A Genome-Wide Phenotypic Analysis of Saccharomyces cerevisiae’s Adaptive Response and Tolerance to Chitosan in Conditions Relevant for Winemaking
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Media
2.2. Susceptibility Assays to Chitosan
2.3. Phenotypic Screening of the S. cerevisiae Deletion Mutant Collection of Chitosan
3. Results
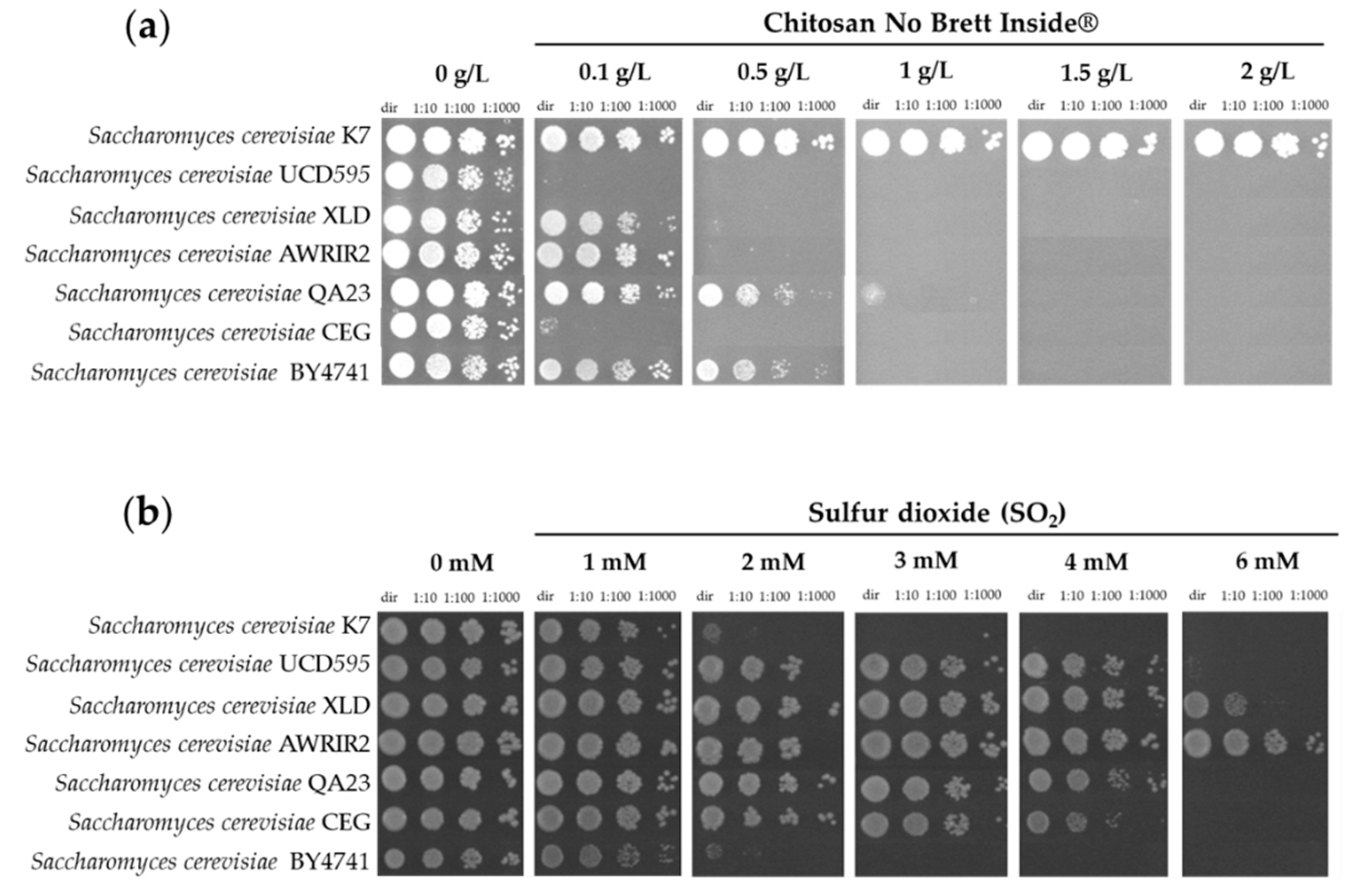
3.1. Effect of Chitosan on Growth of S. cerevisiae and of Wine Spoilage Yeast Strains
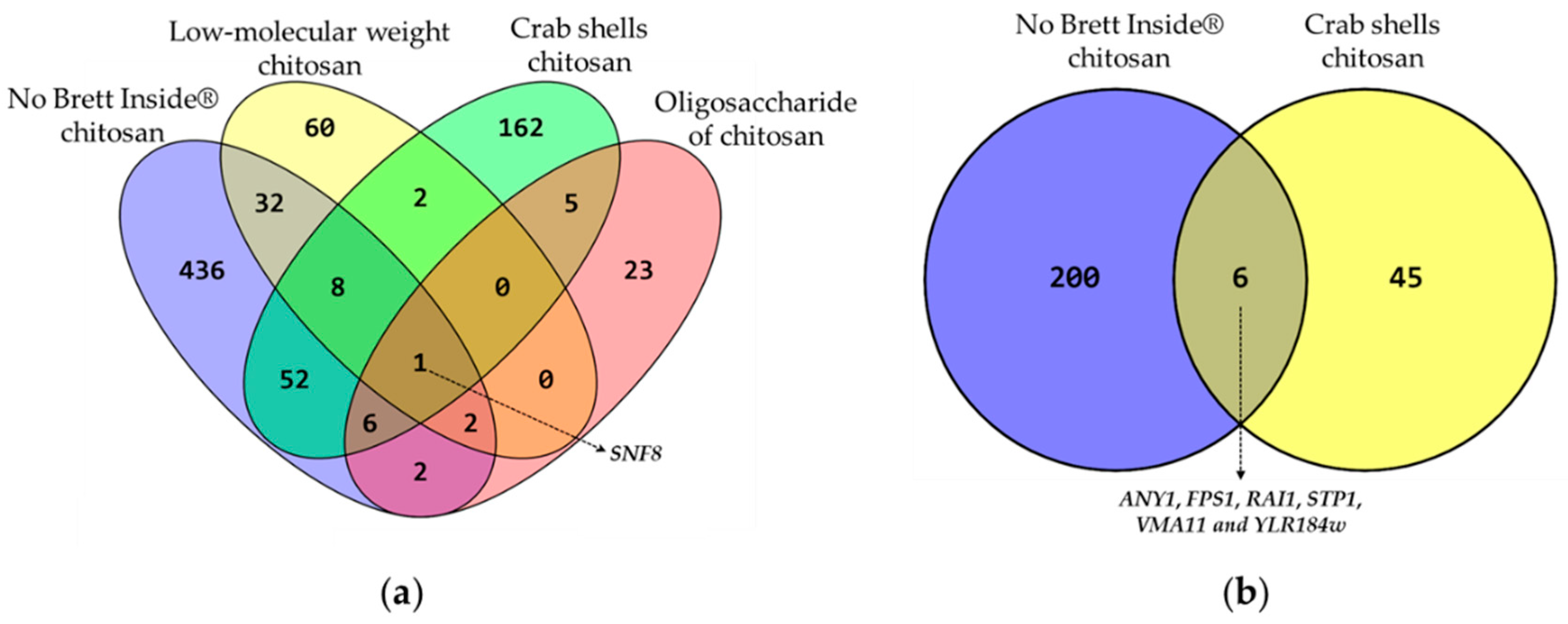
3.2. Chemogenomic Analysis of Chitosan-Stressed S. cerevisiae Cells
3.2.1. Overview
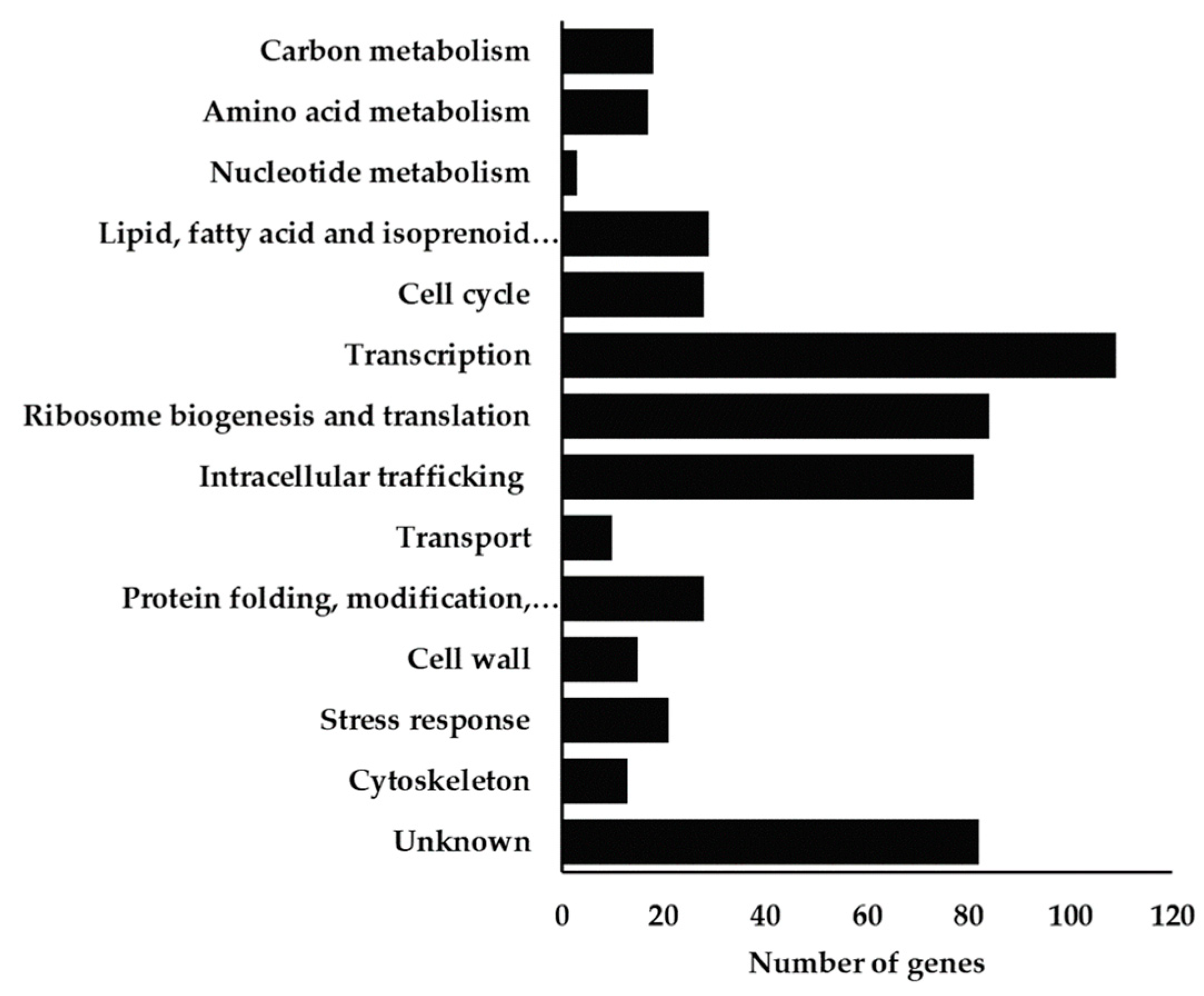
3.2.2. Functional Distribution of S. cerevisiae Genes Contributing to the Tolerance to Chitosan
3.2.3. Functional Distribution of S. cerevisiae Genes Whose Deletion Increases Tolerance to Chitosan
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Renouf, V.; Strehaiano, P.; Lonvaud-Funel, A. Effectiveness of Dimethlydicarbonate to Prevent Brettanomyces bruxellensis Growth in Wine. Food Control 2008, 19, 208–216. [Google Scholar] [CrossRef]
- Vally, H.; Misso, N.L.A.; Madan, V. Clinical Effects of Sulphite Additives. Clin. Exp. Allergy 2009, 39, 1643–1651. [Google Scholar] [CrossRef]
- Costantini, A.; García-Moruno, E.; Moreno-Arribas, M.V. Biochemical Transformations Produced by Malolactic Fermentation. In Wine Chemistry and Biochemistry; Springer: New York, NY, USA, 2009; pp. 27–57. ISBN 9780387741161. [Google Scholar]
- Du Toit, M.; Pretorius, I.S. Microbial Spoilage and Preservation of Wine: Using Weapons from Nature’s Own Arsenal-a Review. S. Afr. J. Enol. Vitic. 2000, 21, 74–96. [Google Scholar] [CrossRef]
- Corte, L.; Roscini, L.; Zadra, C.; Antonielli, L.; Tancini, B.; Magini, A.; Emiliani, C.; Cardinali, G. Effect of PH on Potassium Metabisulphite Biocidic Activity against Yeast and Human Cell Cultures. Food Chem. 2012, 134, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Vally, H.; Thompson, P.J. Role of Sulfite Additives in Wine Induced Asthma: Single Dose and Cumulative Dose Studies. Thorax 2001, 56, 763–769. [Google Scholar] [CrossRef]
- Irwin, S.V.; Fisher, P.; Graham, E.; Malek, A.; Robidoux, A. Sulfites Inhibit the Growth of Four Species of Beneficial Gut Bacteria at Concentrations Regarded as Safe for Food. PLoS ONE 2017, 12, e0186629. [Google Scholar] [CrossRef]
- Divol, B.; Strehaiano, P.; Lonvaud-Funel, A. Effectiveness of Dimethyldicarbonate to Stop Alcoholic Fermentation in Wine. Food Microbiol. 2005, 22, 169–178. [Google Scholar] [CrossRef]
- Castro Marín, A.; Colangelo, D.; Lambri, M.; Riponi, C.; Chinnici, F. Relevance and Perspectives of the Use of Chitosan in Winemaking: A Review. Crit. Rev. Food Sci. Nutr. 2021, 61, 3450–3464. [Google Scholar] [CrossRef]
- Miot-Sertier, C.; Paulin, M.; Dutilh, L.; Ballestra, P.; Albertin, W.; Masneuf-Pomarède, I.; Coulon, J.; Moine, V.; Vallet-Courbin, A.; Maupeu, J.; et al. Assessment of Chitosan Antimicrobial Effect on Wine Microbes. Int. J. Food Microbiol. 2022, 381, 109907. [Google Scholar] [CrossRef]
- Bağder Elmaci, S.; Gülgör, G.; Tokatli, M.; Erten, H.; İşci, A.; Özçelik, F. Effectiveness of Chitosan against Wine-Related Microorganisms. Antonie Van Leeuwenhoek 2015, 107, 675–686. [Google Scholar] [CrossRef]
- Aider, M. Chitosan Application for Active Bio-Based Films Production and Potential in the Food Industry: Review. LWT-Food Sci. Technol. 2010, 43, 837–842. [Google Scholar] [CrossRef]
- Gómez-Rivas, L.; Escudero-Abarca, B.I.; Aguilar-Uscanga, M.G.; Hayward-Jones, P.M.; Mendoza, P.; Ramírez, M. Selective Antimicrobial Action of Chitosan against Spoilage Yeasts in Mixed Culture Fermentations. J. Ind. Microbiol. Biotechnol. 2004, 31, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Portugal, C.; Sáenz, Y.; Rojo-Bezares, B.; Zarazaga, M.; Torres, C.; Cacho, J.; Ruiz-Larrea, F. Brettanomyces Susceptibility to Antimicrobial Agents Used in Winemaking: In Vitro and Practical Approaches. Eur. Food Res. Technol. 2014, 238, 641–652. [Google Scholar] [CrossRef]
- Ferreira, D.; Moreira, D.; Costa, E.M.; Silva, S.; Pintado, M.M.; Couto, J.A. The Antimicrobial Action of Chitosan Against the Wine Spoilage Yeast Brettanomyces/Dekkera. J. Chitin Chitosan Sci. 2013, 1, 240–245. [Google Scholar] [CrossRef]
- Bornet, A.; Teissedre, P.L. Chitosan, Chitin-Glucan and Chitin Effects on Minerals (Iron, Lead, Cadmium) and Organic (Ochratoxin A) Contaminants in Wines. Eur. Food Res. Technol. 2008, 226, 681–689. [Google Scholar] [CrossRef]
- Rabea, E.I.; Badawy, M.E.T.; Stevens, C.V.; Smagghe, G.; Steurbaut, W. Chitosan as Antimicrobial Agent: Applications and Mode of Action. Biomacromolecules 2003, 4, 1457–1465. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Sultan, Y.Y.; Seif, M.M.; Marrez, D.A. Bio-Evaluation of Crustacean and Fungal Nano-Chitosan for Applying as Food Ingredient. Toxicol. Rep. 2018, 5, 348–356. [Google Scholar] [CrossRef]
- Abd El-Hack, M.E.; El-Saadony, M.T.; Shafi, M.E.; Zabermawi, N.M.; Arif, M.; Batiha, G.E.; Khafaga, A.F.; Abd El-Hakim, Y.M.; Al-Sagheer, A.A. Antimicrobial and Antioxidant Properties of Chitosan and Its Derivatives and Their Applications: A Review. Int. J. Biol. Macromol. 2020, 164, 2726–2744. [Google Scholar] [CrossRef]
- Colangelo, D.; Torchio, F.; De Faveri, D.M.; Lambri, M. The Use of Chitosan as Alternative to Bentonite for Wine Fining: Effects on Heat-Stability, Proteins, Organic Acids, Colour, and Volatile Compounds in an Aromatic White Wine. Food Chem. 2018, 264, 301–309. [Google Scholar] [CrossRef]
- Nardi, T.; Vagnoli, P.; Minacci, A.; Gautier, S.; Sieczkowski, N. Evaluating the Impact of a Fungal-Origin Chitosan Preparation on Brettanomyces Bruxellensis in the Context of Wine Aging. Wine Stud. 2014, 3, 4574. [Google Scholar] [CrossRef]
- Zuehlke, J.M.; Petrova, B.; Edwards, C.G. Advances in the Control of Wine Spoilage by Zygosaccharomyces and Dekkera/Brettanomyces. Annu. Rev. Food Sci. Technol. 2013, 4, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Taillandier, P.; Joannis-Cassan, C.; Jentzer, J.B.; Gautier, S.; Sieczkowski, N.; Granes, D.; Brandam, C. Effect of a Fungal Chitosan Preparation on Brettanomyces bruxellensis, a Wine Contaminant. J. Appl. Microbiol. 2015, 118, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Petrova, B.; Cartwright, Z.M.; Edwards, C.G. Effectiveness of Chitosan Preparations against Brettanomyces bruxellensis Grown in Culture Media and Red Wines. J. Int. Sci. Vigne Vin 2016, 50, 49–56. [Google Scholar] [CrossRef]
- Rhoades, J.; Roller, S. Antimicrobial Actions of Degraded and Native Chitosan against Spoilage Organisms in Laboratory Media and Foods. Appl. Environ. Microbiol. 2000, 66, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Roller, S.; Covill, N. The Antifungal Properties of Chitosan in Laboratory Media and Apple Juice. Int. J. Food Microbiol. 1999, 47, 67–77. [Google Scholar] [CrossRef]
- Raafat, D.; Fouad, G. Chitosan as an Antimicrobial Compound: Modes of Action and Resistance Mechanisms. Ph.D. Thesis, University of Bonn, Bonn, Germany, 2008. [Google Scholar]
- Scansani, S.; Rauhut, D.; Brezina, S.; Semmler, H.; Benito, S. The Impact of Chitosan on the Chemical Composition of Wines Fermented with Schizosaccharomyces pombe and Saccharomyces cerevisiae. Foods 2020, 9, 1423. [Google Scholar] [CrossRef]
- Liu, H.; Du, Y.; Wang, X.; Sun, L. Chitosan Kills Bacteria through Cell Membrane Damage. Int. J. Food Microbiol. 2004, 95, 147–155. [Google Scholar] [CrossRef]
- Helander, I.M.; Nurmiaho-Lassila, E.L.; Ahvenainen, R.; Rhoades, J.; Roller, S. Chitosan Disrupts the Barrier Properties of the Outer Membrane of Gram-Negative Bacteria. Int. J. Food Microbiol. 2001, 71, 235–244. [Google Scholar] [CrossRef]
- Kong, M.; Chen, X.G.; Xing, K.; Park, H.J. Antimicrobial Properties of Chitosan and Mode of Action: A State of the Art Review. Int. J. Food Microbiol. 2010, 144, 51–63. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Huang, I.C.; Jansson, H.B.; Salinas, J.; Lopez-Llorca, L.V.; Read, N.D. Chitosan Permeabilizes the Plasma Membrane and Kills Cells of Neurospora crassa in an Energy Dependent Manner. Fungal Genet. Biol. 2009, 46, 585–594. [Google Scholar] [CrossRef]
- Goy, R.C.; de Britto, D.; Assis, O.B.G. A Review of the Antimicrobial Activity of Chitosan. Polímeros 2009, 19, 241–247. [Google Scholar] [CrossRef]
- Hernández, R.B.; Yola, O.R.; Mercê, A.L.R. Chemical Equilibrium in the Complexation of First Transition Series Divalent Cations, Cu2+, Mn2+ and Zn2+, with Chitosan. J. Braz. Chem. Soc. 2007, 18, 1388–1396. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Monteiro, P.T.; Guerreiro, J.F.; Gonçalves, J.P.; Mira, N.P.; Dos Santos, S.C.; Cabrito, T.R.; Palma, M.; Costa, C.; Francisco, A.P.; et al. The YEASTRACT Database: An Upgraded Information System for the Analysis of Gene and Genomic Transcription Regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014, 34, D446–D451. [Google Scholar] [CrossRef]
- Lopez-Moya, F.; Kowbel, D.; Nueda, M.J.; Palma-Guerrero, J.; Glass, N.L.; Lopez-Llorca, L.V. Neurospora crassa Transcriptomics Reveals Oxidative Stress and Plasma Membrane Homeostasis Biology Genes as Key Targets in Response to Chitosan. Mol. BioSyst. 2016, 12, 391–403. [Google Scholar] [CrossRef]
- Zakrzewska, A.; Boorsma, A.; Brul, S.; Hellingwerf, K.J.; Klis, F.M. Transcriptional Response of Saccharomyces cerevisiae to the Plasma Membrane-Perturbing Compound Chitosan. Eukaryot. Cell 2005, 4, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, A.; Boorsma, A.; Delneri, D.; Brul, S.; Oliver, S.G.; Klis, F.M. Cellular Processes and Pathways That Protect Saccharomyces cerevisiae Cells against the Plasma Membrane-Perturbing Compound Chitosan. Eukaryot. Cell 2007, 13, 2–9. [Google Scholar] [CrossRef]
- Bermejo, C.; Rodríguez, E.; García, R.; Rodríguez-Peña, J.M.; Rodríguez de la Concepción, M.L.; Rivas, C.; Arias, P.; Nombela, C.; Posas, F.; Arroyo, J. The Sequential Activation of the Yeast HOG and SLT2 Pathways Is Required for Cell Survival to Cell Wall Stress. Mol. Biol. Cell 2008, 19, 1113–1124. [Google Scholar] [CrossRef]
- Organisation Internationale de la Vigne et du Vin (OIV). International Code of Oenological Practices OIV/OENO, 338A/2009; International Organisation of Vine and Wine: Paris, France, 2009. [Google Scholar]
- Lim, S.H.; Hudson, S.M. Synthesis and Antimicrobial Activity of a Water-Soluble Chitosan Derivative with a Fiber-Reactive Group. Carbohydr. Res. 2004, 339, 313–319. [Google Scholar] [CrossRef]
- Mira, N.P.; Teixeira, M.C.; Sá-Correia, I. Adaptive Response and Tolerance to Weak Acids in Saccharomyces cerevisiae: A Genome-Wide View. Omi. J. Integr. Biol. 2010, 14, 525–540. [Google Scholar] [CrossRef]
- Sagoo, S.K.; Board, R.; Roller, S. Chitosan Potentiates the Antimicrobial Action of Sodium Benzoate on Spoilage Yeasts. Lett. Appl. Microbiol. 2002, 34, 168–172. [Google Scholar] [CrossRef]
- Galván Márquez, I.; Akuaku, J.; Cruz, I.; Cheetham, J.; Golshani, A.; Smith, M.L. Disruption of Protein Synthesis as Antifungal Mode of Action by Chitosan. Int. J. Food Microbiol. 2013, 164, 108–112. [Google Scholar] [CrossRef]
- Lage, P.; Sampaio-Marques, B.; Ludovico, P.; Mira, N.P.; Mendes-Ferreira, A. Transcriptomic and Chemogenomic Analyses Unveil the Essential Role of Com2-Regulon in Response and Tolerance of Saccharomyces cerevisiae to Stress Induced by Sulfur Dioxide. Microb. Cell 2019, 6, 509–523. [Google Scholar] [CrossRef]
- Gaikani, H.; Smith, A.M.; Lee, A.Y.; Giaever, G.; Nislow, C. Systematic Prediction of Antifungal Drug Synergy by Chemogenomic Screening in Saccharomyces cerevisiae. Front. Fungal Biol. 2021, 2, 683414. [Google Scholar] [CrossRef]
- Barazandeh, M.; Kriti, D.; Nislow, C.; Giaever, G. The Cellular Response to Drug Perturbation Is Limited: Comparison of Large-Scale Chemogenomic Fitness Signatures. BMC Genom. 2022, 23, 197. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, S.C.; Sá-Correia, I. Genome-Wide Identification of Genes Required for Yeast Growth under Imatinib Stress: Vacuolar H+-Atpase Function Is an Important Target of This Anticancer Drug. Omi. J. Integr. Biol. 2009, 13, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Hillenmeyer, M.E.; Fung, E.; Wildenhain, J.; Pierce, S.E.; Hoon, S.; Lee, W.; Proctor, M.; St Onge, R.P.; Tyers, M.; Koller, D.; et al. The Chemical Genomic Portrait of Yeast: Uncovering a Phenotype for All Genes. Science 2008, 320, 362–365. [Google Scholar] [CrossRef]
- Jaime, M.D.L.A.; Lopez-Llorca, L.V.; Conesa, A.; Lee, A.Y.; Proctor, M.; Heisler, L.E.; Gebbia, M.; Giaever, G.; Westwood, J.T.; Nislow, C. Identification of Yeast Genes That Confer Resistance to Chitosan Oligosaccharide (COS) Using Chemogenomics. BMC Genom. 2012, 13, 267. [Google Scholar] [CrossRef]
- Friedman, N.; Rando, O.J. Epigenomics and the Structure of the Living Genome. Genome Res. 2015, 25, 1482–1490. [Google Scholar] [CrossRef]
- Magraner-Pardo, L.; Pelechano, V.; Coloma, M.D.; Tordera, V. Dynamic Remodeling of Histone Modifications in Response to Osmotic Stress in Saccharomyces cerevisiae. BMC Genom. 2014, 15, 247. [Google Scholar] [CrossRef]
- De Almeida, R.F.M. A Route to Understanding Yeast Cellular Envelope–Plasma Membrane Lipids Interplaying in Cell Wall Integrity. FEBS J. 2018, 285, 2402–2404. [Google Scholar] [CrossRef]
- Kodedová, M.; Sychrová, H. Changes in the Sterol Composition of the Plasma Membrane Affect Membrane Potential, Salt Tolerance and the Activity of Multidrug Resistance Pumps in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0139306. [Google Scholar] [CrossRef]
- Khakhina, S.; Johnson, S.S.; Manoharlal, R.; Russo, S.B.; Blugeon, C.; Lemoine, S.; Sunshine, A.B.; Dunham, M.J.; Cowart, L.A.; Devaux, F.; et al. Control of Plasma Membrane Permeability by ABC Transporters. Eukaryot. Cell 2015, 14, 442–453. [Google Scholar] [CrossRef]
- Kaur, R.; Bachhawat, A.K. The Yeast Multidrug Resistance Pump, Pdr5p, Confers Reduced Drug Resistance in Erg Mutants of Saccharomyces Cerevisiae. Microbiology 1999, 145, 809–818. [Google Scholar] [CrossRef][Green Version]
- Decottignies, A.; Grant, A.M.; Nichols, J.W.; De Wet, H.; McIntosh, D.B.; Goffeau, A. ATPase and Multidrug Transport Activities of the Overexpressed Yeast ABC Protein Yor1p. J. Biol. Chem. 1998, 273, 12612–12622. [Google Scholar] [CrossRef]
- Mollapour, M.; Shepherd, A.; Piper, P.W. Presence of the Fps1p Aquaglyceroporin Channel Is Essential for Hog1p Activation, but Suppresses Slt2(Mpk1)p Activation, with Acetic Acid Stress of Yeast. Microbiology 2009, 155, 3304–3311. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Lopez-Jimenez, J.A.; Pérez-Berná, A.J.; Huang, I.C.; Jansson, H.B.; Salinas, J.; Villalaín, J.; Read, N.D.; Lopez-Llorca, L.V. Membrane Fluidity Determines Sensitivity of Filamentous Fungi to Chitosan. Mol. Microbiol. 2010, 75, 1021–1032. [Google Scholar] [CrossRef]
- Ikeda, M.; Kihara, A.; Denpoh, A.; Igarashi, Y. The Rim101 Pathway Is Involved in Rsb1 Expression Induced by Altered Lipid Asymmetry. Mol. Biol. Cell 2008, 49, 1922–1931. [Google Scholar] [CrossRef]
- Mattiazzi, M.; Jambhekar, A.; Kaferle, P.; DeRisi, J.L.; Križaj, I.; Petrovič, U. Genetic Interactions between a Phospholipase A2 and the Rim101 Pathway Components in S. cerevisiae Reveal a Role for This Pathway in Response to Changes in Membrane Composition and Shape. Mol. Genet. Genom. 2010, 283, 519–530. [Google Scholar] [CrossRef][Green Version]
- Rockenfeller, P.; Smolnig, M.; Diessl, J.; Bashir, M.; Schmiedhofer, V.; Knittelfelder, O.; Ring, J.; Franz, J.; Foessl, I.; Khan, M.J.; et al. Diacylglycerol Triggers Rim101 Pathway-Dependent Necrosis in Yeast: A Model for Lipotoxicity. Cell Death Differ. 2018, 25, 767–783. [Google Scholar] [CrossRef]
- Obara, K.; Kihara, A. Signaling Events of the Rim101 Pathway Occur at the Plasma Membrane in a Ubiquitination-Dependent Manner. Mol. Cell. Biol. 2014, 34, 3525–3534. [Google Scholar] [CrossRef]
- Mira, N.P.; Palma, M.; Guerreiro, J.F.; Sá-Correia, I. Genome-Wide Identification of Saccharomyces cerevisiae Genes Required for Tolerance to Acetic Acid. Microb. Cell Fact. 2010, 9, 79. [Google Scholar] [CrossRef]
- Johnston, E.J.; Moses, T.; Rosser, S.J. The Wide-Ranging Phenotypes of Ergosterol Biosynthesis Mutants, and Implications for Microbial Cell Factories. Yeast 2020, 37, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Maciaszczyk-Dziubinska, E.; Migdal, I.; Migocka, M.; Bocer, T.; Wysocki, R. The Yeast Aquaglyceroporin Fps1p Is a Bidirectional Arsenite Channel. FEBS Lett. 2010, 584, 726–732. [Google Scholar] [CrossRef]
- Mollapour, M.; Piper, P.W. Hog1 Mitogen-Activated Protein Kinase Phosphorylation Targets the Yeast Fps1 Aquaglyceroporin for Endocytosis, Thereby Rendering Cells Resistant to Acetic Acid. Mol. Cell. Biol. 2007. [Google Scholar] [CrossRef]
- Aouida, M.; Pagé, N.; Leduc, A.; Peter, M.; Ramotar, D. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals Altered Transport As a Mechanism of Resistance to the Anticancer Drug Bleomycin. Cancer Res. 2004, 64, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.L.; Faraj, S.A.S.; Calixto, M.D.; Ayumi, V.A.; Pedro, C.A.; Silva, P.G.; Monique, F.; da Cunha, F.H.M.R.; Hollunder, K.A.; Fernandes, S.S.; et al. Yeast Double Transporter Gene Deletion Library for Identification of Xenobiotic Carriers in Low or High Throughput. MBio 2021, 12, e03221-21. [Google Scholar] [CrossRef]
- Dmitriev, S.E.; Vladimirov, D.O.; Lashkevich, K.A. A Quick Guide to Small-Molecule Inhibitors of Eukaryotic Protein Synthesis. Biochemistry 2020, 85, 1389–1421. [Google Scholar] [CrossRef] [PubMed]



| Species | Strain | Source |
|---|---|---|
| Saccharomyces cerevisiae strains | ||
| Saccharomyces cerevisiae | Lalvin T73 | Lalvin, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Lalvin EC1118 | Lalvin, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Fermivin | DSM, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Zymaflore VL1 | Laffort, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Lalvin QA23 | Lalvin, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Uvaferm CEG | UVAFERM, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | VIN13 | Anchor, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | NT116 | Anchor, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Lalvin BM45 | UVAFERM, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Lalvin BRL97 | UVAFERM, Proenol, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Fermicru XL | DSM, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | Fermicru LVCB | DSM, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | XLD | DSM, Vila Nova de Gaia, Portugal |
| Saccharomyces cerevisiae | UCD522 | Maurivin, Enovitis, Peso da Régua, Portugal |
| Saccharomyces cerevisiae | UCD595 | UC Davis collection, California, USA |
| Saccharomyces cerevisiae | UCD505 | UC Davis collection, California, USA |
| Saccharomyces cerevisiae | AWRI796 | Maurivin, Enovitis, Peso da Régua, Portugal |
| Saccharomyces cerevisiae | AWRI R2 | Maurivin, Enovitis, Peso da Régua, Portugal |
| Saccharomyces cerevisiae | W3 | Brewing Society of Japan (NRIB) |
| Saccharomyces cerevisiae | K7 | Brewing Society of Japan (NRIB) |
| Saccharomyces cerevisiae | BY4741 | Euroscarf collection, Frankfurt, Germany |
| Non-Saccharomyces strains | ||
| Dekkera anomala | IGC5153 | Portuguese Collection of Yeast Cultures |
| Dekkera anomala | IGC5152 | Portuguese Collection of Yeast Cultures |
| Pichia anomala | UTAD37 | University of Trás-os-Montes and Alto Douro, Vila Real, Portugal |
| Pichia anomala | UTAD38 | University of Trás-os-Montes and Alto Douro, Vila Real, Portugal |
| Pichia anomala | UTAD40 | University of Trás-os-Montes and Alto Douro, Vila Real, Portugal |
| Saccharomycodes ludwigii | UTAD17 | University of Trás-os-Montes and Alto Douro, Vila Real, Portugal |
| Zygosaccharomyces bailii | UTAD265 | University of Trás-os-Montes and Alto Douro, Vila Real, Portugal |
| Gene/ORF | Function | Susceptibility to Chitosan |
|---|---|---|
| Cell wall biosynthesis | ||
| BGL2 | Endo-beta-1,3-glucanase, a major protein of the cell wall, involved in cell wall maintenance | ++ |
| GAS1 | Beta-1,3-glucanosyltransferase, required for cell wall assembly and also has a role in transcriptional silencing | ++ |
| GAS2 | 1,3-beta-glucanosyltransferase, involved with Gas4p in spore wall assembly | + |
| Lipid metabolism | ||
| CHO1 | Phosphatidylserine synthase, functions in phospholipid biosynthesis | ++ |
| CHO2 | Phosphatidylethanolamine methyltransferase (PEMT), catalyzes the first step in the conversion of phosphatidylethanolamine to phosphatidylcholine during the methylation pathway of phosphatidylcholine biosynthesis | ++ |
| OPI1 | Transcriptional regulator of a variety of genes; phosphorylation by protein kinase A stimulates Opi1p function in negative regulation of phospholipid biosynthetic genes | ++ |
| OPI3 | Methylene-fatty-acyl-phospholipid synthase; catalyzes the last two steps in phosphatidylcholine biosynthesis | ++ |
| SUR1 | Mannosylinositol phosphorylceramide (MIPC) synthase catalytic subunit | ++ |
| SUR4 | Elongase, involved in fatty acid and sphingolipid biosynthesis; synthesizes very long chain 20-26-carbon fatty acids from C18-CoA primers | + |
| Ribosome biosynthesis | ||
| RPS24B | Protein component of the small (40S) ribosomal subunit | + |
| RPS26B | Protein component of the small (40S) ribosomal subunit | ++ |
| RPS27A | Protein component of the small (40S) ribosomal subunit | ++ |
| RPS28A | Protein component of the small (40S) ribosomal subunit | ++ |
| RPL31B | Protein component of the large (60S) ribosomal subunit | ++ |
| RPL38 | Protein component of the large (60S) ribosomal subunit | + |
| Rim101 pathway | ||
| RIM101 | Transcriptional repressor involved in response to pH and in cell wall construction | ++ |
| RIM13 | Protein involved in proteolytic activation of Rim101p; part of response to alkaline pH | ++ |
| RIM20 | Protein involved in proteolytic activation of Rim101p; part of response to alkaline pH | ++ |
| RIM21 | pH sensor molecule, component of the RIM101 pathway; has a role in cell wall construction and alkaline pH response | ++ |
| RIM8 | Protein involved in proteolytic activation of Rim101p in response to alkaline pH | ++ |
| RIM9 | Protein involved in the proteolytic activation of Rim101p in response to alkaline pH | ++ |
| Ion transport | ||
| FTR1 | High-affinity iron permease involved in the transport of iron across the plasma membrane | + |
| CTR1 | High-affinity copper transporter of the plasma membrane; mediates nearly all copper uptake under low copper conditions | + |
| PDR5 | Plasma membrane ATP-binding cassette (ABC) transporter, multidrug transporter actively regulated by Pdr1p | + |
| YOR1 | Plasma membrane ATP-binding cassette (ABC) transporter, multidrug transporter mediates export of many different organic anions including oligomycin | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lage, P.; Coelho, B.B.; Mira, N.P.; Mendes-Ferreira, A. A Genome-Wide Phenotypic Analysis of Saccharomyces cerevisiae’s Adaptive Response and Tolerance to Chitosan in Conditions Relevant for Winemaking. Fermentation 2023, 9, 172. https://doi.org/10.3390/fermentation9020172
Lage P, Coelho BB, Mira NP, Mendes-Ferreira A. A Genome-Wide Phenotypic Analysis of Saccharomyces cerevisiae’s Adaptive Response and Tolerance to Chitosan in Conditions Relevant for Winemaking. Fermentation. 2023; 9(2):172. https://doi.org/10.3390/fermentation9020172
Chicago/Turabian StyleLage, Patrícia, Bárbara B. Coelho, Nuno P. Mira, and Ana Mendes-Ferreira. 2023. "A Genome-Wide Phenotypic Analysis of Saccharomyces cerevisiae’s Adaptive Response and Tolerance to Chitosan in Conditions Relevant for Winemaking" Fermentation 9, no. 2: 172. https://doi.org/10.3390/fermentation9020172
APA StyleLage, P., Coelho, B. B., Mira, N. P., & Mendes-Ferreira, A. (2023). A Genome-Wide Phenotypic Analysis of Saccharomyces cerevisiae’s Adaptive Response and Tolerance to Chitosan in Conditions Relevant for Winemaking. Fermentation, 9(2), 172. https://doi.org/10.3390/fermentation9020172





