In Pursuit of Understanding the Rumen Microbiome
Abstract
1. Introduction
2. Rumen Development
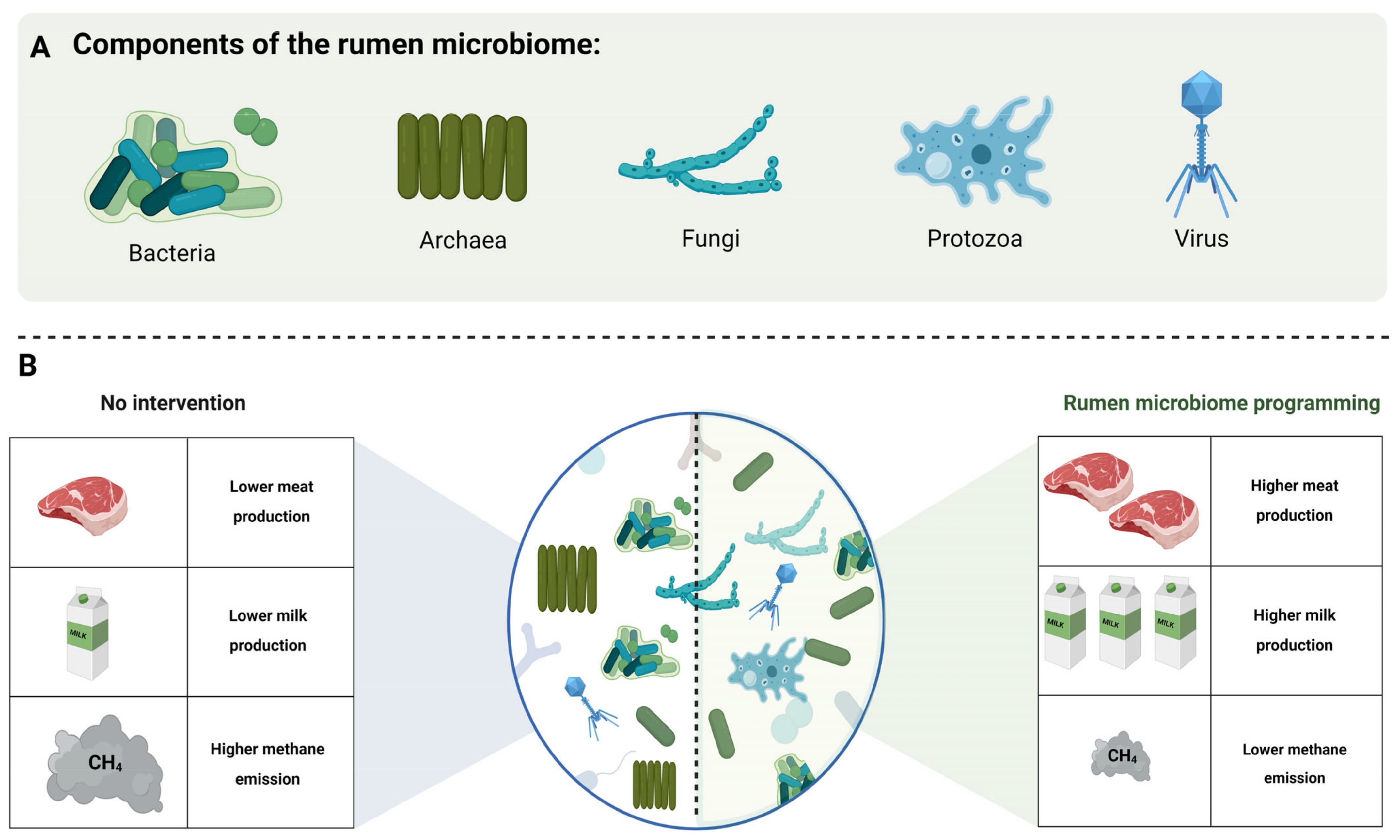
3. Rumen Microbiome
3.1. Bacteria
3.2. Archaea
3.3. Fungi
3.4. Protozoa
3.5. Virus
4. Rumen Fermentation-Metabolic Cascades
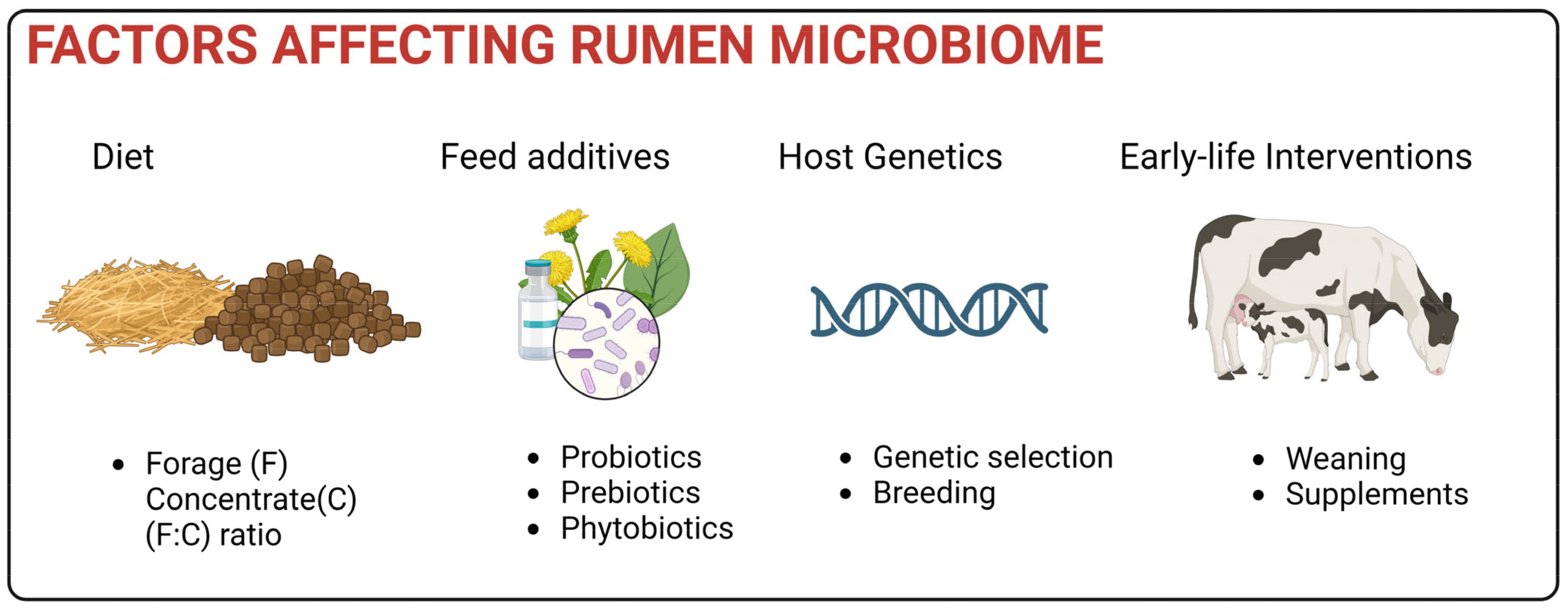
5. Factors Affecting Rumen Microbiome
5.1. Impact of Diet
5.2. Impact of host
5.3. Feed Additives
5.4. Early Life Interventions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hackmann, T.J.; Spain, J.N. Invited Review: Ruminant Ecology and Evolution: Perspectives Useful to Ruminant Livestock Research and Production. J. Dairy Sci. 2010, 93, 1320–1334. [Google Scholar] [CrossRef]
- Castillo-González, A.R.; Burrola-Barraza, M.E.; Domínguez-Viveros, J.; Chávez-Martínez, A. Rumen microorganisms and fermentation. Arch. Med. Vet. 2014, 46, 349–361. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; Ramos-Morales, E. Review: Ruminal Microbiome and Microbial Metabolome: Effects of Diet and Ruminant Host. Animal 2020, 14, s78–s86. [Google Scholar] [CrossRef] [PubMed]
- Gruninger, R.J.; Ribeiro, G.O.; Cameron, A.; McAllister, T.A. Invited Review: Application of Meta-Omics to Understand the Dynamic Nature of the Rumen Microbiome and How It Responds to Diet in Ruminants. Animal 2019, 13, 1843–1854. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy Contributions of Volatile Fatty Acids from the Gastrointestinal Tract in Various Species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Hackmann, T.J.; Firkins, J.L. Maximizing Efficiency of Rumen Microbial Protein Production. Front. Microbiol. 2015, 6, 465. [Google Scholar] [CrossRef]
- Tseten, T.; Sanjorjo, R.A.; Kwon, M.; Kim, S.W. Strategies to Mitigate Enteric Methane Emissions from Ruminant Animals. J. Microbiol. Biotechnol. 2022, 32, 269–277. [Google Scholar] [CrossRef]
- Jangir, K.; Gujar, B. Role of Livestock in Global Warming. Pharma Innov. J. 1934, 11, 1934–1938. [Google Scholar]
- Arndt, C.; Hristov, A.N.; Price, W.J.; McClelland, S.C.; Pelaez, A.M.; Cueva, S.F.; Oh, J.; Dijkstra, J.; Bannink, A.; Bayat, A.R.; et al. Full Adoption of the Most Effective Strategies to Mitigate Methane Emissions by Ruminants Can Help Meet the 1.5 °C Target by 2030 but Not 2050. Proc. Natl. Acad. Sci. USA 2022, 119, e2111294119. [Google Scholar] [CrossRef]
- Lan, X.; Thoning, K.W.; Dlugokencky, E.J. Trends in Globally-Averaged CH4, N2O, and SF6; Determined from NOAA Global Monitoring Laboratory Measurements; NOAA Global Monitoring Laboratory Measurements: Boulder, CO, USA, 2022. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency Office of Atmospheric Protection Greenhouse Gas Reporting Program (GHGRP). Climate Change Indicators: Atmospheric Concentrations of Greenhouse Gases. 2022. Available online: https://www.epa.gov/climate-indicators (accessed on 6 November 2022).
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The Rumen Microbiome: A Crucial Consideration When Optimising Milk and Meat Production and Nitrogen Utilisation Efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Pereira, L.G.; Vet, M.; Machado, F.S.; Campos, M.M.; Guimaraes Júnior, R.; Tomich, T.R.; Reis, L.G.; Coombs, C.; Vet St, M. Enteric methane mitigation strategies in ruminants: A review. Rev. Colomb. Cienc. Pecu. 2015, 28, 124–143. [Google Scholar] [CrossRef]
- Rate, N.M. World Population Prospects: The 2017 Revision; United Nations: New York, NY, USA, 2017. [Google Scholar]
- Hatab, A.A.; Cavinato, M.E.R.; Lagerkvist, C.J. Urbanization, Livestock Systems and Food Security in Developing Countries: A Systematic Review of the Literature. Food Secur. 2019, 11, 279–299. [Google Scholar] [CrossRef]
- Melletti, M.; Burton, J. Ecology, Evolution and Behaviour of Wild Cattle: Implications for Conservation; Cambridge University Press: Cambridge, UK, 2014; ISBN 9781139568098. [Google Scholar]
- Pradesh, M.; Baghel, I.R.; Thakur, D.; Govil, K.; Yadav, D.S.; Patil, A.K.; Nayak, S.; Baghel, R.; Yadav, P.K. Feeding Management for Early Rumen Development in Calves. J. Entomol. Zool. Stud. 2017, 5, 1132–1139. [Google Scholar]
- Górka, P.; Kowalski, Z.M.; Pietrzak, P.; Kotunia, A.; Jagusiak, W.; Zabielski, R. Is Rumen Development in Newborn Calves Affected by Different Liquid Feeds and Small Intestine Development? J. Dairy Sci. 2011, 94, 3002–3013. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L.; McLeod, K.R.; Klotz, J.L.; Heitmann, R.N. Rumen Development, Intestinal Growth and Hepatic Metabolism in the Pre- and Postweaning Ruminant. J. Dairy Sci. 2004, 87, E55–E65. [Google Scholar] [CrossRef]
- Heinrichs, A.J.; Lesmeister, K.E. Rumen Development in the Dairy Calf. Calf and heifer rearing: Principles of rearing the modern dairy heifer from calf to calving. In Proceedings of the 60th University of Nottingham Easter School in Agricultural Science, Nottingham, UK, 23–24 March 2004; pp. 53–65. [Google Scholar]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating Rumen Microbiome and Fermentation through Interventions during Early Life: A Review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef]
- Diao, Q.; Zhang, R.; Fu, T. Review of Strategies to Promote Rumen Development in Calves. Animals 2019, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Choi, S.H.; Nogoy, K.M.; Liang, S. Review: The Development of the Gastrointestinal Tract Microbiota and Intervention in Neonatal Ruminants. Animal 2021, 15, 100316. [Google Scholar] [CrossRef]
- Jiao, J.; Li, X.; Beauchemin, K.A.; Tan, Z.; Tang, S.; Zhou, C. Rumen Development Process in Goats as Affected by Supplemental Feeding v. Grazing: Age-Related Anatomic Development, Functional Achievement and Microbial Colonisation. Br. J. Nutr. 2015, 113, 888–900. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Liang, G.; Guan, L.L. Regulation of Rumen Development in Neonatal Ruminants through Microbial Metagenomes and Host Transcriptomes. Genome Biol. 2019, 20, 172. [Google Scholar] [CrossRef]
- Khalil, A.; Batool, A.; Arif, S. Healthy Cattle Microbiome and Dysbiosis in Diseased Phenotypes. Ruminants 2022, 2, 134–156. [Google Scholar] [CrossRef]
- Zou, X.; Liu, G.; Meng, F.; Hong, L.; Li, Y.; Lian, Z.; Yang, Z.; Luo, C.; Liu, D. Exploring the Rumen and Cecum Microbial Community from Fetus to Adulthood in Goat. Animals 2020, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Guzman, C.E.; Wood, J.L.; Egidi, E.; White-Monsant, A.C.; Semenec, L.; Grommen, S.V.H.; Hill-Yardin, E.L.; de Groef, B.; Franks, A.E. A Pioneer Calf Foetus Microbiome. Sci. Rep. 2020, 10, 17712. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.O.; Nagaraja, T.G.; Wright, A.D.G.; Callaway, T.R. Board-Invited Review: Rumen Microbiology: Leading the Way in Microbial Ecology1,2. J. Anim. Sci. 2013, 91, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.-T.; Bryant, M.P.; Robert, E. Hungate: Pioneer of Anaerobic Microbial Ecology. Anaerobe 1997, 3, 213–217. [Google Scholar] [CrossRef]
- Deusch, S.; Camarinha-Silva, A.; Conrad, J.; Beifuss, U.; Rodehutscord, M.; Seifert, J. A Structural and Functional Elucidation of the Rumen Microbiome Influenced by Various Diets and Microenvironments. Front. Microbiol. 2017, 8, 1605. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.; Yilmaz, P.; Kumar, S.; Forster, R.J.; Kelly, W.J.; Leahy, S.C.; Guan, L.L.; Janssen, P.H. Improved Taxonomic Assignment of Rumen Bacterial 16S RRNA Sequences Using a Revised SILVA Taxonomic Framework. PeerJ 2019, 2019, e6496. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.L.A.; Li, F.; Ghoshal, B.; McAllister, T.; Guan, L.L. Enhancing the Resolution of Rumen Microbial Classification from Metatranscriptomic Data Using Kraken and Mothur. Front. Microbiol. 2017, 8, 2445. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.Y.; Ji, S.K.; Yan, H.; Wang, Y.J.; Liu, J.J.; Cao, Z.J.; Yang, H.J.; Zhang, W.J.; Li, S.L. Dynamic Change of the Gastrointestinal Bacterial Ecology in Cows from Birth to Adulthood. Microbiol. Open 2020, 9, e1119. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Ossa, F. REVIEW: The Rumen Microbiome: Composition, Abundance, Diversity, and New Investigative Tools. Prof. Anim. Sci. 2014, 30, 1–12. [Google Scholar] [CrossRef]
- Hennessy, M.L.; Indugu, N.; Vecchiarelli, B.; Bender, J.; Pappalardo, C.; Leibstein, M.; Toth, J.; Katepalli, A.; Garapati, S.; Pitta, D. Temporal Changes in the Fecal Bacterial Community in Holstein Dairy Calves from Birth through the Transition to a Solid Diet. PLoS ONE 2020, 15, e0238882. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Guan, L.L.; Liu, J.X. Assessment of Rumen Bacteria in Dairy Cows with Varied Milk Protein Yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef] [PubMed]
- Koringa, P.G.; Thakkar, J.R.; Pandit, R.J.; Hinsu, A.T.; Parekh, M.J.; Shah, R.K.; Jakhesara, S.J.; Joshi, C.G. Metagenomic Characterisation of Ruminal Bacterial Diversity in Buffaloes from Birth to Adulthood Using 16S RRNA Gene Amplicon Sequencing. Funct. Integr. Genom. 2019, 19, 237–247. [Google Scholar] [CrossRef]
- Rabee, A.E.; Kewan, K.Z.; Sabra, E.A.; el Shaer, H.M.; Lamara, M. Rumen Bacterial Community Profile and Fermentation in Barki Sheep Fed Olive Cake and Date Palm Byproducts. PeerJ 2021, 9, e12447. [Google Scholar] [CrossRef]
- Guo, W.; Li, Y.; Wang, L.; Wang, J.; Xu, Q.; Yan, T.; Xue, B. Evaluation of Composition and Individual Variability of Rumen Microbiota in Yaks by 16S RRNA High-Throughput Sequencing Technology. Anaerobe 2015, 34, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Gruninger, R.J.; Sensen, C.W.; McAllister, T.A.; Forster, R.J. Diversity of Rumen Bacteria in Canadian Cervids. PLoS ONE 2014, 9, e89682. [Google Scholar] [CrossRef]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Hadjithomas, M.; Varghese, N.J.; et al. Cultivation and Sequencing of Rumen Microbiome Members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H.; Abecia, L.; Angarita, E.; Aravena, P.; Arenas, G.N.; et al. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Bhujbal, S.K.; Ghosh, P.; Vijay, V.K.; Rathour, R.; Kumar, M.; Singh, L.; Kapley, A. Biotechnological Potential of Rumen Microbiota for Sustainable Bioconversion of Lignocellulosic Waste to Biofuels and Value-Added Products. Sci. Total Environ. 2022, 814, 152773. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Hao, Z.; Shi, H.; Li, T.; Wang, H.; Li, Q.; Zhang, Y.; Ma, Y. Metagenomic Analysis Revealed Differences in Composition and Function Between Liquid-Associated and Solid-Associated Microorganisms of Sheep Rumen. Front. Microbiol. 2022, 13, 851567. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen Methanogens and Mitigation of Methane Emission by Anti-Methanogenic Compounds and Substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, C.O.; Olaniyan, O.T.; Dash, R.; Varma, A. The Process of Methanogenesis by Rumen Microorganisms: State of Art. In Animal Manure; Soil Biology; Springer: Cham, Switzerland, 2022; pp. 13–20. [Google Scholar]
- Glasson, C.R.K.; Kinley, R.D.; de Nys, R.; King, N.; Adams, S.L.; Packer, M.A.; Svenson, J.; Eason, C.T.; Magnusson, M. Benefits and Risks of Including the Bromoform Containing Seaweed Asparagopsis in Feed for the Reduction of Methane Production from Ruminants. Algal Res. 2022, 64, 102673. [Google Scholar] [CrossRef]
- Bauchop, T. Inhibition of Rumen Methanogenesis by Methane Analogues. J. Bacteriol. 1967, 94, 171–175. [Google Scholar] [CrossRef]
- Janssen, P.H.; Kirs, M. Structure of the Archaeal Community of the Rumen. Appl. Environ. Microbiol. 2008, 74, 3619–3625. [Google Scholar] [CrossRef]
- Levy, B.; Jami, E. Exploring the Prokaryotic Community Associated with the Rumen Ciliate Protozoa Population. Front. Microbiol. 2018, 9, 2526. [Google Scholar] [CrossRef]
- Wang, Z.; Elekwachi, C.O.; Jiao, J.; Wang, M.; Tang, S.; Zhou, C.; Tan, Z.; Forster, R.J. Investigation and Manipulation of Metabolically Active Methanogen Community Composition during Rumen Development in Black Goats. Sci. Rep. 2017, 7, 422. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty Years of Research on Rumen Methanogenesis: Lessons Learned and Future Challenges for Mitigation. Animal 2020, 14, S2–S16. [Google Scholar] [CrossRef]
- Kittelmann, S.; Seedorf, H.; Walters, W.A.; Clemente, J.C.; Knight, R.; Gordon, J.I.; Janssen, P.H. Simultaneous Amplicon Sequencing to Explore Co-Occurrence Patterns of Bacterial, Archaeal and Eukaryotic Microorganisms in Rumen Microbial Communities. PLoS ONE 2013, 8, e47879. [Google Scholar] [CrossRef]
- Aller, J.Y.; Kemp, P.F. Are Archaea Inherently Less Diverse than Bacteria in the Same Environments? FEMS Microbiol. Ecol. 2008, 65, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Vanwonterghem, I.; Evans, P.N.; Parks, D.H.; Jensen, P.D.; Woodcroft, B.J.; Hugenholtz, P.; Tyson, G.W. Methylotrophic Methanogenesis Discovered in the Archaeal Phylum Verstraetearchaeota. Nat. Microbiol. 2016, 1, 16170. [Google Scholar] [CrossRef] [PubMed]
- Sundset, M.A.; Edwards, J.E.; Cheng, Y.F.; Senosiain, R.S.; Fraile, M.N.; Northwood, K.S.; Praesteng, K.E.; Glad, T.; Mathiesen, S.D.; Wright, A.-D.G. Rumen Microbial Diversity in Svalbard Reindeer, with Particular Emphasis on Methanogenic Archaea. FEMS Microbiol. Ecol. 2009, 70, 553–562. [Google Scholar] [CrossRef]
- Hook, S.E.; Wright, A.D.G.; McBride, B.W. Methanogens: Methane Producers of the Rumen and Mitigation Strategies. Archaea 2010, 2010, 945785. [Google Scholar] [CrossRef] [PubMed]
- Guzman, C.E.; Bereza-Malcolm, L.T.; de Groef, B.; Franks, A.E. Presence of Selected Methanogens, Fibrolytic Bacteria, and Proteobacteria in the Gastrointestinal Tract of Neonatal Dairy Calves from Birth to 72 Hours. PLoS ONE 2015, 10, e0133048. [Google Scholar] [CrossRef]
- Akin, D.E.; Borneman, W.S. Role of Rumen Fungi in Fiber Degradation. J. Dairy Sci. 1990, 73, 3023–3032. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.W.; Khoo, I.Y.S.; Tan, S.G.; Abdullah, N.; Jalaludin, S.; Kudo, H. Isozyme Analysis of Anaerobic Rumen Fungi and Their Relationship to Aerobic Chytrids. Microbiology 1994, 140, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Hagen, L.H.; Brooke, C.G.; Shaw, C.A.; Norbeck, A.D.; Piao, H.; Arntzen, M.; Olson, H.M.; Copeland, A.; Isern, N.; Shukla, A.; et al. Proteome Specialization of Anaerobic Fungi during Ruminal Degradation of Recalcitrant Plant Fiber. ISME J. 2021, 15, 421–434. [Google Scholar] [CrossRef]
- Elekwachi, C.O.; Wang, Z.; Wu, X.; Rabee, A.; Forster, R.J. Total RRNA-Seq Analysis Gives Insight into Bacterial, Fungal, Protozoal and Archaeal Communities in the Rumen Using an Optimized RNA Isolation Method. Front. Microbiol. 2017, 8, 1814. [Google Scholar] [CrossRef]
- Guo, W.; Wang, W.; Bi, S.; Long, R.; Ullah, F.; Shafiq, M.; Zhou, M.; Zhang, Y. Characterization of Anaerobic Rumen Fungal Community Composition in Yak, Tibetan Sheep and Small Tail Han Sheep Grazing on the Qinghai-Tibetan Plateau. Animals 2020, 10, 144. [Google Scholar] [CrossRef]
- Rabee, A.E.; Forster, R.J.; Elekwachi, C.O.; Kewan, K.Z.; Sabra, E.A.; Shawket, S.M.; Mahrous, H.A.; Khamiss, O.A. Community Structure and Fibrolytic Activities of Anaerobic Rumen Fungi in Dromedary Camels. J. Basic Microbiol. 2019, 59, 101–110. [Google Scholar] [CrossRef]
- Wang, H.; Li, P.; Liu, X.; Zhang, C.; Lu, Q.; Xi, D.; Yang, R.; Wang, S.; Bai, W.; Yang, Z.; et al. The Composition of Fungal Communities in the Rumen of Gayals (Bos Frontalis), Yaks (Bos Grunniens), and Yunnan and Tibetan Yellow Cattle (Bos Taurs). Pol. J. Microbiol. 2019, 68, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Elshahed, M.S.; Hanafy, R.A.; Cheng, Y.; Dagar, S.S.; Edwards, J.E.; Flad, V.; Fliegerová, K.O.; Griffith, G.W.; Kittelmann, S.; Lebuhn, M.; et al. Characterization and Rank Assignment Criteria for the Anaerobic Fungi (Neocallimastigomycota). Int. J. Syst. Evol. Microbiol. 2022, 72, 005449. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E.; Kingston-Smith, A.H.; Jimenez, H.R.; Huws, S.A.; Skøt, K.P.; Griffith, G.W.; McEwan, N.R.; Theodorou, M.K. Dynamics of Initial Colonization of Nonconserved Perennial Ryegrass by Anaerobic Fungi in the Bovine Rumen. FEMS Microbiol. Ecol. 2008, 66, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vallvé, S.; Romeu, A.; Palau, J. Horizontal Gene Transfer of Glycosyl Hydrolases of the Rumen Fungi. Mol. Biol. Evol. 2000, 17, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Gruninger, R.J.; Nguyen, T.T.M.; Reid, I.D.; Yanke, J.L.; Wang, P.; Abbott, D.W.; Tsang, A.; McAllister, T. Application of Transcriptomics to Compare the Carbohydrate Active Enzymes That Are Expressed by Diverse Genera of Anaerobic Fungi to Degrade Plant Cell Wall Carbohydrates. Front. Microbiol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Duarte, E.R.; Abrão, F.O.; Ribeiro, I.C.O.; Vieira, E.A.; Nigri, A.C.; Silva, K.L.; Júnior, G.F.V.; Barreto, S.M.P.; Geraseev, L.C. Rumen Protozoa of Different Ages of Beef Cattle Raised in Tropical Pastures during the Dry Season. J. Appl. Anim. Res. 2018, 46, 1457–1461. [Google Scholar] [CrossRef]
- Newbold, C.J.; de la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The Role of Ciliate Protozoa in the Rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef]
- dos Reis, C.C.; Maeda, E.M.; Cedrola, F.; Martins, E.N.; de Paula, F.M.; Martinele, I. Diet and Breed Alter Community Structures of Rumen Protozoa in Cattle Subjected to Different Feeding Systems. Semin. Ciênc. Agrár. 2019, 40, 909–918. [Google Scholar] [CrossRef]
- Francisco, A.E.; Santos-Silva, J.M.; Portugal, A.P.v.; Alves, S.P.; Bessa, R.J.B. Relationship between Rumen Ciliate Protozoa and Biohydrogenation Fatty Acid Profile in Rumen and Meat of Lambs. PLoS ONE 2019, 14, e0221996. [Google Scholar] [CrossRef]
- Ayemele, A.G.; Ma, L.; Park, T.; Xu, J.; Yu, Z.; Bu, D. Giant Milkweed (Calotropis Gigantea): A New Plant Resource to Inhibit Protozoa and Decrease Ammoniagenesis of Rumen Microbiota in Vitro without Impairing Fermentation. Sci. Total Environ. 2020, 743, 140665. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Ramírez-Restrepo, C.A.; Shah, A.M.; Hu, R.; Bell, M.; Wang, Z.; McSweeney, C. The Community Structure and Microbial Linkage of Rumen Protozoa and Methanogens in Response to the Addition of Tea Seed Saponins in the Diet of Beef Cattle. J. Anim. Sci. Biotechnol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Coleman, G.S. Rumen ciliate protozoa. Adv. Parasitol. 1980, 18, 121–173. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.G.; Williams, A.G.; Butler, R.D. Hydrogenosomes in the Rumen Entodiniomorphid Ciliate Polyplastron Multivesiculatum. J. Gen. Microbiol. 1990, 136, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Park, T.; Yu, Z. Aerobic Cultivation of Anaerobic Rumen Protozoa, Entodinium Caudatum and Epidinium Caudatum. J. Microbiol. Methods 2018, 152, 186–193. [Google Scholar] [CrossRef]
- Williams, C.L.; Thomas, B.J.; McEwan, N.R.; Rees Stevens, P.; Creevey, C.J.; Huws, S.A. Rumen Protozoa Play a Significant Role in Fungal Predation and Plant Carbohydrate Breakdown. Front. Microbiol. 2020, 11, 720. [Google Scholar] [CrossRef]
- Findley, S.D.; Mormile, M.R.; Sommer-Hurley, A.; Zhang, X.-C.; Tipton, P.; Arnett, K.; Porter, J.H.; Kerley, M.; Stacey, G. Activity-Based Metagenomic Screening and Biochemical Characterization of Bovine Ruminal Protozoan Glycoside Hydrolases. Appl. Environ. Microbiol. 2011, 77, 8106–8113. [Google Scholar] [CrossRef]
- Ricard, G.; McEwan, N.R.; Dutilh, B.E.; Jouany, J.-P.; Macheboeuf, D.; Mitsumori, M.; McIntosh, F.M.; Michalowski, T.; Nagamine, T.; Nelson, N.; et al. Horizontal Gene Transfer from Bacteria to Rumen Ciliates Indicates Adaptation to Their Anaerobic, Carbohydrates-Rich Environment. BMC Genom. 2006, 7, 22. [Google Scholar] [CrossRef]
- Adams, J.C.; Gazaway, J.A.; Brailsford, M.D.; Hartman, P.A.; Jacobson, N.L. Isolation of Bacteriophages from the Bovine Rumen. Experientia 1966, 22, 717–718. [Google Scholar] [CrossRef]
- Paynter, M.J.B.; Ewert, D.L.; Chalupa, W. Some Morphological Types of Bacteriophages in Bovine Rumen Contents. Appl. Microbiol. 1969, 18, 942–943. [Google Scholar] [CrossRef]
- Gilbert, R.A.; Townsend, E.M.; Crew, K.S.; Hitch, T.C.A.; Friedersdorff, J.C.A.; Creevey, C.J.; Pope, P.B.; Ouwerkerk, D.; Jameson, E. Rumen Virus Populations: Technological Advances Enhancing Current Understanding. Front. Microbiol. 2020, 11, 450. [Google Scholar] [CrossRef]
- Lobo, R.R.; Faciola, A.P. Ruminal Phages—A Review. Front. Microbiol. 2021, 12, 763416. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.A.; Klieve, A.V. Ruminal Viruses (Bacteriophages, Archaeaphages). In Rumen Microbiology: From Evolution to Revolution; Springer: New Delhi, India, 2015; pp. 121–141. [Google Scholar]
- Alemneh, T.; Getabalew, M. Strategies to Reduce Methane Emission in Ruminants. Int. J. Ecol. 2019, 6, 16–22. [Google Scholar]
- Gilbert, R.; Ouwerkerk, D. The Genetics of Rumen Phage Populations. Proceedings 2019, 36, 165. [Google Scholar] [CrossRef]
- Berg Miller, M.E.; Yeoman, C.J.; Chia, N.; Tringe, S.G.; Angly, F.E.; Edwards, R.A.; Flint, H.J.; Lamed, R.; Bayer, E.A.; White, B.A. Phage-Bacteria Relationships and CRISPR Elements Revealed by a Metagenomic Survey of the Rumen Microbiome. Environ. Microbiol. 2012, 14, 207–227. [Google Scholar] [CrossRef] [PubMed]
- Friedersdorff, J.C.A.; Kingston-Smith, A.H.; Pachebat, J.A.; Cookson, A.R.; Rooke, D.; Creevey, C.J. The Isolation and Genome Sequencing of Five Novel Bacteriophages From the Rumen Active Against Butyrivibrio Fibrisolvens. Front. Microbiol. 2020, 11, 1588. [Google Scholar] [CrossRef]
- Rohwer, F.; Thurber, R.V. Viruses Manipulate the Marine Environment. Nature 2009, 459, 207–212. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P. Genetics of Rumen Microorganisms: Gene Transfer, Genetic Analysis and Strain Manipulation. In Ruminant Physiology: Digestion, Metabolism, Growth and Reproduction; CABI: Oxfordshire, UK, 2000; pp. 389–408. [Google Scholar]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The Rumen Microbiome: Balancing Food Security and Environmental Impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. The Road Not Taken: The Rumen Microbiome, Functional Groups, and Community States. Trends Microbiol. 2019, 27, 538–549. [Google Scholar] [CrossRef]
- Gleason, C.B.; Beckett, L.M.; White, R.R. Rumen Fermentation and Epithelial Gene Expression Responses to Diet Ingredients Designed to Differ in Ruminally Degradable Protein and Fiber Supplies. Sci. Rep. 2022, 12, 2933. [Google Scholar] [CrossRef]
- Russell, J.B.; Muck, R.E.; Weimer, P.J. Quantitative Analysis of Cellulose Degradation and Growth of Cellulolytic Bacteria in the Rumen. FEMS Microbiol. Ecol. 2009, 67, 183–197. [Google Scholar] [CrossRef]
- Russell, J.B.; Hespell, R.B. Microbial Rumen Fermentation. J. Dairy Sci. 1981, 64, 1153–1169. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, X.; Dong, W.; Huang, B.; Wang, Y.; Zhu, M.; Wang, C. Plant Cell Wall Breakdown by Hindgut Microorganisms: Can We Get Scientific Insights From Rumen Microorganisms? J. Equine Vet. Sci. 2022, 115, 104027. [Google Scholar] [CrossRef]
- Krause, D.O.; Denman, S.E.; Mackie, R.I.; Morrison, M.; Rae, A.L.; Attwood, G.T.; McSweeney, C.S. Opportunities to Improve Fiber Degradation in the Rumen: Microbiology, Ecology, and Genomics. FEMS Microbiol. Rev. 2003, 27, 663–693. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J. Cellulose Degradation by Ruminal Microorganisms. Crit. Rev. Biotechnol. 1992, 12, 189–223. [Google Scholar] [CrossRef]
- Bainbridge, M.L.; Saldinger, L.K.; Barlow, J.W.; Alvez, J.P.; Roman, J.; Kraft, J. Alteration of Rumen Bacteria and Protozoa through Grazing Regime as a Tool to Enhance the Bioactive Fatty Acid Content of Bovine Milk. Front. Microbiol. 2018, 9, 904. [Google Scholar] [CrossRef]
- Kelly, W.J.; Mackie, R.I.; Attwood, G.T.; Janssen, P.H.; McAllister, T.A.; Leahy, S.C. Hydrogen and Formate Production and Utilisation in the Rumen and the Human Colon. Anim. Microbiome 2022, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Buan, N.R.; Metcalf, W.W. Methanogenesis by Methanosarcina Acetivorans Involves Two Structurally and Functionally Distinct Classes of Heterodisulfide Reductase. Mol. Microbiol. 2010, 75, 843–853. [Google Scholar] [CrossRef]
- Leahy, S.C.; Janssen, P.H.; Attwood, G.T.; Mackie, R.I.; Mcallister, T.A.; Kelly, W.J. Electron Flow: Key to Mitigating Ruminant Methanogenesis. Trends Microbiol. 2022, 30, 209–212. [Google Scholar] [CrossRef]
- McAllister, T.A.; Newbold, C.J. Redirecting Rumen Fermentation to Reduce Methanogenesis. Aust. J. Exp. Agric. 2008, 48, 7–13. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic Archaea: Ecologically Relevant Differences in Energy Conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef]
- Dias, J.; Marcondes, M.I.; Noronha, M.F.; Resende, R.T.; Machado, F.S.; Mantovani, H.C.; Dill-McFarland, K.A.; Suen, G. Effect of Pre-Weaning Diet on the Ruminal Archaeal, Bacterial, and Fungal Communities of Dairy Calves. Front. Microbiol. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Dijkstra, J.; Tamminga, S. Simulation of the Effects of Diet on the Contribution of Rumen Protozoa to Degradation of Fibre in the Rumen. Br. J. Nutr. 1995, 74, 617–634. [Google Scholar] [CrossRef]
- Rayburn, E.B.; Sharpe, P. Introduction to Pasture Ecology. In Horse Pasture Management; Academic Press: Cambridge, MA, USA, 2019; pp. 81–91. [Google Scholar]
- Diatta, A.A.; Min, D.; Jagadish, S.V.K. Drought Stress Responses in Non-Transgenic and Transgenic Alfalfa—Current Status and Future Research Directions. Adv. Agron. 2021, 170, 35–100. [Google Scholar]
- Belanche, A.; Doreau, M.; Edwards, J.E.; Moorby, J.M.; Pinloche, E.; Newbold, C.J. Shifts in the Rumen Microbiota Due to the Type of Carbohydrate and Level of Protein Ingested by Dairy Cattle Are Associated with Changes in Rumen Fermentation. J. Nutr. 2012, 142, 1684–1692. [Google Scholar] [CrossRef]
- Neubauer, V.; Petri, R.M.; Humer, E.; Kröger, I.; Reisinger, N.; Baumgartner, W.; Wagner, M.; Zebeli, Q. Starch-Rich Diet Induced Rumen Acidosis and Hindgut Dysbiosis in Dairy Cows of Different Lactations. Animals 2020, 10, 1727. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Wang, Y.; Li, S.; Cao, Z.; Ji, S.; He, Y.; Zhang, H. Effect of Dietary Forage to Concentrate Ratios on Dynamic Profile Changes and Interactions of Ruminal Microbiota and Metabolites in Holstein Heifers. Front. Microbiol. 2017, 8, 2206. [Google Scholar] [CrossRef]
- Pang, K.; Chai, S.; Yang, Y.; Wang, X.; Liu, S.; Wang, S. Dietary Forage to Concentrate Ratios Impact on Yak Ruminal Microbiota and Metabolites. Front. Microbiol. 2022, 13, 964564. [Google Scholar] [CrossRef]
- Chen, H.; Wang, C.; Huasai, S.; Chen, A. Effects of Dietary Forage to Concentrate Ratio on Nutrient Digestibility, Ruminal Fermentation and Rumen Bacterial Composition in Angus Cows. Sci. Rep. 2021, 11, 17023. [Google Scholar] [CrossRef]
- Chen, G.J.; Song, S.D.; Wang, B.X.; Zhang, Z.F.; Peng, Z.L.; Guo, C.H.; Zhong, J.C.; Wang, Y. Effects of Forage:Concentrate Ratio on Growth Performance, Ruminal Fermentation and Blood Metabolites in Housing-Feeding Yaks. Asian Australas. J. Anim. Sci. 2015, 28, 1736–1741. [Google Scholar] [CrossRef]
- Wang, L.; Li, Y.; Zhang, Y.; Wang, L. The Effects of Different Concentrate-to-Forage Ratio Diets on Rumen Bacterial Microbiota and the Structures of Holstein Cows during the Feeding Cycle. Animals 2020, 10, 957. [Google Scholar] [CrossRef]
- Meijerink, E.; Neuenschwander, S.; Fries, R.; Dinter, A.; Bertschinger, H.U.; Stranzinger, G.; Vögeli, P. A DNA Polymorphism Influencing α(1,2)Fucosyltransferase Activity of the Pig FUT1 Enzyme Determines Susceptibility of Small Intestinal Epithelium to Escherichia Coli F18 Adhesion. Immunogenetics 2000, 52, 129–136. [Google Scholar] [CrossRef]
- McKnite, A.M.; Perez-Munoz, M.E.; Lu, L.; Williams, E.G.; Brewer, S.; Andreux, P.A.; Bastiaansen, J.W.M.; Wang, X.; Kachman, S.D.; Auwerx, J.; et al. Murine Gut Microbiota Is Defined by Host Genetics and Modulates Variation of Metabolic Traits. PLoS ONE 2012, 7, e39191. [Google Scholar] [CrossRef]
- Suzuki, T.A.; Phifer-Rixey, M.; Mack, K.L.; Sheehan, M.J.; Lin, D.; Bi, K.; Nachman, M.W. Host Genetic Determinants of the Gut Microbiota of Wild Mice. Mol. Ecol. 2019, 28, 3197–3207. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host Genetics Influence the Rumen Microbiota and Heritable Rumen Microbial Features Associate with Feed Efficiency in Cattle. Microbiome 2019, 7, 92. [Google Scholar] [CrossRef]
- Fan, P.; Bian, B.; Teng, L.; Nelson, C.D.; Driver, J.; Elzo, M.A.; Jeong, K.C. Host Genetic Effects upon the Early Gut Microbiota in a Bovine Model with Graduated Spectrum of Genetic Variation. ISME J. 2020, 14, 302–317. [Google Scholar] [CrossRef]
- Li, Z.; Wright, A.-D.G.; Si, H.; Wang, X.; Qian, W.; Zhang, Z.; Li, G. Changes in the Rumen Microbiome and Metabolites Reveal the Effect of Host Genetics on Hybrid Crosses. Environ. Microbiol. Rep. 2016, 8, 1016–1023. [Google Scholar] [CrossRef]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A Heritable Subset of the Core Rumen Microbiome Dictates Dairy Cow Productivity and Emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef]
- Mukhopadhya, I.; Moraïs, S.; Laverde-Gomez, J.; Sheridan, P.O.; Walker, A.W.; Kelly, W.; Klieve, A.V.; Ouwerkerk, D.; Duncan, S.H.; Louis, P.; et al. Sporulation Capability and Amylosome Conservation among Diverse Human Colonic and Rumen Isolates of the Keystone Starch-Degrader Ruminococcus bromii. Environ. Microbiol. 2018, 20, 324–336. [Google Scholar] [CrossRef]
- Li, R.W.; Wu, S.; Baldwin, R.L.; Li, W.; Li, C. Perturbation Dynamics of the Rumen Microbiota in Response to Exogenous Butyrate. PLoS ONE 2012, 7, e29392. [Google Scholar] [CrossRef]
- Liu, X.; Nagy, P.; Bonfini, A.; Houtz, P.; Bing, X.-L.; Yang, X.; Buchon, N. Microbes Affect Gut Epithelial Cell Composition through Immune-Dependent Regulation of Intestinal Stem Cell Differentiation. Cell Rep. 2022, 38, 110572. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Zocco, M.A.; Ainora, M.E.; Gasbarrini, G.; Gasbarrini, A. Bacteroides Thetaiotaomicron in the Gut: Molecular Aspects of Their Interaction. Dig. Liver Dis. 2007, 39, 707–712. [Google Scholar] [CrossRef]
- Nishihara, K.; Suzuki, Y.; Roh, S. Ruminal Epithelial Insulin-like Growth Factor-binding Proteins 2, 3, and 6 Are Associated with Epithelial Cell Proliferation. Anim. Sci. J. 2020, 91, e13422. [Google Scholar] [CrossRef]
- Markandey, M.; Bajaj, A.; Ilott, N.E.; Kedia, S.; Travis, S.; Powrie, F.; Ahuja, V. Gut Microbiota: Sculptors of the Intestinal Stem Cell Niche in Health and Inflammatory Bowel Disease. Gut Microbes 2021, 13, 1990827. [Google Scholar] [CrossRef]
- Ma, N.; Chen, X.; Johnston, L.J.; Ma, X. Gut Microbiota-stem Cell Niche Crosstalk: A New Territory for Maintaining Intestinal Homeostasis. iMeta 2022, 1, e54. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Michalak, M.; Wojnarowski, K.; Cholewińska, P.; Szeligowska, N.; Bawej, M.; Pacoń, J. Selected Alternative Feed Additives Used to Manipulate the Rumen Microbiome. Animals 2021, 11, 1542. [Google Scholar] [CrossRef]
- Mani, S.; Aiyegoro, O.A.; Adeleke, M.A. Characterization of Rumen Microbiota of Two Sheep Breeds Supplemented with Direct-Fed Lactic Acid Bacteria. Front. Vet. Sci. 2021, 7, 570074. [Google Scholar] [CrossRef]
- Kulkarni, N.A.; Chethan, H.S.; Srivastava, R.; Gabbur, A.B. Role of Probiotics in Ruminant Nutrition as Natural Modulators of Health and Productivity of Animals in Tropical Countries: An Overview. Trop. Anim. Health Prod. 2022, 54, 110. [Google Scholar] [CrossRef]
- Maake, T.W.; Aiyegoro, O.A.; Adeleke, M.A. Effects of Lactobacillus Rhamnosus and Enterococcus Faecalis Supplementation as Direct-Fed Microbials on Rumen Microbiota of Boer and Speckled Goat Breeds. Vet. Sci. 2021, 8, 103. [Google Scholar] [CrossRef]
- Pinloche, E.; McEwan, N.; Marden, J.-P.; Bayourthe, C.; Auclair, E.; Newbold, C.J. The Effects of a Probiotic Yeast on the Bacterial Diversity and Population Structure in the Rumen of Cattle. PLoS ONE 2013, 8, e67824. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, L.; Luo, R.; Chen, H.; Nie, C.; Niu, J.; Chen, C.; Xu, Y.; Li, X.; Zhang, W. Effect of a Multispecies Probiotic Mixture on the Growth and Incidence of Diarrhea, Immune Function, and Fecal Microbiota of Pre-Weaning Dairy Calves. Front. Microbiol. 2021, 12, 681014. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Estrada-Angulo, A.; Zapata-Ramírez, O.; Castro-Pérez, B.I.; Urías-Estrada, J.D.; Gaxiola-Camacho, S.; Angulo-Montoya, C.; Ríos-Rincón, F.G.; Barreras, A.; Zinn, R.A.; Leyva-Morales, J.B.; et al. The Effects of Single or Combined Supplementation of Probiotics and Prebiotics on Growth Performance, Dietary Energetics, Carcass Traits, and Visceral Mass in Lambs Finished under Subtropical Climate Conditions. Biology 2021, 10, 1137. [Google Scholar] [CrossRef]
- Lucey, P.M.; Lean, I.J.; Aly, S.S.; Golder, H.M.; Block, E.; Thompson, J.S.; Rossow, H.A. Effects of Mannan-Oligosaccharide and Bacillus Subtilis Supplementation to Preweaning Holstein Dairy Heifers on Body Weight Gain, Diarrhea, and Shedding of Fecal Pathogens. J. Dairy Sci. 2021, 104, 4290–4302. [Google Scholar] [CrossRef]
- Baker, L.M.; Kraft, J.; Karnezos, T.P.; Greenwood, S.L. Review: The Effects of Dietary Yeast and Yeast-Derived Extracts on Rumen Microbiota and Their Function. Anim. Feed Sci. Technol. 2022, 294, 115476. [Google Scholar] [CrossRef]
- Calsamiglia, S.; Busquet, M.; Cardozo, P.W.; Castillejos, L.; Ferret, A. Invited Review: Essential Oils as Modifiers of Rumen Microbial Fermentation. J. Dairy Sci. 2007, 90, 2580–2595. [Google Scholar] [CrossRef]
- Oh, J.; Wall, E.H.; Bravo, D.M.; Hristov, A.N. Host-Mediated Effects of Phytonutrients in Ruminants: A Review. J. Dairy Sci. 2017, 100, 5974–5983. [Google Scholar] [CrossRef]
- Ku-Vera, J.C.; Jiménez-Ocampo, R.; Valencia-Salazar, S.S.; Montoya-Flores, M.D.; Molina-Botero, I.C.; Arango, J.; Gómez-Bravo, C.A.; Aguilar-Pérez, C.F.; Solorio-Sánchez, F.J. Role of Secondary Plant Metabolites on Enteric Methane Mitigation in Ruminants. Front. Vet. Sci. 2020, 7, 584. [Google Scholar] [CrossRef]
- Orzuna-Orzuna, J.; Dorantes-Iturbide, G.; Lara-Bueno, A.; Mendoza-Martínez, G.; Miranda-Romero, L.; Hernández-García, P. Effects of Dietary Tannins’ Supplementation on Growth Performance, Rumen Fermentation, and Enteric Methane Emissions in Beef Cattle: A Meta-Analysis. Sustainability 2021, 13, 7410. [Google Scholar] [CrossRef]
- Orzuna-Orzuna, J.F.; Dorantes-Iturbide, G.; Lara-Bueno, A.; Miranda-Romero, L.A.; Mendoza-Martínez, G.D.; Santiago-Figueroa, I. A Meta-Analysis of Essential Oils Use for Beef Cattle Feed: Rumen Fermentation, Blood Metabolites, Meat Quality, Performance and, Environmental and Economic Impact. Fermentation 2022, 8, 254. [Google Scholar] [CrossRef]
- Kong, F.; Wang, S.; Dai, D.; Cao, Z.; Wang, Y.; Li, S.; Wang, W. Preliminary Investigation of the Effects of Rosemary Extract Supplementation on Milk Production and Rumen Fermentation in High-Producing Dairy Cows. Antioxidants 2022, 11, 1715. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.; Khas, E.; Ao, C.; Bai, C. Effects of Allium Mongolicum Regel Ethanol Extract on Three Flavor-Related Rumen Branched-Chain Fatty Acids, Rumen Fermentation and Rumen Bacteria in Lambs. Front. Microbiol. 2022, 13, 978057. [Google Scholar] [CrossRef]
- Stefańska, B.; Sroka, J.; Katzer, F.; Goliński, P.; Nowak, W. The Effect of Probiotics, Phytobiotics and Their Combination as Feed Additives in the Diet of Dairy Calves on Performance, Rumen Fermentation and Blood Metabolites during the Preweaning Period. Anim. Feed Sci. Technol. 2021, 272, 114738. [Google Scholar] [CrossRef]
- Steele, M.A.; Penner, G.B.; Chaucheyras-Durand, F.; Guan, L.L. Development and Physiology of the Rumen and the Lower Gut: Targets for Improving Gut Health. J. Dairy Sci. 2016, 99, 4955–4966. [Google Scholar] [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef]
- Arshad, M.A.; Hassan, F.; Rehman, M.S.; Huws, S.A.; Cheng, Y.; Din, A.U. Gut Microbiome Colonization and Development in Neonatal Ruminants: Strategies, Prospects, and Opportunities. Anim. Nutr. 2021, 7, 883–895. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, M.; Loor, J.J.; Elolimy, A.; Li, L.; Xu, C.; Wang, W.; Yin, S.; Qu, Y. Analysis of Cow-Calf Microbiome Transfer Routes and Microbiome Diversity in the Newborn Holstein Dairy Calf Hindgut. Front. Nutr. 2021, 8, 736270. [Google Scholar] [CrossRef]
- Hummel, G.; Woodruff, K.; Austin, K.; Knuth, R.; Lake, S.; Cunningham-Hollinger, H. Late Gestation Maternal Feed Restriction Decreases Microbial Diversity of the Placenta While Mineral Supplementation Improves Richness of the Fetal Gut Microbiome in Cattle. Animals 2021, 11, 2219. [Google Scholar] [CrossRef]
- Elolimy, A.; Alharthi, A.; Zeineldin, M.; Parys, C.; Helmbrecht, A.; Loor, J.J. Supply of Methionine during Late-Pregnancy Alters Fecal Microbiota and Metabolome in Neonatal Dairy Calves Without Changes in Daily Feed Intake. Front. Microbiol. 2019, 10, 2159. [Google Scholar] [CrossRef]
- Amin, N.; Schwarzkopf, S.; Kinoshita, A.; Tröscher-Mußotter, J.; Dänicke, S.; Camarinha-Silva, A.; Huber, K.; Frahm, J.; Seifert, J. Evolution of Rumen and Oral Microbiota in Calves Is Influenced by Age and Time of Weaning. Anim. Microbiome 2021, 3, 31. [Google Scholar] [CrossRef]
- Bi, Y.; Cox, M.S.; Zhang, F.; Suen, G.; Zhang, N.; Tu, Y.; Diao, Q. Feeding Modes Shape the Acquisition and Structure of the Initial Gut Microbiota in Newborn Lambs. Environ. Microbiol. 2019, 21, 2333–2346. [Google Scholar] [CrossRef]
- Palma-Hidalgo, J.M.; Jiménez, E.; Popova, M.; Morgavi, D.P.; Martín-García, A.I.; Yáñez-Ruiz, D.R.; Belanche, A. Inoculation with Rumen Fluid in Early Life Accelerates the Rumen Microbial Development and Favours the Weaning Process in Goats. Anim. Microbiome 2021, 3, 11. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanjorjo, R.A.; Tseten, T.; Kang, M.-K.; Kwon, M.; Kim, S.-W. In Pursuit of Understanding the Rumen Microbiome. Fermentation 2023, 9, 114. https://doi.org/10.3390/fermentation9020114
Sanjorjo RA, Tseten T, Kang M-K, Kwon M, Kim S-W. In Pursuit of Understanding the Rumen Microbiome. Fermentation. 2023; 9(2):114. https://doi.org/10.3390/fermentation9020114
Chicago/Turabian StyleSanjorjo, Rey Anthony, Tenzin Tseten, Min-Kyoung Kang, Moonhyuk Kwon, and Seon-Won Kim. 2023. "In Pursuit of Understanding the Rumen Microbiome" Fermentation 9, no. 2: 114. https://doi.org/10.3390/fermentation9020114
APA StyleSanjorjo, R. A., Tseten, T., Kang, M.-K., Kwon, M., & Kim, S.-W. (2023). In Pursuit of Understanding the Rumen Microbiome. Fermentation, 9(2), 114. https://doi.org/10.3390/fermentation9020114






