Abstract
Novacetimonas cocois WE7 (formally named Komagataeibacter cocois WE7) is a strain isolated from contaminated coconut milk, capable of producing bacterial cellulose (BC). We sequenced its genome to investigate why WE7 cannot synthesize BC from glucose efficiently. It contains about 3.5 Mb and six plasmid DNAs. N. cocois WE7 contains two bcs operons (bacterial cellulose operon, bcs I and bcs II); the absence of bcs III operons may lead to reduced BC production. From genome predictions, glucose, sucrose, fructose, maltose, and glycerol can be utilized to generate BC, with WE7 unable to metabolize carbohydrate carbon sources through the Embden–Meyerhof–Parnas (EMP) pathway, but rather through the Hexose Monophosphate Pathway (HMP) and tricarboxylic acid (TCA) pathways. It has a complete gluconic acid production pathway, suggesting that BC yield might be very low when glucose, maltose, and trehalose are used as carbon sources. This study represents the first genome analysis of N. cocois. This information is crucial for understanding BC production and regulation mechanisms in N. cocois and lays a foundation for constructing engineered strains tailored for diverse BC application purposes.
1. Introduction
Bacterial cellulose (BC) is an insoluble glucan chain that microorganisms produce and has the same chemical structure as plant cellulose [1]. The strains that can produce BC currently include the genus Novacetimonas, Komagataeibacter, Acetobacter, Glucoacetobacter, and Sarcina [2,3,4]. Strains in the genus Novacetimonas (formally called Komagataeibacter and earlier Gluconacetobacter) and Komagataeibacte are the most efficient producers of BC [5]. On the other hand, the number and types of the critical enzymes (bcs) for the synthesis of BC are varied in different genera of microorganisms, leading to differences in BC synthesis ability among strains [6].
In addition, the utilization and metabolic pathways of carbon sources (glucose, mannitol, glycerol, sucrose, fructose, mannitol) for the synthesis of BC are different in strains [7], resulting in the yield and properties of BC differing substantially. The properties of BC produced by different carbon sources include tensile property, tensile strength, elastic modulus, stress, Young’s modulus, and porosity [8,9,10]. These properties affect the mechanical properties of BC. Therefore, the culture of BC-producing strains under different carbon source conditions is a potential method to obtain BC with different textures.
A comprehensive understanding of the genome characteristics of strains helps study fermentation conditions, genetic modifications, and metabolic mechanisms of BC-producing strains [11]. Many researchers have analyzed strains from the genome perspective of BC production and studied the mechanism of BC synthesis. Twenty strains of Novacetimonas underwent genome sequencing, including three from Novacetimonas pomaceti (frame diagrams), 14 from Novacetimonas hansenii (two completed diagrams), two from Novacetimonas maltaceti (frame diagrams), and one from N. cocois (frame diagram, drawn during strain identification in our laboratory). The genomes of 60 strains from Komagataeibacter have been sequenced (data from the NCBI database). Novacetimonas cocois WE7 was a strain isolated from gel membrane samples of naturally fermented coconut milk in our laboratory [12], adding a new member to the microorganism that produces BC.
To fully understand the production and genetic characteristics of BC-producing strain N. cocois WE7, we performed genome sequencing and comparative genomics analysis of N. cocois WE7. The carbon metabolism characteristics and metabolic pathway of synthetic BC produced by N. cocois WE7 were analyzed from the gene level according to the carbon source preference suggested by genomics analysis. The changes in pH value and organic acid content in the fermentation process using different carbon sources were measured to clarify the metabolic characteristics of different carbon sources in the production and synthesis of BC. The data from this type of strain of N. cocois can increase the understanding of the genetic diversity in carbon metabolic genes and pathways during BC synthesis. This approach lays a foundation for the research on BC synthesis-related gene modification, gene editing, and other molecular levels.
2. Materials and Methods
2.1. Strain and Cultivation Conditions
N. cocois sp. nov. WE7T (CGMCC 1.15338T, JCM 31140T) was stored in our laboratory. The primary and seed medium was the HS medium [13].
Hestrin–Schramm (HS) medium (yeast extract 20 g/L; 5 g/L peptone; 2.7 g/L Na2HPO·12H2O and 1.2 g/L citric acid monohydrate) [13]. The carbon source, glucose, fructose, or sucrose, was added at a final concentration of 20 g/L. The medium pH was adjusted to 6.0 using 1.0 M NaOH or HCl.
BC-producing strains were first cultivated in HS medium to prepare seed cultures at 30 °C for 48 h under static conditions. These seed cultures were filtered through sterile absorbent cotton to remove cellulose [14]. Then, 2 mL of the filtered seed culture was transferred into a 100 mL flask containing 40 mL of different medium and cultured at 30 °C for up to seven days for BC production [15].
2.2. Genome Sequencing and De Novo Assembly
The cells were collected in the logarithmic growth phase, washed twice with 1× phosphate-buffered saline, centrifuged at 8000× g for 10 min at 4 °C, frozen in liquid nitrogen, and stored at −80 °C. Genomes were extracted by Meiji Biotech (Shanghai, China) and sequenced on PacBio and Illumina Hiseq 2000 platforms. We generated high-quality data sets with 100× sequencing depth [16].
SOAPdenovo 2.04 was used for de novo assembly. GapCloser 1.12 was used for gap filling. Glimmer 3.02 and GeneMarkS 4.3 were used to predict coding sequence (CDS) and plasmid genes, respectively. GO, KEGG, COG, Non-Redundant Protein, Swiss-Prot, and Pfam were used to annotate the strain’s gene and protein sequences, and the sequences’ functions were speculated [17]. The complete genomic sequences of N. cocois WE7 obtained in this study can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/, accessed on 15 November 2022) under accession numbers CP110924 (Chromosome), CP110925 (pKWE7-1), CP110926 (pKWE7-2), CP110927 (pKWE7-3), CP110928 (pKWE7-4), CP110929 (pKWE7-5), and CP110930 (pKWE7-6).
2.3. Phylogenetic Analysis
We downloaded the genome DNA of closely related species from NCBI. The downloaded genome was preferentially selected from the genome completion map to ensure the genome assembly level. To identify the core genome from the assembled genome and downloaded genome, pan-genome analysis (GET_HOMOLOGUS v3.2.3) was performed. Using these core gene sets, a maximum-likelihood phylogenetic tree was constructed (bootstrap: 1000 replicates; best fit alternative model: TIM2+F+I+G4 model) [18,19].
2.4. Fermentation Profile Analysis under Different Carbon Source
For analysis of the fermentation profiles of WE7 under HS medium with a different carbon source, the yield of BC after different fermentation times (1, 2, 3, 4, 5, 6, and 7 days) was purified and determined as the method described by Cannazza, using the following formula [15,20]:
Yield of BC (g/L) = Dry weight of BC (g) × 1000/volume of fermentation (mL)
The fermentation broth was centrifuged (4 °C, 8000× g, 10 min) to obtain supernatant and precipitate. The precipitate was centrifuged with distilled water at 8000× g for 10 min at 4 °C and washed twice to monitor the biomass. The precipitate was soaked in 1% cellulase solution (pH 5.0) prepared with citric acid buffer (19% 0.1 mol/L C6H8O7, 81% 0.1 mol/L Na3C6H5O7, pH 6.0) in a water bath at 50 °C to hydrolyzes cellulose. After the BC membrane was dissolved, the precipitate was placed in an oven (Shanghai Yuejin Medical Instruments CO., Ltd., Shanghai, China) at 80 °C to dry until it achieved constant weight, weighing as biomass [21].
The supernatant was used for the determination of organic acid content. Before determination, the supernatant was mixed with potassium ferricyanide trihydrate (36 g/L) and zinc sulfate (72 g/L) (v:v:v = 1:1:1) and centrifuged after complete reaction (setting parameters: 4 °C, 8000× g, 8 min). Finally, the supernatant was filtered through a 0.22 μm stream filter membrane and set aside for use.
Organic acids were determined by Agilent 1200 liquid chromatography with a VWD detector (Agilent, Santa Clara, CA, USA). The separation was carried out on a Zorbax SB-AQ (250 × 4.6 mm) column at a 0.6 mL/min flow rate. Mobile phase A consisted of aqueous perchloric acid, liquid B consisted of methanol:water = 98:2, the column temperature was 30 °C, the wavelength was 210 nm, and the injection volume was 10 μL [22].
2.5. Statistical Analysis
SPSS 22.0 (IBM Corporation, Armonk, NY, USA) was used to analyze the data. The LSD and Duncan tests were used to test the significance (p < 0.05). The trial was repeated three times. GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, USA) and Excel software 2021 (Microsoft, Washington, DC, USA)were used for plotting.
3. Results and Discussion
3.1. Genome Features of N. cocois WE7
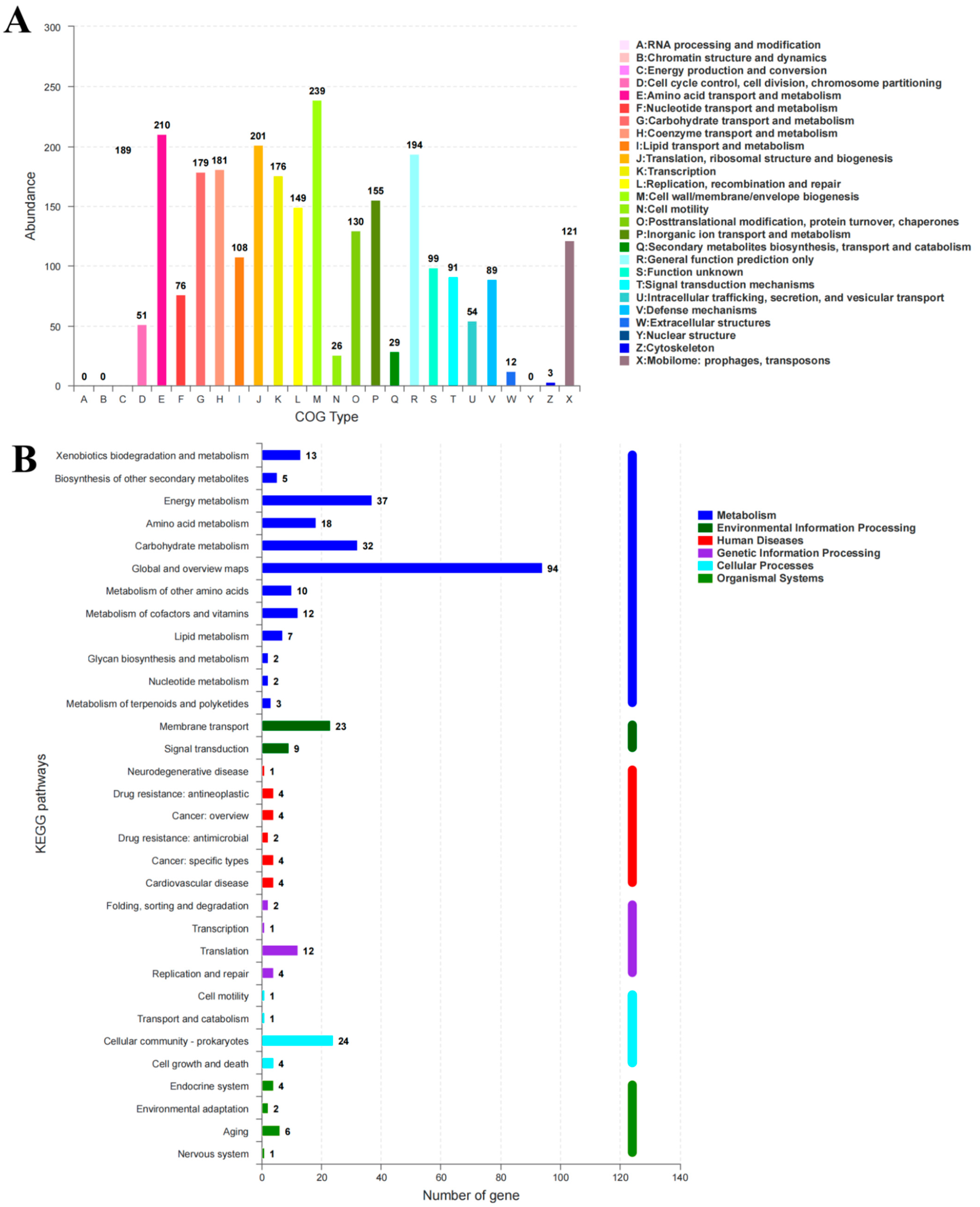
The genome was sequenced to explore the genetic structure of the BC-producing N. cocois WE7. Sequence analysis revealed that the genome consisted of one chromosome and six plasmids, with a total length of 3,492,580 bp and a GC content of 62.20%. We predicted 3106 genes in the genome, including 56 tRNA, 15 rRNA, 21 sRNA, 42 repetitive DNA, one prophage, and two CRISPR-Cas sequences (Table 1). Among the predicted genes, 11.1% were successfully assigned to the KEGG pathway. We annotated 32 genes on KEGG (Figure 1B) and classified them into six categories: metabolism, environmental information processing, human diseases, genetic information processing, cellular processes, and biological systems. Of these, 7.57% were involved in metabolism, including two operons and 11 genes related to BC synthesis (Figure 2). According to the COG classification, 2496 coding genes were allocated to 23 classes (Figure 1A). The most significant functional group was “cell wall/membrane/envelope biogenesis”, with 239 genes, while 210 genes were for amino acid transport and metabolism.

Table 1.
General features of the N. cocois WE7 genome.
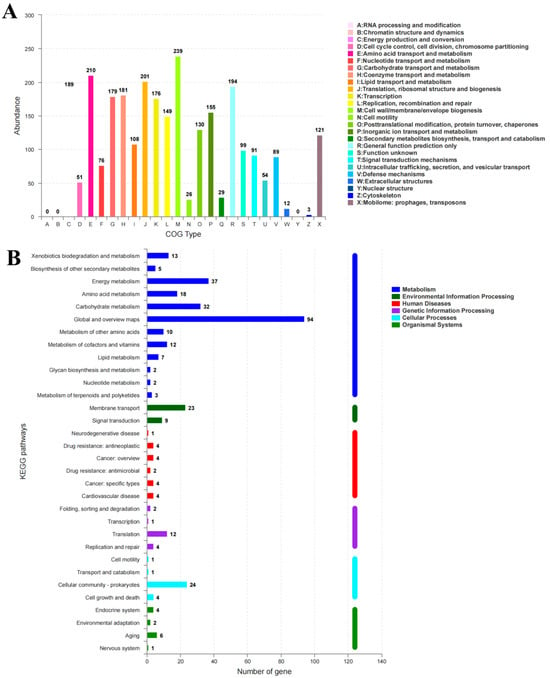
Figure 1.
The COG (A) and KEGG (B) classification and functional categories of N. cocois WE7.
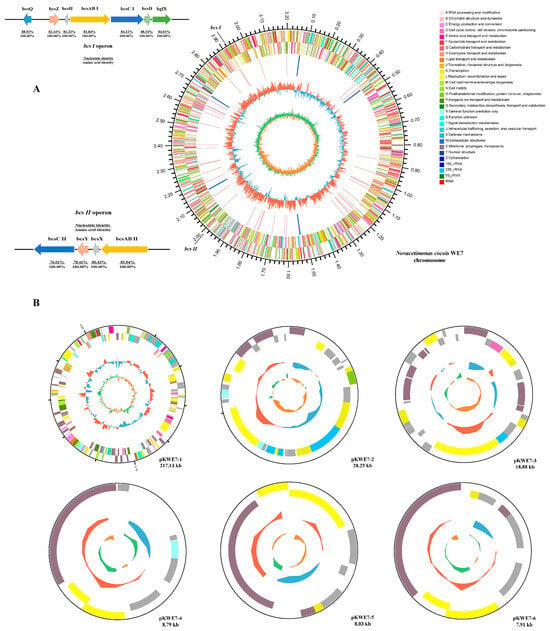
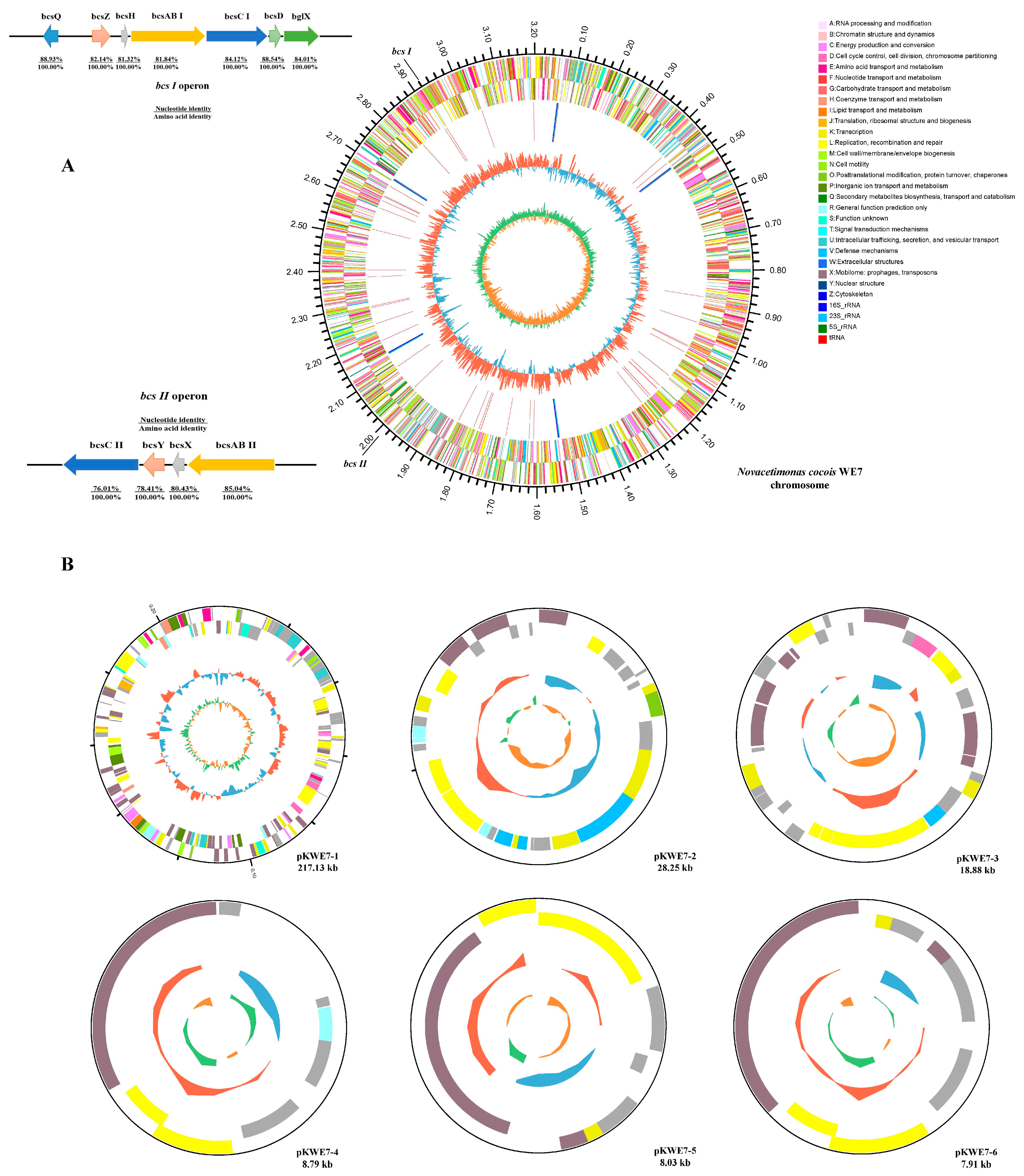
Figure 2.
Genome structure of the N. cocois WE7 strain. One circular chromosome (A) and six putative plasmids ((B), pKWE7-1, pKWE7-2, pKWE7-3, pKWE7-4, pKWE7-5, and pKWE7-6) are shown. The outermost circle marks the size of the genome; the second circle and the third circle are CDS on the positive and negative chains. Different colors indicate the functional classification of different COGs of CDS. The fourth circle is rRNA and tRNA. The fifth circle is the GC content. The outward red part indicates that the GC content of this region is higher than the average GC content of the whole genome; the inward blue part indicates that the GC content of this region is lower than the average GC content of the whole genome. The inner circle is the GC-SKEW value, and the specific algorithm is G-C/G+C, which can assist in judging the leading chain and the lagging chain.
3.2. Phylogenetic Analysis of WE7 Strain
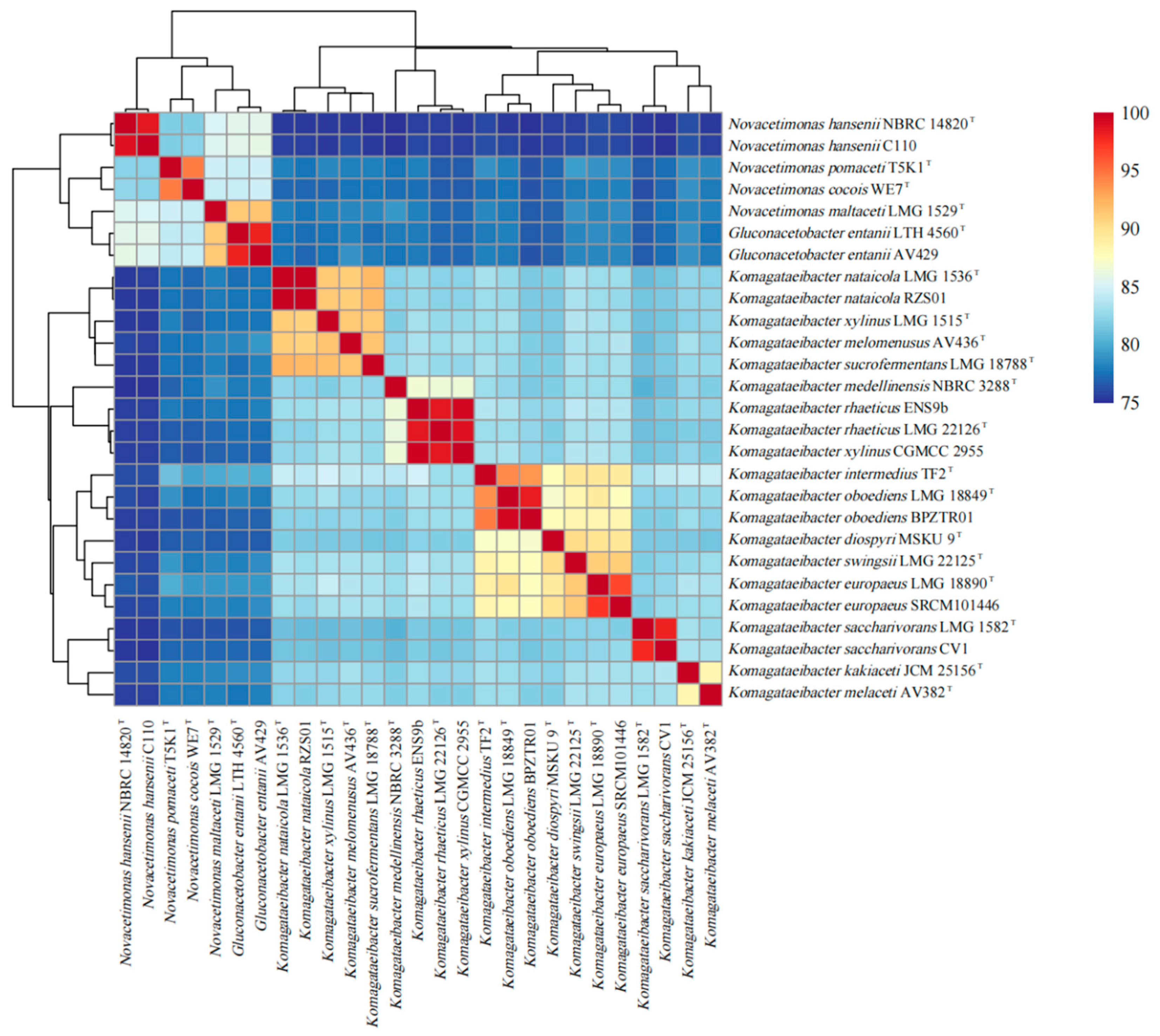
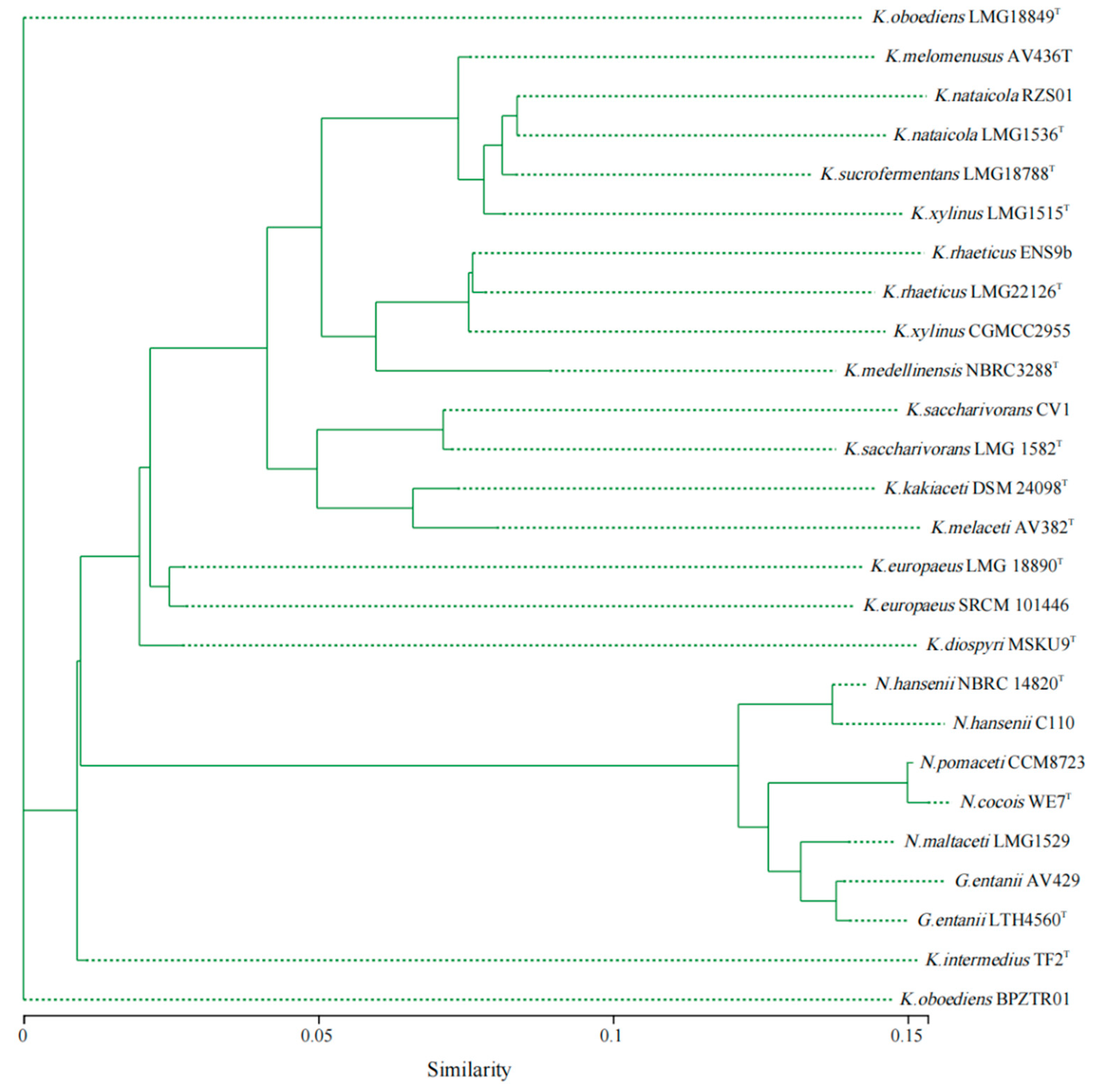
As shown in Figure 3, compared with Komagataeibacter strains, the ANI (average nucleotide identity) values of Novacetimonas strains are below 80%, and the ANI values of strains of Novacetimonas species are between 81.96% and 94.49%. In the cluster analysis, the strains of Novacetimonas and Komagataeibacter are divided into different branches, consistent with the ANI data analyzed when the genus Novacetimonas was proposed [2]. The ANI of N. cocois WE7 was less than 95% compared with other strains, consistent with ANI comparison data when WE7 was reported [3]. The results of the phylogenetic tree based on the core gene set of the pan-genome analysis showed similar rules to the results of the ANI cluster analysis (Figure 4).
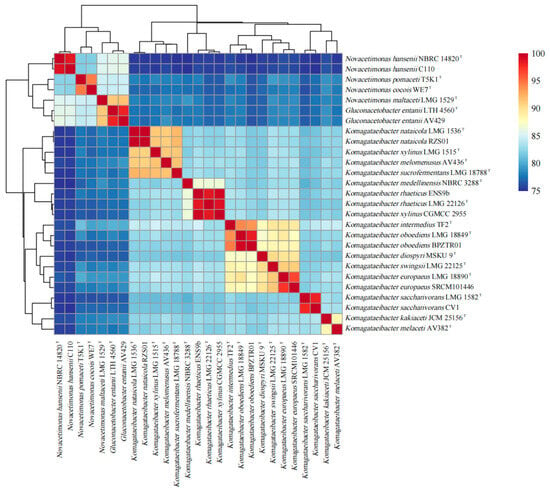
Figure 3.
ANIb (average nucleotide identity blast) values of 26 Novacetimonas, Komagataeibacter, and Gluconacetobacter strains pairwise comparisons. Values are represented in the central bi-color gradient heatmap.
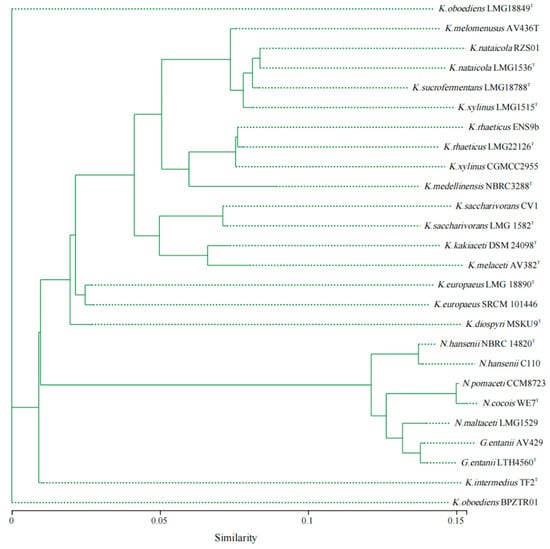
Figure 4.
Phylogenetic analysis based on the concatenated sequences of core genes (nucleotide sequence) from Novacetimonas, Komagataeibacter, and Gluconacetobacter strains (n = 26). The evolutionary history was inferred using the FastTree approximately-maximum-likelihood method, the Bayesian Information Criterion model, and a bootstrap of 1000 replicates.
In addition to the significant differences between Komagataeibacter and Novacetimonas in ANI cluster analysis and phylogenetic tree, the genomes of different species within Novacetimonas also showed some differences. ANI data and phylogenetic tree revealed that Novacetimonas strains were divided into three genomic groups. Group I included N. cocois WE7T and N. pomaceti T5K1T, group II included N. maltaceti LMG 1529T, G. entanii LHT 4560T, and G. entanii AV429. Group III included N. hansenii NBRC 14820T and N. hansenii C110 [2]. These strains with different genome groups have different gene structures, which increases the diversity of BC synthesis and regulation mechanisms.
Based on the current analysis results, it was found that several strains within the genus Komagataeibacter could be reclassified according to ANI value (95%) and clustering. The ANI value between K. xylinus CGMCC 2955 and K. xylinus LMG 1515T was 82.62%, far lower than the ANI threshold (>95%) for species definition. The ANI values of K. xylinus CGMCC 2955 between K. rhaeticus LMG 22126T and K. rhaeticus ENS 9b were 98.72% and 99.54%, respectively. CGMCC 2955 appears to be a strain of K. rhaeticus. The phylogenetic tree constructed based on the core gene set of pan-genome analysis (Figure 4) shows that (similar to the ANI data) strain CGMCC 2955, K. rhaeticus LMG 22126T, and K. rhaeticus ENS 9b strain were classified into the same clade. It was supported that CGMCC 2955 was a member of K. rhaeticus.
3.3. The Compositions of bcs Operons in WE7
Cellulose synthase is a critical enzyme in the BC biosynthesis pathway [23]. Four bcs operons were found in BC-producing strains, and most strains of Komagataeibacter contained these four. In Novacetimonas, the fourth operon was not found, and most strains contained three [2]. In Novacetimonas, the bcsABCD gene cluster contains a smaller bcsA gene that does not present a stop codon. The bcsA and bcsB are fused to form a long gene bcsAB. The bcsⅡ and bcsⅢ operons are essential during BC synthesis in Komagataeibacter sp. CGMCC 17276, especially bcsⅢ. Two bcs persons were annotated in WE7 (Figure 2, bcs I and bcsⅡ), which lacks the bcsⅢ operon. This finding may be related to the low BC production in WE7. The first operon contains three genes (bcsAB, bcsC, and bcsD) that encode subunits of the BCS complex. Wong first reported that bcsI operon is essential in cellulose synthesis [24,25]. Deleting bcsAB1 resulted in a non-fiber phenotype in K.hansenii ATCC 23769 [26]. The bcsC subunit is closely related to the outer membrane transport of cellulose and contains ten amino acids at the boundary between the proximal domains of the membrane, forming flexible ligaments [15,27]. The bcsD gene is related to BC porosity and crystallinity. When the bcsD gene’s expression level decreased, BC’s porosity increased significantly, and the crystallinity decreased [28]. The four genes were annotated in the bcs type I operon, in which bcsQ, bcsZ, and bcsH were upstream of the operon, and bglX was downstream. The bcsQ gene has been reported to be associated with cell division in Escherichia coli, suggesting that it may play a role in the cellular localization of bcs complexes [29]. BcsH protein, also known as cellulose complementing factor (ccpA), can affect the expression levels of bcsB and bcsC and also interact with bcsD, helping the glucan chains align into crystal bands [6,12]. BcsZ encodes endogenous -1,4-glucanase, effectively degrading amorphous glucan chains [20]. BglX encodes a glucosidase that affects the expression levels of bcsB and bcsC. The bcs type II operon in WE7 contains bcsAB, bcsX, bcsY, and bcsC. Bimmer found that the bcsAB2 formed other nonfibrous EPS structures [26]. Szymzack presumed that the bcs II operon, especially the bcsX, was involved in synthesizing the acylated cellulose [5]. The bcsY gene may be involved in synthesizing transacylase, acetylcellulose, or modification of the cellulose hydroxyl group; however, the product has not been identified and needs study [30].
3.4. Metabolic Pathways and the Critical Gene for BC Production
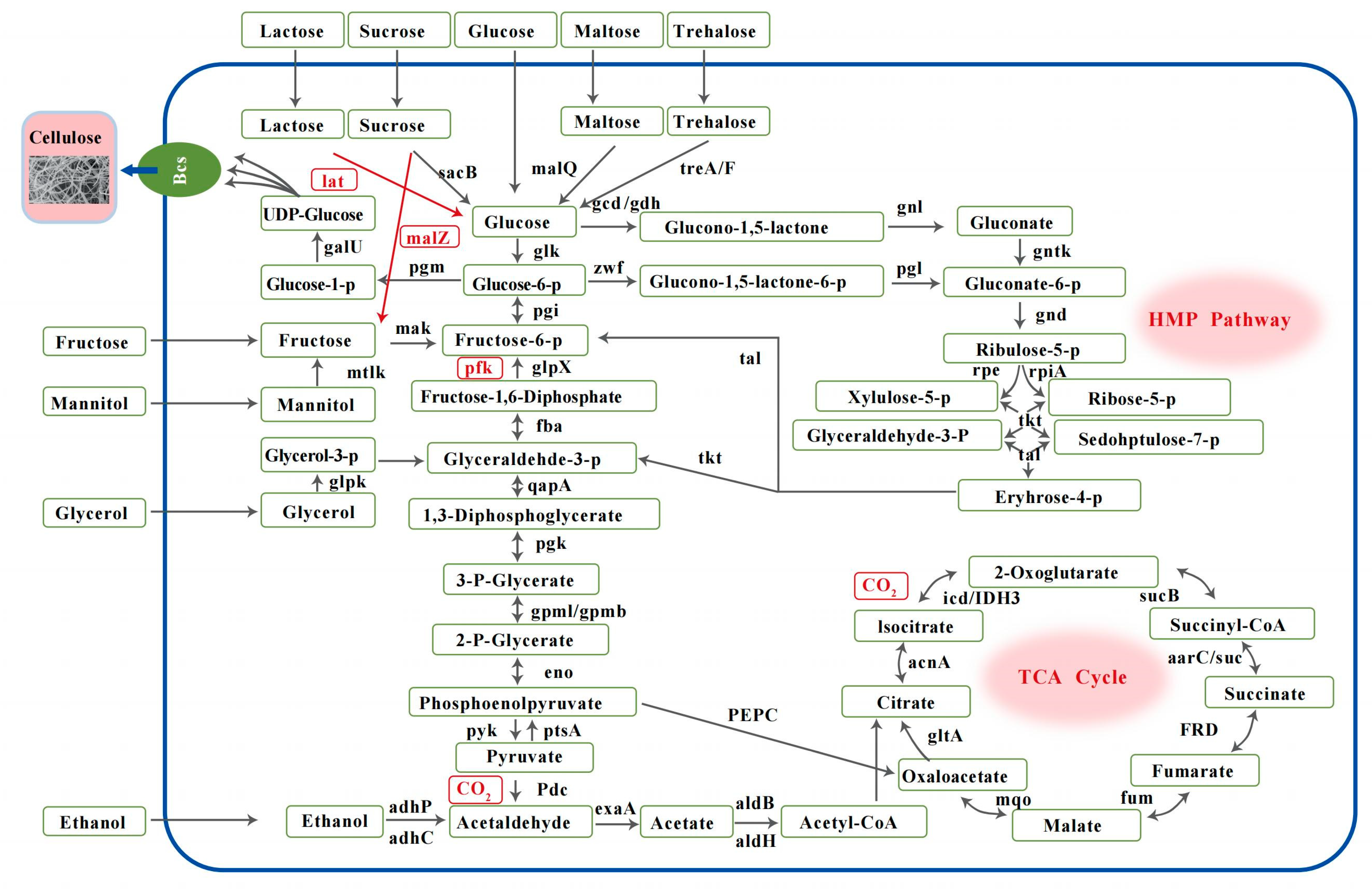
The BC synthesis gene classifications of WE7 are shown in Figure 5. WE7 does not possess phosphofructokinase (pfk), mannitol 2-dehydrogenase (mtlk), sucrose phosphorylase (gtfA), and lactate (lat) genes. Pfk, a rate-limiting enzyme in the EMP pathway [15], is lacking in most BC-producing bacteria. The absence of pfk indicated that WE7 could not metabolize carbohydrate carbon sources through the EMP pathway but metabolized carbon sources through the HMP and TCA pathways to maintain cell growth and BC synthesis. Mtlk and lat are critical enzymes for mannitol and lactose metabolism, respectively. The absence of these enzymes indicates that WE7 cannot use mannitol and lactose as the sole carbon source, consistent with the results reported in the literature [3]. Three genes encoding glucokinase (glk), glucose-6-phosphate isomerase (pgi), and phosphoglucomutase (pgm) were identified in this strain; these enzymes transfer glucose and fructose to glucose-1-P, thus smoothly into the BC synthesis pathway. We also identified a gene encoding UTP-glucose-1-phosphate uridyltransferase galU, which converts glucose-1-P to UDP-glucose, a substrate for BC synthesis.
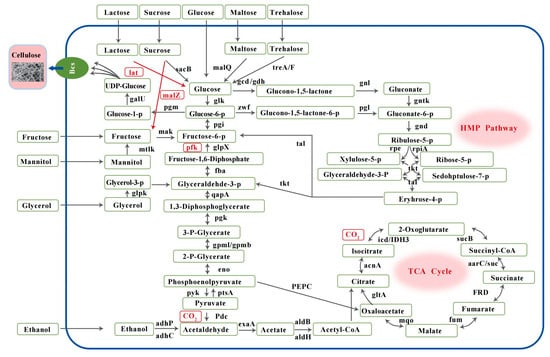
Figure 5.
The metabolic pathways of BC synthesis in N. cocois WE7T. Gene names and gene products. The black arrows mark genes or pathways in the genome. The red arrows and gene names marked genes or pathways not found in the genome. glk: glucokinase; pgm: phosphoglucomutase; galU: UTP-glucose-1-phosphate uridylyl transferase; gcd: quinoprotein glucose dehydrogenase; gdh: glucose dehydrogenase; gnl: gluconolactonase; gntK: gluconokinase; zwf: glucose-6-phosphate 1-dehydrogenase; pgl: 6-phosphogluco-nolactonase; gnd: 6-phosphogluconate dehydrogenase; rpe: ribulose-phosphate 3-epimerase; rpiA: ribose 5-phosphate isomerase A; glpK: glycerol kinase; mtlk: mannitol 2-dehydrogenase; HK: hexokinase; pgi: glucose-6-phosphate isomerase; glpX: fructose-1,6-bisphosphatase II; fba: fructose-bisphosphate aldolase, class II; gapA: glyceraldehyde 3-phosphate dehydrogenase; pgk: phosphoglycerate kinase; gpmI: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase; gpmB: probable phosphoglycerate mutase; neo: enolase; pyk: pyruvate kinase; PtsA: phosphate ABC transporter permease; Pdc: pyruvate dehydrogenase; suc: succinyl-CoA synthetase; adhP: alcohol dehydrogenase, propanol-preferring; adhC: alcohol dehydrogenase; exaA: alcohol dehydrogenase (cytochrome c); aldH: aldehyde dehydrogenase (NAD+); aldB: aldehyde dehydrogenase; sacB: levansucrase; malQ: 4-alpha-glucanotransferase; treA: alpha-trehalase; malZ: alpha-glucosidase.
The energy metabolism system regulates the biosynthetic process of BC. The relevant site is on nicotinamide adenine-dependent glucose dehydrogenase (gdp), which is related to NAD and sensitive to ATP. When the ATP content in the cell is low, glucose is converted into glucose-6-phosphate (G-6-P), which enters the EMP pathway and enhances the energy-producing metabolic efficiency of the strain. When there is high intracellular ATP content, ATP inhibits G-6-P synthesis, and glucose metabolism is directed to the BC pathway [31]. Compared with other carbon sources, BC-producing strains using glucose as the sole carbon source need to accumulate sufficient ATP in the cell before the metabolic flow is shifted to BC synthesis; therefore, BC production is generally lower. As shown in Figure 5, the genes for gdh and pyrroloquinolin quinone-dependent glucose dehydrogenase (gcd) in the gluconic acid production pathway were predicted in the WE7 genome, and gcd requires pyrroloquinolin quinone (PQQ) as a redox cofactor for its enzymatic activity [32]. The PQQ operon is composed of six genes, and at least four (PqqA, PqqC, PqqD, and PqqE) are required to catalyze the biosynthesis of PQQ [33]. Five genes (PqqA, PqqB, PqqC, PqqD, and PqqE) were predicted to be involved in the biosynthesis of PQQ in the genome of WE7, which means that WE7 can synthesize PQQ [34]. The PQQ gene is critical for E. coli to secrete gluconic acid [23,32]. In summary, WE7 has all the genes required to produce gluconic acid. When glucose is used as the carbon source, it consumes some of the carbon sources to produce gluconic acid, causing the accumulation of gluconic acid, significantly reducing pH, and resulting in low levels of BC synthesis.
3.5. Effect of Carbon Source on BC Synthesis
3.5.1. Effect of Different Carbon Sources on BC Yield and Biomass
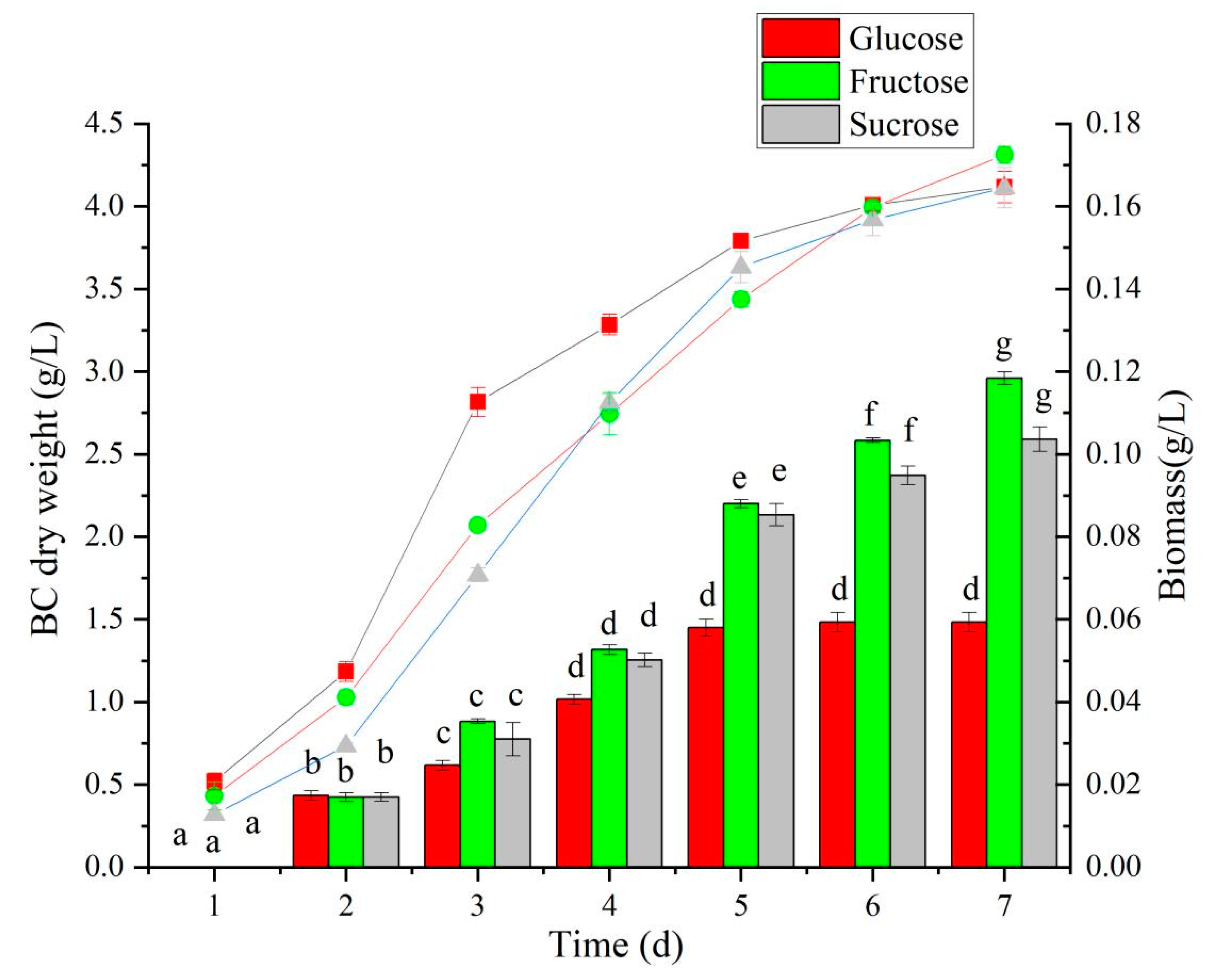
BC production is related to the strain type and carbon source used in the medium. HS medium is a universal medium for BC production. Using glucose as a carbon source, gluconic acid will be produced in the process of BC production, resulting in a decrease in the pH of the medium and affecting the efficiency of BC production. Therefore, based on the results of genomic analysis, fructose and sucrose were selected as carbon sources instead of glucose in this study to study the effects of different carbon sources on the production of BC. When glucose was used as a carbon source, the BC yield peaked on the fifth fermentation day (1.45 g/L), and the yield did not significantly increase on the sixth day (Figure 6). With fructose as a carbon source, BC production increased steadily over seven days of fermentation and reached 2.96 g/L on the seventh day. When sucrose was used as a carbon source, the BC yield was higher than that of glucose, but there was no significant difference with fructose, suggesting that fructose could be used for N. cocois WE7.
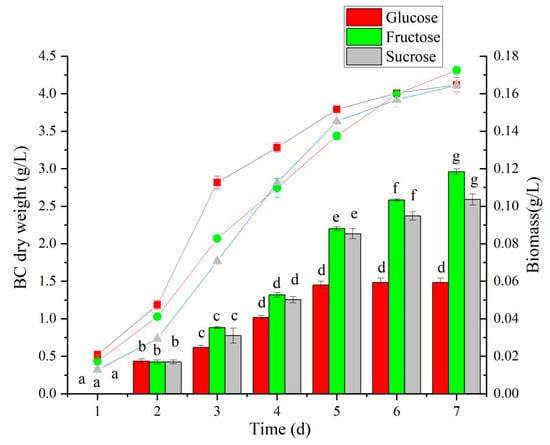
Figure 6.
The effect of different carbon sources on BC yield and biomass. Note: The BC yield is marked by bars; lines mark the biomass. Different lowercase letters indicate a significant difference (p < 0.05).
The cell biomass of N. cocois WE7 during fermentation with different carbon sources is shown in Figure 6. In the early stage of fermentation culture, the biomass of those with glucose as a carbon source was the highest (p < 0.05). This effect may be because glucose is more easily utilized by cells. In a medium with glucose as a carbon source, N. cocois WE7 had higher biomass; however, its BC yield was the lowest among the three sugars.
3.5.2. Relationship between the Yield of BC Synthesized by Different Carbon Sources and Organic Acid
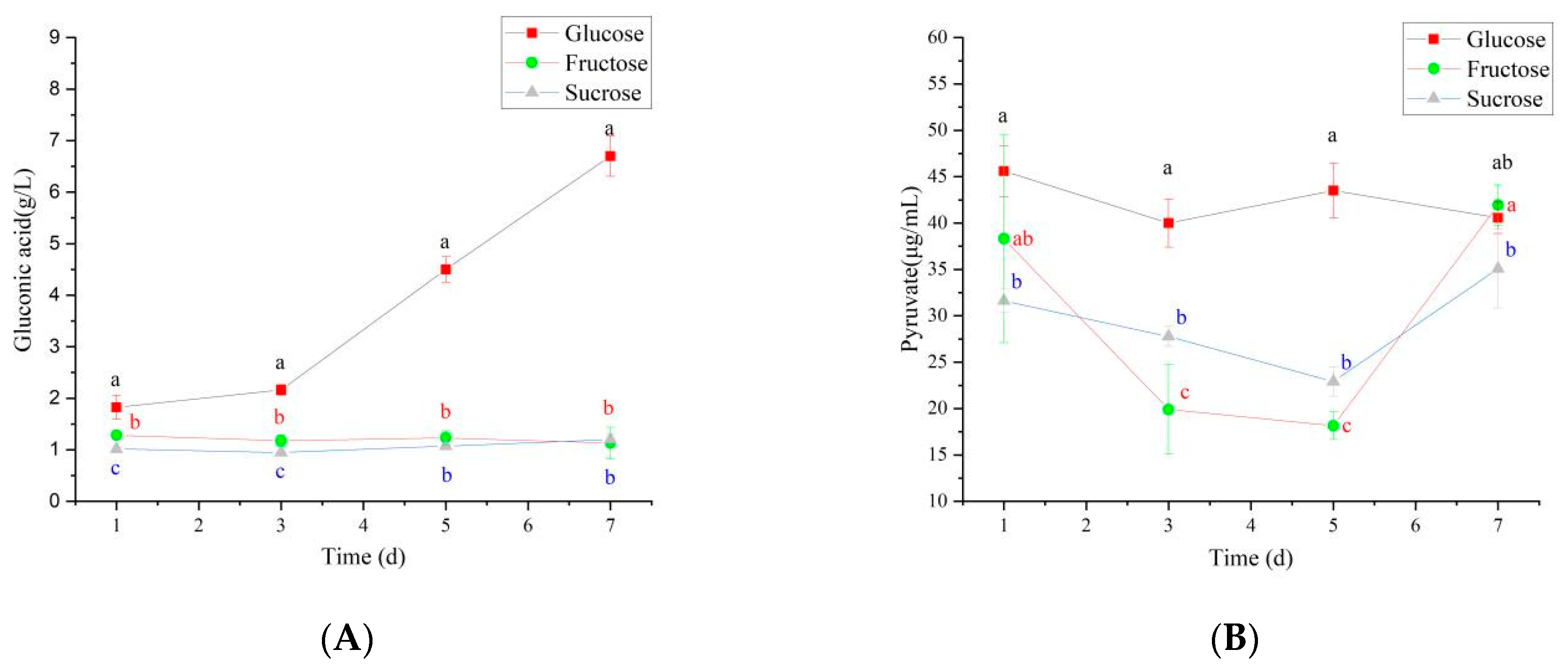
It is reported that if gluconic acid is produced too much or too early in the system, it will directly affect the supply level of UDPGlc, the cellulose synthesis precursor, and the yield of BC [35]. Therefore, it is necessary to fully consider effectively avoiding gluconic acid accumulation when selecting carbon sources.
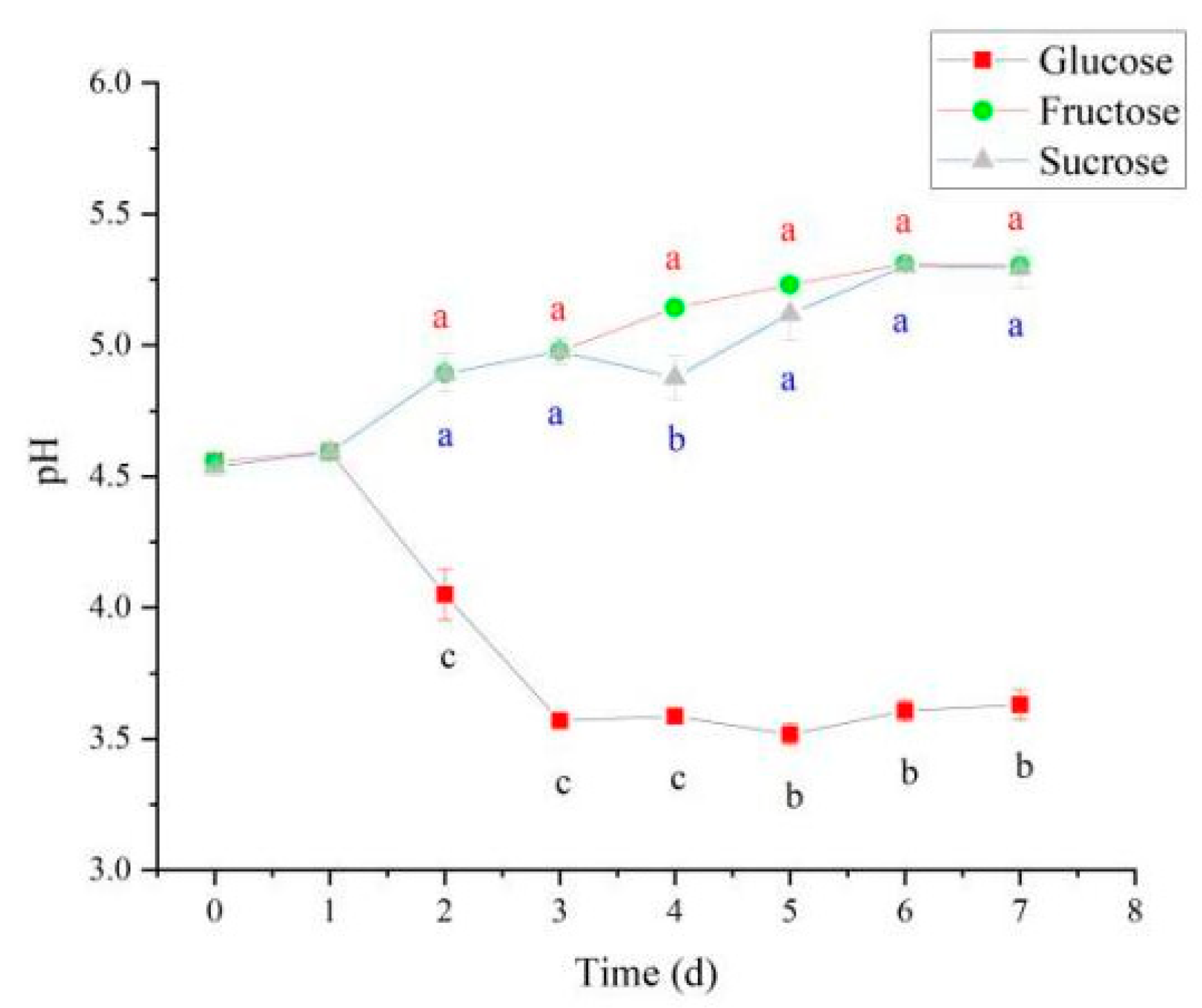
Under different carbon source conditions, N. cocois WE7 substantially differs in the metabolic pathway. When glucose is used as a carbon source, some glucose will enter the gluconic acid production pathway in addition to energy metabolism through the HMP pathway. Thus, on the third day of fermentation, gluconic acid content increased rapidly (Figure 7A) and was significantly higher than sucrose and fructose as carbon sources, resulting in a significant decrease in pH (Figure 8). In the middle stage of fermentation, when glucose was used as a carbon source, pyruvate content was significantly higher than that of sucrose and fructose as carbon sources (Figure 7B), indicating that more metabolic flow turned to energy generation-related metabolic pathways, and the BC synthesis pathway was weakened. Therefore, BC yield was the lowest when glucose was used as a carbon source. When fructose is used as a carbon source, in addition to maintaining the growth of bacteria, the metabolic flow enters the BC production pathway. Therefore, gluconic acid and pyruvate contents do not increase significantly during the metabolic process, and the BC production is higher than that when glucose and sucrose are used as carbon sources. When sucrose is used as the carbon source, due to the slow release of glucose and fructose in the fermentation process, more glucose metabolic flow is diverted to HMP and other energy generation pathways, and gluconic acid and pyruvate do not increase significantly. The release of fructose increases BC production to a certain extent, but it is lower than fructose as a carbon source.
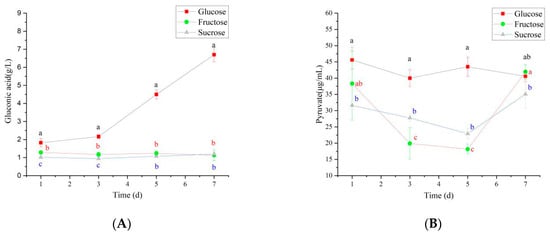
Figure 7.
Changes of organic acids during the synthesis of BC fermentation by different carbon sources ((A): gluconic acid content and (B): pyruvate content). Different lowercase letters indicate a significant difference (p < 0.05).
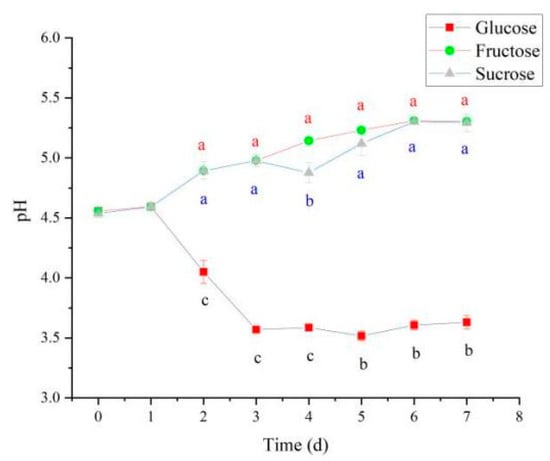
Figure 8.
Changes in pH during BC synthesis from various carbon sources. Different lowercase letters indicate a significant difference (p < 0.05).
Considering the changes in BC yield, biomass, pH, and organic acids during fermentation, the biomass of fermentation broth was not directly proportional to BC production. The key is determining whether more metabolic flux is diverted to BC synthesis. BC production was low when glucose was used as the carbon source, possibly because part of glucose is used for energy production through the TCA cycle and the HMP pathway. In the early fermentation stage, glucose may synthesize gluconic acid in large amounts, leading to decreased pH of the fermentation broth. These results are consistent with metabolic flux analysis during BC synthesis [35]. When fructose and sucrose were used as carbon sources, the amount of gluconic acid produced by bacterial metabolism was less, which is favorable for metabolic flux diverted to BC synthesis, increasing the BC production of N. cocois WE7.
4. Conclusions
Genomic analysis showed WE7 contained one chromosome and six plasmids, predicted 3106 genes, two BC production-related operons (bcs I and bcs II), and 11 genes encoding BC production components. The lack of bcs III operons in WE7 may result in lower production of BC. WE7 had an intact metabolic pathway for secreting gluconic acid, affecting BC production. WE7 has enzymes that use glucose, fructose, sucrose, glycerol, maltose, and trehalose as the sole carbon source but lacks enzymes to metabolize carbohydrate carbon source through the EMP pathway, so it is speculated that WE7 metabolizes carbon source through the HMP and TCA pathways. When glucose is used as a carbon source, it will enter the gluconic acid production pathway, decreasing BC production. However, when fructose is used as a carbon source, they do not directly enter the gluconic acid production pathway, and more metabolic flow flows to the BC synthesis pathway, so higher BC yield can be obtained. These predictions were confirmed in experiments on carbon source utilization.
When glucose is used as the carbon source, the metabolic flow will turn more toward the energy production pathway, and gluconic acid will be synthesized in large quantities in the early stage of fermentation, resulting in decreased pH of fermentation liquid and decreased BC production. With fructose and sucrose as carbon sources, the amount of gluconic acid produced by bacteria metabolism is less, and the metabolic flow of bacteria turns more to the metabolic pathway of BC synthesis to improve the production of bacterial cellulose synthesized by N. cocois WE7. This is the first genome study of N. cocois. It is of great theoretical significance to study the regulatory characteristics and mechanisms of BC synthase genes using strain WE7 and lays a foundation for the construction of engineering strains adapted to different BC applications.
Author Contributions
Conceptualization, S.L. and L.L.; Methodology and Investigation, Y.Z., M.C., J.L. and S.S.; Writing—original draft, Y.Z.; Writing—review and editing, M.C. and S.F.; Funding acquisition and Supervision, C.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Hainan Province Key R & D Project (ZDYF2020102), the National Natural Science Foundation of China (32060529 and 31660458), the Taishan Industrial Experts Program, and Science and Technology Support Plan for Youth Innovation of Colleges and Universities in Shandong Province (2022KJ088).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the NCBI GenBank (https://www.ncbi.nlm.nih.gov/, accessed on 15 November 2022) under accession numbers CP110924 (Chromosome), CP110925 (pKWE7-1), CP110926 (pKWE7-2), CP110927 (pKWE7-3), CP110928 (pKWE7-4), CP110929 (pKWE7-5), and CP110930 (pKWE7-6).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lahiri, D.; Nag, M.; Dutta, B.; Dey, A.; Sarkar, T.; Pati, S.; Edinur, H.A.; Abdul, K.Z.; Mohd, N.N.; Ray, R.R. Bacterial Cellulose: Production, Characterization, and Application as Antimicrobial Agent. Int. J. Mol. Sci. 2021, 22, 12984. [Google Scholar] [CrossRef] [PubMed]
- Brandao, P.R.; Crespo, M.T.B.; Nascimento, F.X. Phylogenomic and comparative analyses support the reclassification of several Komagataeibacter species as novel members of the Novacetimonas gen. nov. and bring new insights into the evolution of cellulose synthase genes. Int. J. Syst. Evol. Microbiol. 2022, 72, 005252. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.X.; Liu, S.X.; Wang, Y.M.; Bi, J.C.; Chen, H.M.; Deng, J.; Zhang, C.; Hu, Q.S.; Li, C.F. Komagataeibacter cocois sp. nov., a novel cellulose-producing strain isolated from coconut milk. Int. J. Syst. Evol. Microbiol. 2018, 68, 3125–3131. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Han, Y.H.; Ye, Y.X.; Shi, X.X.; Xiang, P.; Chen, D.L.; Li, M. Physicochemical characterization of high-quality bacterial cellulose produced by Komagataeibacter sp. strain W1 and identification of the associated genes in bacterial cellulose production. RSC Adv. 2017, 7, 45145–45155. [Google Scholar] [CrossRef]
- Szymczak, I.; Pietrzyk-Brzezinska, A.J.; Duszynski, K.; Ryngajllo, M. Characterization of the Putative Acylated Cellulose Synthase Operon in Komagataeibacter xylinus E25. Int. J. Mol. Sci. 2022, 23, 7851. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef]
- Wang, S.S.; Han, Y.H.; Chen, J.L.; Zhang, D.C.; Shi, X.X.; Ye, Y.X.; Chen, D.L.; Liu, M. Insights into Bacterial Cellulose Biosynthesis from Different Carbon Sources and the Associated Biochemical Transformation Pathways in Komagataeibacter sp. W1. Polymer 2018, 10, 963–985. [Google Scholar] [CrossRef]
- Fernandes, I.; Pedro, A.C.; Ribeiro, V.R.; Bortolini, D.G.; Ozaki, M.; Maciel, G.M.; Haminiuk, C. Bacterial cellulose: From production optimization to new applications. Int. J. Biol. Macromol. 2020, 164, 2598–2611. [Google Scholar] [CrossRef]
- Rai, R.; Dhar, P. Biomedical engineering aspects of nanocellulose: A review. Nanotechnology 2022, 33, 2001. [Google Scholar] [CrossRef]
- Randhawa, A.; Dutta, S.D.; Ganguly, K.; Patil, T.V.; Patel, D.K.; Lim, K.T. A Review of Properties of Nanocellulose, Its Synthesis, and Potential in Biomedical Applications. Appl. Sci. 2022, 12, 7090. [Google Scholar] [CrossRef]
- Mohd Amin, M.C.I.; Ahmad, N.; Halib, N.; Ahmad, I. Synthesis and characterization of thermo- and pH-responsive bacterial cellulose/acrylic acid hydrogels for drug delivery. Carbohydr. Polym. 2012, 88, 465–473. [Google Scholar] [CrossRef]
- Nakai, T.; Sugano, Y.; Shoda, M.; Sakakibara, H.; Oiwa, K.; Tuzi, S.; Imai, T.; Sugiyama, J.; Takeuchi, M.; Yamauchi, D.; et al. Formation of highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J. Bacteriol. 2013, 195, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Schramm, M.; Hestrin, S. Factors affecting production of cellulose at the air/liquid interface of a culture of Acetobacter xylinum. J. Gen. Microbiol. 1954, 11, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Sun, Q.; Han, Z.; Li, J.; Liao, B.; Hu, L.; Huang, J.; Zou, C.; Jia, C.; Huang, J.; et al. Comparison of bacterial nanocellulose produced by different strains under static and agitated culture conditions. Carbohydr. Polym. 2020, 227, 115323. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Gao, H.; Liao, B.; Wu, J.; Zhang, W.; Huang, J.; Liu, M.; Huang, J.; Chang, Z.; Jin, M.; et al. Characterization and optimization of production of bacterial cellulose from strain CGMCC 17276 based on whole-genome analysis. Carbohydr. Polym. 2020, 232, 115788. [Google Scholar] [CrossRef]
- La China, S.; Bezzecchi, A.; Moya, F.; Petroni, G.; Di Gregorio, S.; Gullo, M. Genome sequencing and phylogenetic analysis of K1G4: A new Komagataeibacter strain producing bacterial cellulose from different carbon sources. Biotechnol. Lett. 2020, 42, 807–818. [Google Scholar] [CrossRef]
- Hernandez-Arriaga, A.M.; Del, C.C.; Urbina, L.; Eceiza, A.; Corcuera, M.A.; Retegi, A.; Auxiliadora, P.M. Genome sequence and characterization of the bcs clusters for the production of nanocellulose from the low pH resistant strain Komagataeibacter medellinensis ID13488. Microb. Biotechnol. 2019, 12, 620–632. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Cannazza, P.; Rissanen, A.J.; Guizelini, D.; Losoi, P.; Sarlin, E.; Romano, D.; Santala, V.; Mangayil, R. Characterization of Komagataeibacter isolate reveals new prospects in waste stream valorization for bacterial cellulose production. Microorganisms 2021, 9, 2230. [Google Scholar] [CrossRef]
- Tseng, Y.S.; Patel, A.K.; Chen, C.; Dong, C.; Singhania, R.R. Improved production of bacterial cellulose by Komagataeibacter europaeus employing fruit extract as carbon source. J. Food Sci. Technol. 2023, 60, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Du, J.; Li, J.; Li, M. Quantification of the Organic Acids in Hawthorn Wine: A Comparison of Two HPLC Methods. Molecules 2019, 24, 2150. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Azi, F.; Ge, Z.W.; Liu, Y.F.; Yin, X.T.; Dong, M.S. Bio-conversion of kitchen waste into bacterial cellulose using a new multiple carbon utilizing Komagataeibacter rhaeticus: Fermentation profiles and genome-wide analysis. Int. J. Biol. Macromol. 2021, 191, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Saxena, I.M.; Lin, F.C.; Brown, R.J. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol. Biol. 1990, 15, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.C.; Fear, A.L.; Calhoon, R.D.; Eichinger, G.H.; Mayer, R.; Amikam, D.; Benziman, M.; Gelfand, D.H.; Meade, J.H.; Emerick, A.W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 1990, 87, 8130–8134. [Google Scholar] [CrossRef]
- Bimmer, M.; Mientus, M.; Klingl, A.; Ehrenreich, A.; Liebl, W. The roles of the various cellulose biosynthesis operons in Komagataeibacter hansenii atcc 23769. Appl. Environ. Microbiol. 2022, 88, e246021. [Google Scholar] [CrossRef]
- Krasteva, P.V.; Bernal-Bayard, J.; Travier, L.; Martin, F.A.; Kaminski, P.A.; Karimova, G.; Fronzes, R.; Ghigo, J.M. Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun. 2017, 8, 2065. [Google Scholar] [CrossRef]
- Huang, L.H.; Li, X.J.; Sun, X.W.; Wang, X.; Wang, Y.T.; Jia, S.R.; Zhong, C. Regulating the structure of bacterial cellulose by altering the expression of bcsD using CRISPR/dCas9. Chin. J. Biotech. 2022, 38, 772–779. [Google Scholar]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Umeda, Y.; Hirano, A.; Ishibashi, M.; Akiyama, H.; Onizuka, T.; Ikeuchi, M.; Inoue, Y. Cloning of cellulose synthase genes from Acetobacter xylinum JCM 7664: Implication of a novel set of cellulose synthase genes. DNA Res. 1999, 6, 109–115. [Google Scholar] [CrossRef]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 1991, 55, 35–58. [Google Scholar] [CrossRef]
- Kim, C.H.; Han, S.H.; Kim, K.Y.; Cho, B.H.; Kim, Y.H.; Koo, B.S.; Kim, Y.C. Cloning and expression of pyrroloquinoline quinone (PQQ) genes from a phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 2003, 47, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Y.; Xue, S.T.; Zhan, Y.C.; Shen, J.J.; Wu, L.T.; Jin, J.; Wang, Z.; Li, Z.R. Design, synthesis and antiproliferative activity of a novel class of indole-2-carboxylate derivatives. Eur. J. Med. Chem. 2014, 83, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Han, Z.; Ge, X.; Tian, P. Distinct promoters affect pyrroloquinoline quinone production in recombinant Escherichia coli and Klebsiella pneumoniae. Curr. Microbiol. 2014, 69, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Zhang, G.C.; Liu, M.; Zheng, X.T.; Han, P.P.; Jia, S.R. Metabolic flux analysis of Gluconacetobacter xylinus for bacterial cellulose production. Appl. Microbiol. Biotechnol. 2013, 97, 6189–6199. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).