeuwenhoek 2019, 112, 461–469. [Google Scholar] [CrossRef]
- West, T.P. Regulation of the pyrimidine biosynthetic pathway in Pseudomonas mucidolens. Antonie Leeuwenhoek 2005, 88, 181–186. [Google Scholar] [CrossRef] [PubMed]
- West, T.P. Regulation of pyrimidine nucleotide formation in Pseudomonas reptilivora. Lett. Appl. Microbiol. 2004, 38, 81–86. [Google Scholar] [CrossRef]
- West, T.P. Regulation of pyrimidine nucleotide biosynthesis in Pseudomonas synxantha. Antonie Leeuwenhoek 2007, 92, 353–358. [Google Scholar] [CrossRef]
- West, T.P. Pyrimidine biosynthesis in Pseudomonas veronii and its regulation by pyrimidines. Microbiol. Res. 2012, 167, 306–310. [Google Scholar] [CrossRef]
- Condon, S.; Collins, J.K.; O’Donovan, G.A. Regulation of arginine and pyrimidine biosynthesis in Pseudomonas putida. J. Gen. Microbiol. 1976, 92, 375–383. [Google Scholar] [CrossRef]
- Santiago, M.F.; West, T.P. Control of pyrimidine formation in Pseudomonas putida ATCC 17536. Can. J. Microbiol. 2002, 48, 1076–1081. [Google Scholar] [CrossRef]
- Schurr, M.J.; Vickrey, J.F.; Kumar, A.P.; Campbell, A.L.; Cunin, R.; Benjamin, R.C.; Shanley, M.S.; O’Donovan, G.A. Aspartate transcarbamoylase genes of Pseudomonas putida: Requirement for an inactive dihydroorotase for assembly into the dodecameric holoenzyme. J. Bacteriol. 1995, 177, 1753–1759. [Google Scholar] [CrossRef]
- West, T.P. Control of pyrimidine nucleotide formation in Pseudomonas fulva. Antonie Leeuwenhoek 2010, 97, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Chunduru, J.; West, T.P. Pyrimidine nucleotide synthesis in the emerging pathogen Pseudomonas monteilii. Can. J. Microbiol. 2018, 64, 432–438. [Google Scholar] [CrossRef] [PubMed]
- West, T.P. Regulation of pyrimidine formation in Pseudomonas oryzihabitans. J. Basic Microbiol. 2007, 47, 440–443. [Google Scholar] [CrossRef]
- West, T.P. Effect of carbon source on pyrimidine biosynthesis in Pseudomonas oryzihabitans. J. Basic Microbiol. 2010, 50, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Yu, T.; Omata, S. Distribution of enzymes related to cytidine degradation in bacteria. Agric. Biol. Chem. 1976, 40, 1893–1895. [Google Scholar]
- Beck, D.A.; O’Donovan, G.A. Pathways of pyrimidine salvage in Pseudomonas and former Pseudomonas: Detection of recycling enzymes using high-performance liquid chromatography. Curr. Microbiol. 2008, 56, 162–167. [Google Scholar] [CrossRef]
- Shaposhnikov, L.A.; Savin, S.S.; Tishkov, V.I.; Pometun, A.A. Ribonucleoside hydrolases—Structure, functions, physiological role and practical uses. Biomolecules 2023, 13, 1375. [Google Scholar] [CrossRef]
- West, T.P. Degradation of pyrimidine ribonucleosides by Pseudomonas aeruginosa. Antonie Leeuwenhoek 1996, 69, 331–335. [Google Scholar] [CrossRef]
- West, T.P. Utilization of pyrimidine bases and nucleosides by the Pseudomonas stutzeri group. Microbios 1990, 61, 71–81. [Google Scholar]
- West, T.P. Pyrimidine base and ribonucleoside utilization by the Pseudomonas alcaligenes group. Antonie Leeuwenhoek 1991, 59, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Watanabe, T.; Chibata, I. Metabolism of pyrimidine nucleotides in a microorganism. III. Enzymatic production of ribose-5-phosphate from uridine-5′-monophosphate by Pseudomonas oleovorans. Appl. Microbiol. 1971, 22, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.S. Optimization of culture conditions for the production of pyrimidine nucleotide N-ribosidase from Pseudomonas oleovorans. J. Life Sci. 2004, 14, 608–613. [Google Scholar]
- Yu, T.S. Purification and characterization of pyrimidine nucleotide N-ribosidase from Pseudomonas oleovorans. J. Microbiol. Biotechnol. 2005, 15, 573–578. [Google Scholar]
- Gill, R.; West, T.P. Control of a pyrimidine ribonucleotide salvage pathway in Pseudomonas oleovorans. Arch. Microbiol. 2022, 204, 383. [Google Scholar] [CrossRef] [PubMed]
- Terada, M.; Tatibana, M.; Hayaishi, O. Purification and properties of nucleoside hydrolase from Pseudomonas fluorescens. J. Biol. Chem. 1967, 242, 5578–5585. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.P.; West, T.P. Pyrimidine ribonucleoside catabolism in Pseudomonas fluorescens biotype A. Antonie Leeuwenhoek 1990, 57, 253–257. [Google Scholar] [CrossRef]
- West, T.P. Metabolism of pyrimidine bases and nucleosides by Pseudomonas fluorescens biotype F. Microbios 1988, 56, 27–36. [Google Scholar]
- Liu, X.; Wood, P.L.; Parales, J.V.; Parales, R.E. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 2009, 191, 2909–2916. [Google Scholar] [CrossRef]
- Parales, R.E.; Nesteryuk, V.; Hughes, J.G.; Luu, R.A.; Ditty, J.L. Cytosine chemoreceptor McpC in Pseudomonas putida F1 also detects nicotinic acid. Microbiology 2014, 160, 2661–2669. [Google Scholar] [CrossRef]
- West, T.P. Pyrimidine base catabolism in species of Pseudomonas and Burkholderia. Res. J. Microbiol. 2011, 6, 172–181. [Google Scholar] [CrossRef]
- Kim, S.; West, T.P. Pyrimidine catabolism in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 1991, 77, 175–179. [Google Scholar] [CrossRef]
- West, T.P. Isolation and characterization of a dihydropyrimidine dehydrogenase mutant of Pseudomonas chlororaphis. Arch. Microbiol. 1991, 156, 513–516. [Google Scholar] [CrossRef]
- Santiago, M.F.; West, T.P. Effect of nitrogen source on pyrimidine catabolism by Pseudomonas fluorescens. Microbiol. Res. 1999, 154, 221–224. [Google Scholar] [CrossRef]
- West, T.P. Pyrimidine base catabolism in Pseudomonas putida biotype B. Antonie Leeuwenhoek 2001, 80, 163–167. [Google Scholar] [CrossRef]
- Hidese, R.; Mihara, H.; Kurihara, T.; Esaki, N. Pseudomonas putida PydR, a RutR-like transcriptional regulator, represses the dihydropyrimidine dehydrogenase gene in the pyrimidine reductive catabolic pathway. J. Biochem. 2012, 152, 341–346. [Google Scholar] [CrossRef]
- Matcher, G.F.; Jiwaji, M.; de la Mare, J.; Dorrington, R.A. Complex pathways for regulation of pyrimidine metabolism by carbon catabolite repression and quorum sensing in Pseudomonas putida RU-KM3s. Appl. Microbiol. Biotechnol. 2013, 97, 5993–6007. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).