Evaluation of Fermented Turmeric Milk by Lactic Acid Bacteria to Prevent UV-Induced Oxidative Stress in Human Fibroblast Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Microorganisms and Medium
2.3. LAB-Fermented Turmeric Milk Preparation
2.4. Antioxidant Activity Assays
2.5. Fermented Turmeric Milk Extract Preparation
2.6. Phenolic Compounds Content Analysis
2.7. Cell Culture and UVA Irradiation
2.8. Cell Viability
2.9. Measurement of ROS Production
2.10. Detection of Senescence-Associated β-Galactosidase (SA-β-Gal) Activity
2.11. Skin-Associated Gene Expression by qPCR
2.12. Statistical Analysis
3. Results and Discussion
3.1. Strain Selection for Turmeric Fermentation
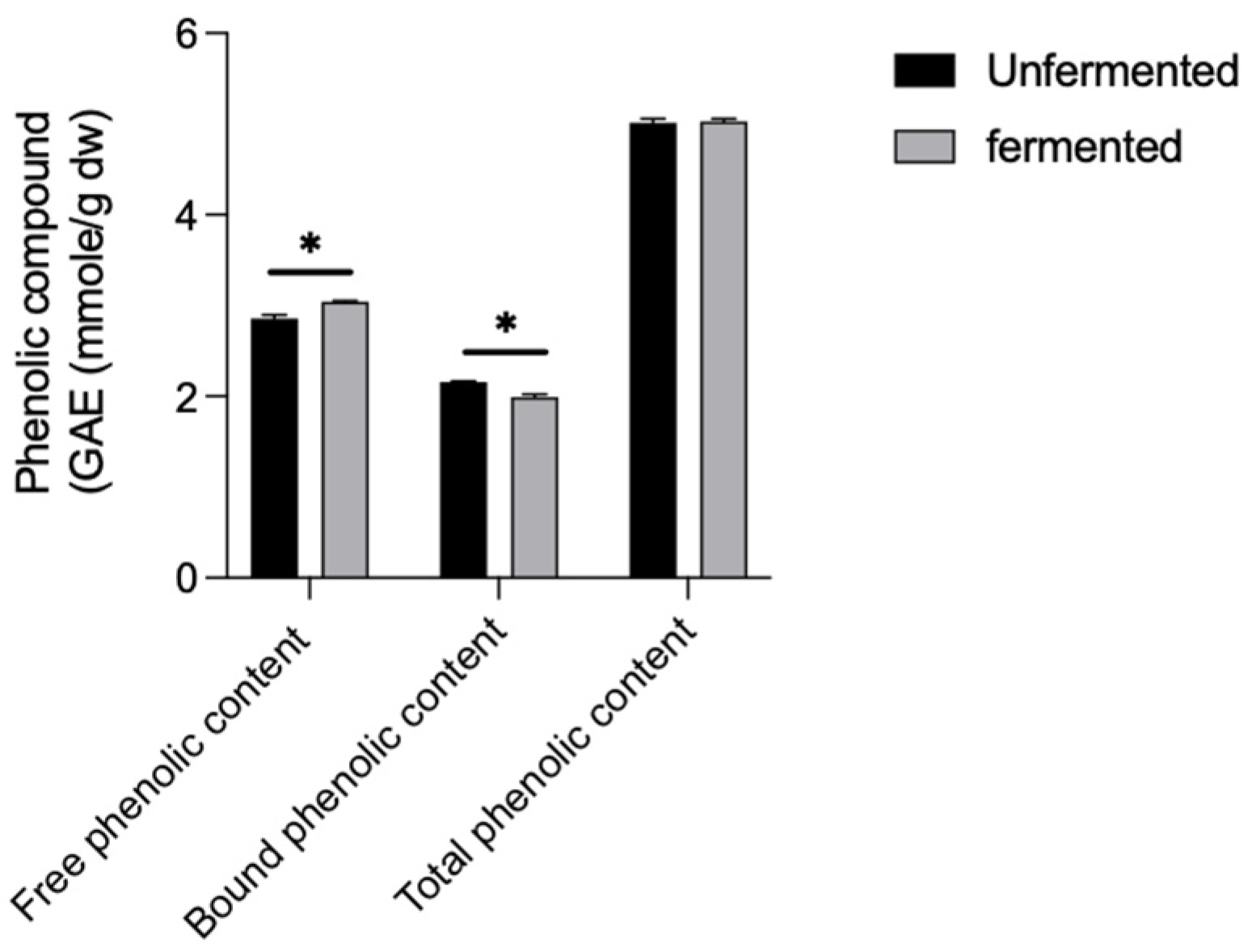
3.2. Influence of L. brevis BCRC12247 Fermentation on Total Phenol and ROS Accumulation
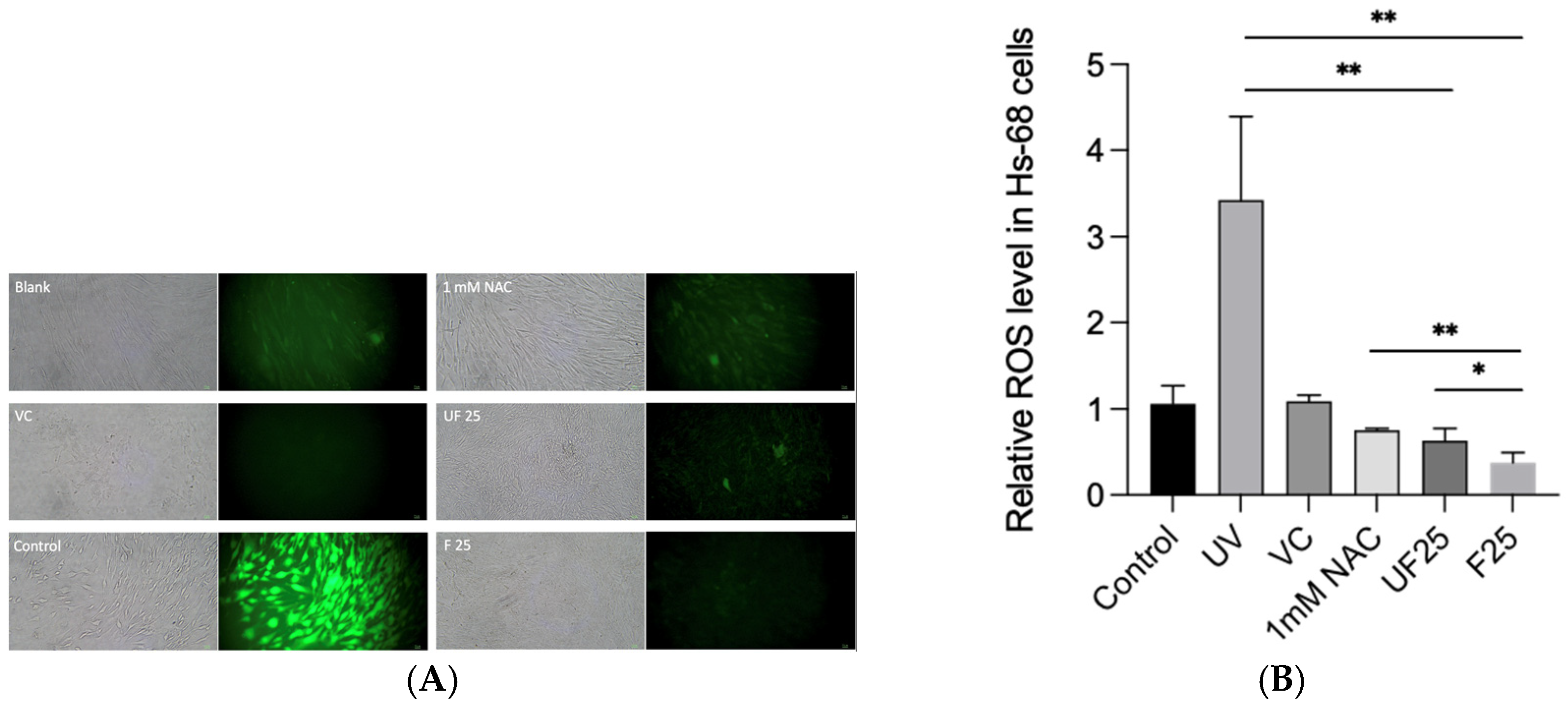
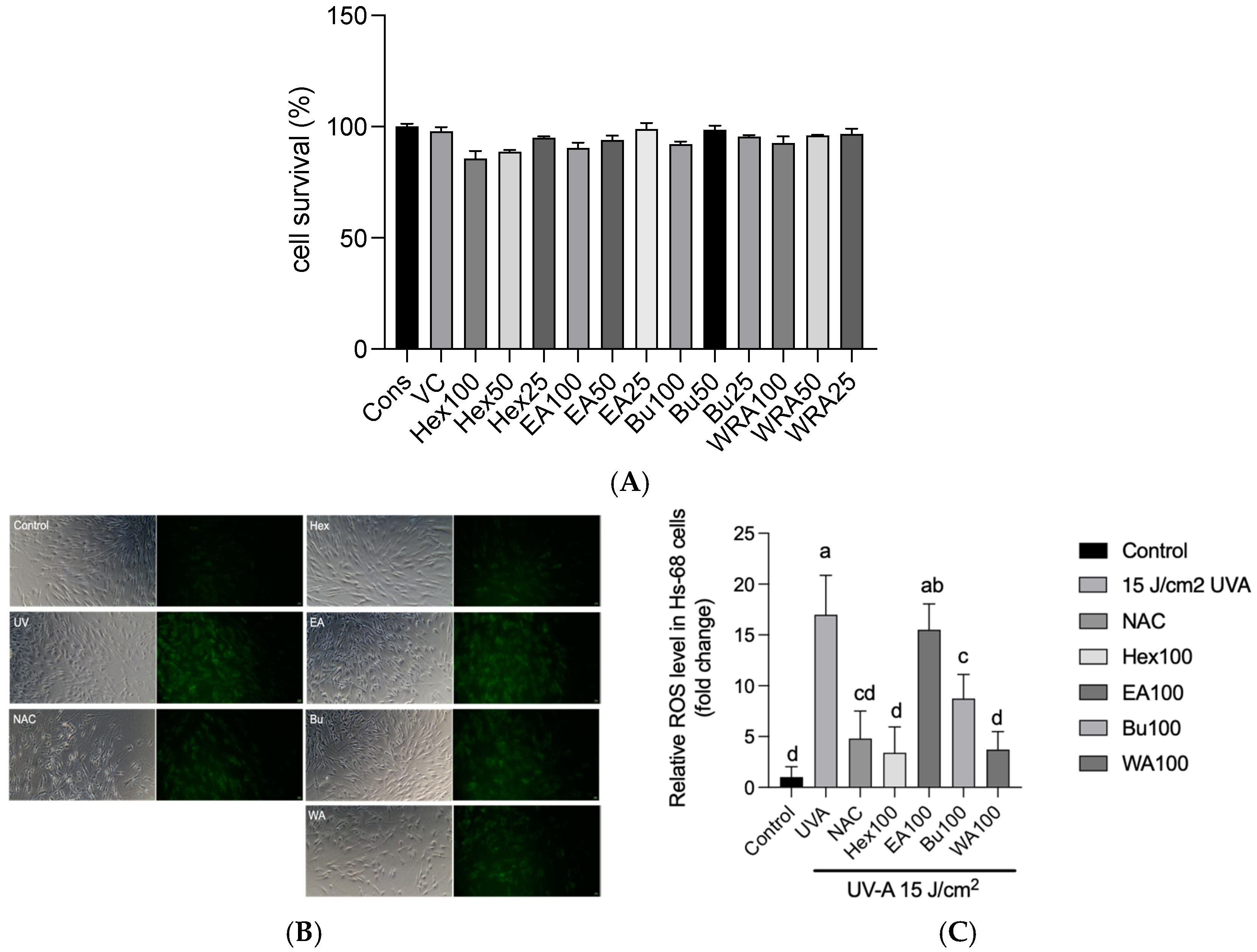
3.3. Effects of Fractions of Fermented Turmeric Milk Extract on ROS Inhibition and Cell Viability in UVA-Irradiated Hs68 Cells
3.4. Effects of Fractions of Fermented Turmeric Milk Extract on Senescence-Associated β-Galactosidase (SA-β-Gal) Activity
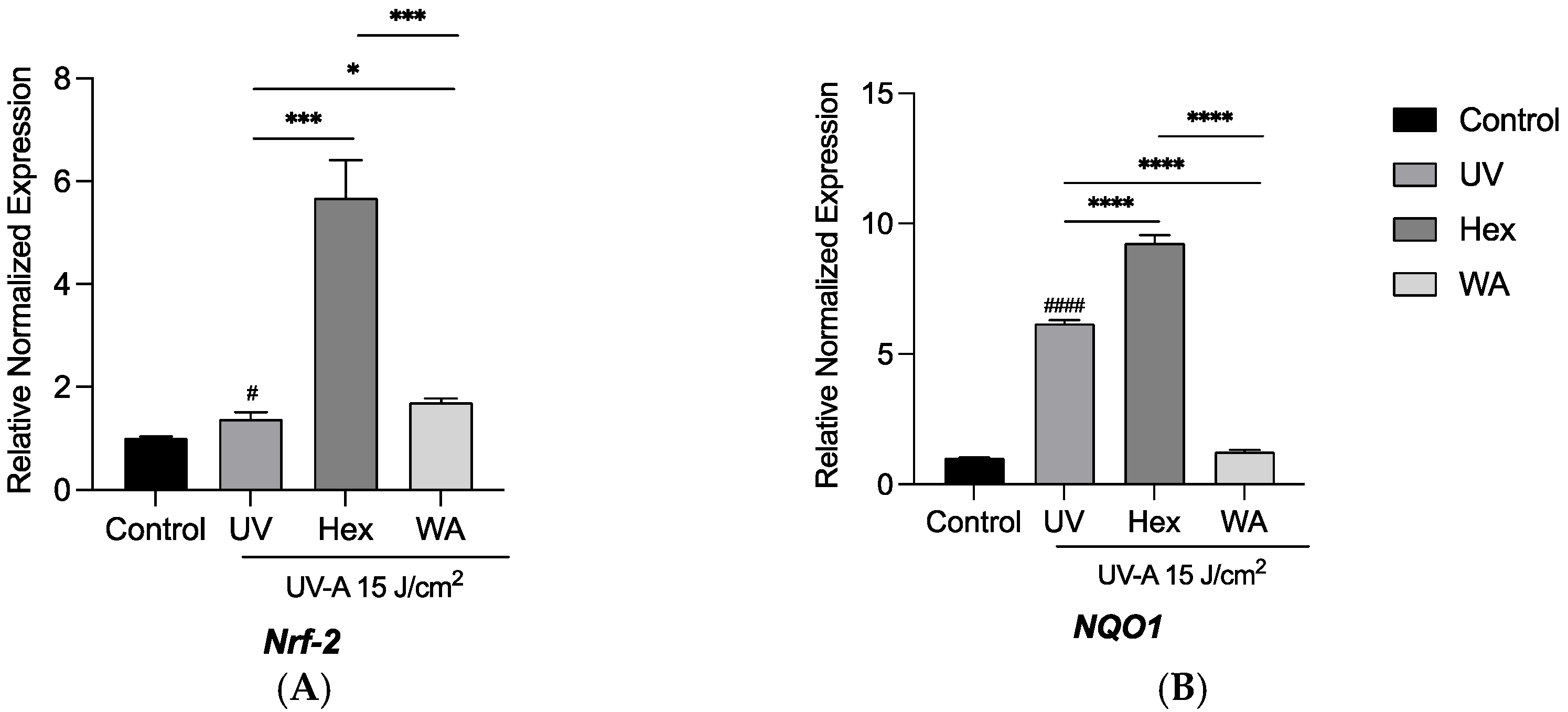
3.5. Effects of Fractions of Fermented Turmeric Milk on Antioxidant Genes Expression in UVA-Exposed Hs68 Cells
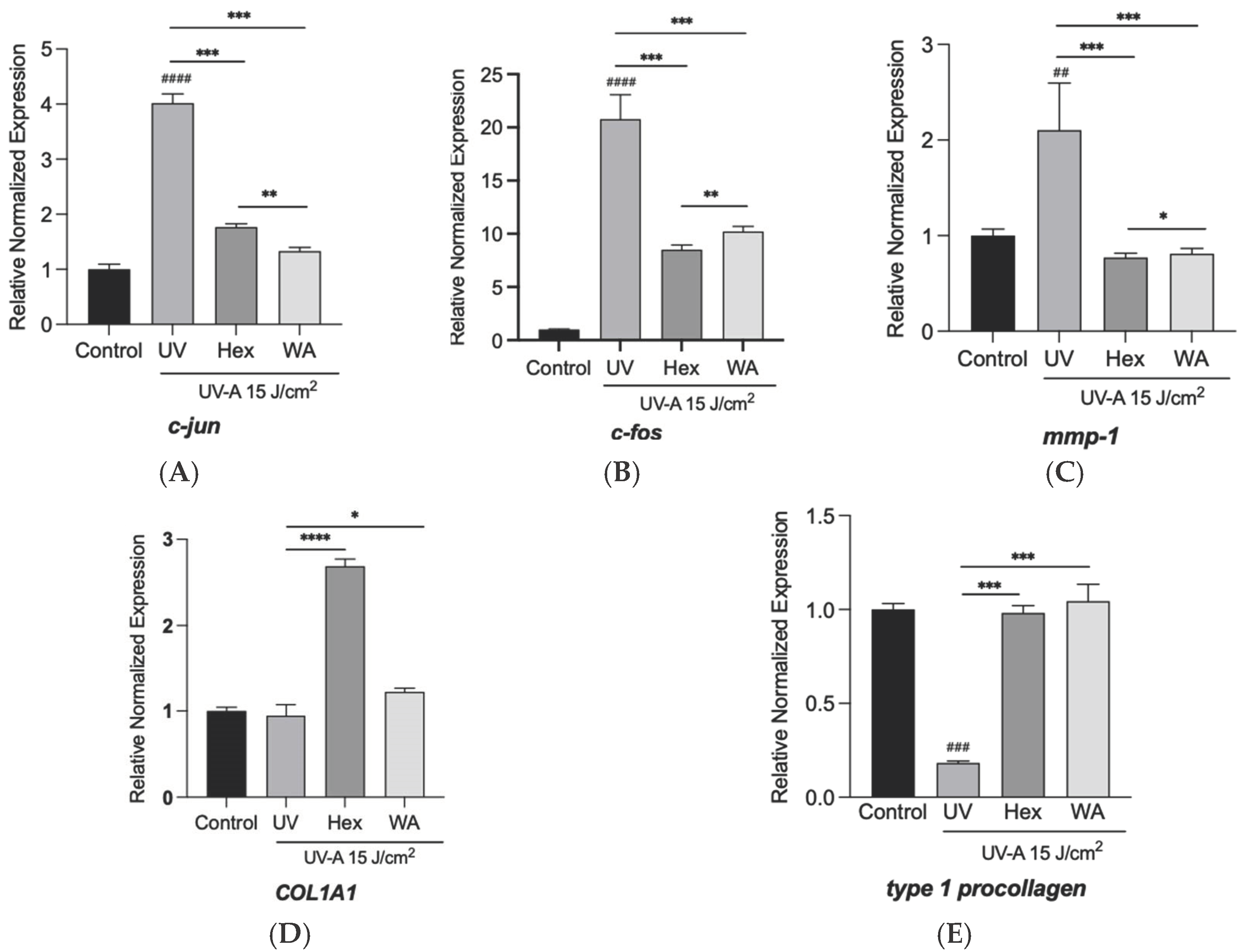
3.6. Effects of Fractions of Fermented Turmeric Milk on Skin-Associated Genes Expression in UVA-Exposed Hs68 Cells
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Parisi, A.V.; Turner, J. Variations in the short wavelength cut-off of the solar UV spectra. Photochem. Photobiol. Sci. 2006, 5, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Rabbi, M.; Kim, M.; Ke, C.; Lee, W.; Clark, R.L.; Mieczkowski, P.A.; Marszalek, P.E. UVA generates pyrimidine dimers in DNA directly. Biophys. J. 2009, 96, 1151–1158. [Google Scholar] [CrossRef] [Green Version]
- Cavinato, M.; Jansen-Dürr, P. Molecular mechanisms of UVB-induced senescence of dermal fibroblasts and its relevance for photoaging of the human skin. Exp. Gerontol. 2017, 94, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Han, J.; Jiang, C.; Zhang, Y. Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res. Rev. 2020, 59, 101036. [Google Scholar] [CrossRef] [PubMed]
- Farage, M.A.; Miller, K.W.; Elsner, P.; Maibach, H.I. Intrinsic and extrinsic factors in skin ageing: A review. Int. J. Cosmet. Sci. 2008, 30, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Rittié, L.; Fisher, G.J. Natural and sun-induced aging of human skin. Cold Spring Harb. Perspect. Med. 2015, 5, a015370. [Google Scholar] [CrossRef] [PubMed]
- Lai-Cheong, J.E.; McGrath, J.A. Structure and function of skin, hair and nails. Medicine 2013, 41, 317–320. [Google Scholar] [CrossRef]
- Blume-Peytavi, U.; Kottner, J.; Sterry, W.; Hodin, M.W.; Griffiths, T.W.; Watson, R.E.B.; Hay, R.J.; Griffiths, C.E.M. Age-Associated Skin Conditions and Diseases: Current Perspectives and Future Options. Gerontologist 2016, 56 (Suppl. 2), S230–S242. [Google Scholar] [CrossRef] [Green Version]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “Curecumin”: From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef] [Green Version]
- Sharifi-Rad, J.; Rayess, Y.E.; Rizk, A.A.; Sadaka, C.; Zgheib, R.; Zam, W.; Sestito, S.; Rapposelli, S.; Neffe-Skocińska, K.; Zielińska, D.; et al. Turmeric and Its Major Compound Curcumin on Health: Bioactive Effects and Safety Profiles for Food, Pharmaceutical, Biotechnological and Medicinal Applications. Front. Pharmacol. 2020, 11, 01021. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Lin, Y.-S.; Huang, Y.-W.; Fang, S.-U.; Lin, S.-Y.; Hou, W.-C. Protective Effects of Minor Components of Curcuminoids on Hydrogen Peroxide-Treated Human HaCaT Keratinocytes. J. Agric. Food Chem. 2016, 64, 3598–3608. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wan, M.; Li, H.; Chen, Q.; Li, R.; Liang, B.; Zhu, H. Curcumin protection against ultraviolet-induced photo-damage in Hacat cells by regulating nuclear factor erythroid 2-related factor 2. Bioengineered 2021, 12, 9993–10006. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, R.; Shi, H.; Li, X.; Li, Y.; Taha, A.; Xu, C. Protective effect of curcumin against ultraviolet A irradiation-induced photoaging in human dermal fibroblasts. Mol. Med. Rep. 2018, 17, 7227–7237. [Google Scholar] [CrossRef] [Green Version]
- Thomas, B. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef]
- Azam, M.; Mohsin, M.; Ijaz, H.; Tulain, R.; Ashraf, M.; Fayyaz, A.; Ul Abadeen, Z.; Kamran, Q. Lactic acid bacteria in traditional fermented Asian foods. Pak. J. Pharm. Sci. 2017, 30, 1803–1814. [Google Scholar]
- Albano, C.; Morandi, S.; Silvetti, T.; Casiraghi, M.C.; Manini, F.; Brasca, M. Lactic acid bacteria with cholesterol-lowering properties for dairy applications: In vitro and in situ activity. J. Dairy Sci. 2018, 101, 10807–10818. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.H.; Lee, C.Y.; Chung, D.K. Probiotic Lactic Acid Bacteria and Skin Health. Crit. Rev. Food Sci. Nutr. 2016, 56, 2331–2337. [Google Scholar] [CrossRef]
- Di Caro, S.; Tao, H.; Grillo, A.; Elia, C.; Gasbarrini, G.; Sepulveda, A.R.; Gasbarrini, A. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig. Liver Dis. 2005, 37, 320–329. [Google Scholar] [CrossRef]
- El Mecherfi, K.-E.; Todorov, S.D.; Cavalcanti de Albuquerque, M.A.; Denery-Papini, S.; Lupi, R.; Haertlé, T.; Dora Gombossy de Melo Franco, B.; Larré, C. Allergenicity of Fermented Foods: Emphasis on Seeds Protein-Based Products. Foods 2020, 9, 792. [Google Scholar] [CrossRef]
- Wedajo, B. Lactic Acid Bacteria: Benefits, Selection Criteria and Probiotic Potential in Fermented Food. J. Probiotics Health 2015, 3, 1–9. [Google Scholar] [CrossRef]
- Pianpumepong, P.; Noomhorm, A. Isolation of probiotic bacteria from turmeric (Curcuma longa Linn.) and its application in enriched beverages. Int. J. Food Sci. Technol. 2010, 45, 2456–2462. [Google Scholar] [CrossRef]
- Yong, C.; Yoon, Y.; Yoo, H.; Oh, S. Effect of Lactobacillus Fermentation on the Anti-Inflammatory Potential of Turmeric. J. Microbiol. Biotechnol. 2019, 29, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, J.; Zou, L.; Fu, C.; Li, P.; Zhao, G. Chemical characterization, antioxidant, immune-regulating and anticancer activities of a novel bioactive polysaccharide from Chenopodium quinoa seeds. Int. J. Biol. Macromol. 2017, 99, 622–629. [Google Scholar] [CrossRef]
- Moein, M.R.; Moein, S.; Ahmadizadeh, S. Radical Scavenging and Reducing Power of Salvia mirzayanii Subfractions. Molecules 2008, 13, 2804–2813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.; Ra, J.-H.; Jee, Y.; Kim, J.-S. Impact of different partitioned solvents on chemical composition and bioavailability of Sasa quelpaertensis Nakai leaf extract. J. Food Drug Anal. 2017, 25, 316–326. [Google Scholar] [CrossRef] [Green Version]
- Abderrahim, F.; Huanatico, E.; Segura, R.; Arribas, S.; Gonzalez, M.C.; Condezo-Hoyos, L. Physical features, phenolic compounds, betalains and total antioxidant capacity of coloured quinoa seeds (Chenopodium quinoa Willd.) from Peruvian Altiplano. Food Chem. 2015, 183, 83–90. [Google Scholar] [CrossRef]
- Calò, R.; Marabini, L. Protective effect of Vaccinium myrtillus extract against UVA- and UVB-induced damage in a human keratinocyte cell line (HaCaT Cells). J. Photochem. Photobiol. B Biol. 2014, 132C, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.C.; Korivi, M.; Lin, F.Y.; Li, M.L.; Lin, R.W.; Wu, J.J.; Yang, H.L. Trans-cinnamic acid attenuates UVA-induced photoaging through inhibition of AP-1 activation and induction of Nrf2-mediated antioxidant genes in human skin fibroblasts. J. Derm. Sci. 2018, 90, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Makpol, S.; Durani, L.W.; Chua, K.H.; Mohd Yusof, Y.A.; Ngah, W.Z. Tocotrienol-rich fraction prevents cell cycle arrest and elongates telomere length in senescent human diploid fibroblasts. J. Biomed. Biotechnol. 2011, 2011, 506171. [Google Scholar] [CrossRef] [Green Version]
- Han, B.-I.; Hwang, S.-H.; Lee, M. A progressive reduction in autophagic capacity contributes to induction of replicative senescence in Hs68 cells. Int. J. Biochem. Cell Biol. 2017, 92, 18–25. [Google Scholar] [CrossRef]
- Sun, X.; Zuo, H.; Liu, C.; Yang, Y. Overexpression of miR-200a protects cardiomyocytes against hypoxia-induced apoptosis by modulating the kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 signaling axis. Int. J. Mol. Med. 2016, 38, 1303–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavrogonatou, E.; Konstantinou, A.; Kletsas, D. Long-term exposure to TNF-α leads human skin fibroblasts to a p38 MAPK- and ROS-mediated premature senescence. Biogerontology 2018, 19, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S. Antimicrobial Property of Turmeric (Curcuma longa L.) Rhizome-Derived ar-Turmerone and Curcumin. Food Sci. Biotechnol. 2006, 15, 559–563. [Google Scholar]
- Chen, J.; Liu, Y.; Zhao, Z.; Qiu, J. Oxidative stress in the skin: Impact and related protection. Int. J. Cosmet. Sci. 2021, 43, 495–509. [Google Scholar] [CrossRef]
- Bento-Silva, A.; Koistinen, V.M.; Mena, P.; Bronze, M.R.; Hanhineva, K.; Sahlstrøm, S.; Kitrytė, V.; Moco, S.; Aura, A.-M. Factors affecting intake, metabolism and health benefits of phenolic acids: Do we understand individual variability? Eur. J. Nutr. 2020, 59, 1275–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynh, N.T.; Van Camp, J.; Smagghe, G.; Raes, K. Improved release and metabolism of flavonoids by steered fermentation processes: A review. Int. J. Mol. Sci. 2014, 15, 19369–19388. [Google Scholar] [CrossRef]
- Yi, Y.; Xie, H.; Xiao, X.; Wang, B.; Du, R.; Liu, Y.; Li, Z.; Wang, J.; Sun, L.; Deng, Z.; et al. Ultraviolet A irradiation induces senescence in human dermal fibroblasts by down-regulating DNMT1 via ZEB1. Aging 2018, 10, 212–228. [Google Scholar] [CrossRef] [Green Version]
- Zhong, J.L.; Edwards, G.P.; Raval, C.; Li, H.; Tyrrell, R.M. The role of Nrf2 in ultraviolet A mediated heme oxygenase 1 induction in human skin fibroblasts. Photochem. Photobiol. Sci. 2010, 9, 18–24. [Google Scholar] [CrossRef]
- Khan, N.M.; Haseeb, A.; Ansari, M.Y.; Devarapalli, P.; Haynie, S.; Haqqi, T.M. Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free Radic. Biol. Med. 2017, 106, 288–301. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Zhang, Y.; An, Q.; Wang, D.; You, S.; Zhao, D.; Zhang, J.; Wang, C.; Li, M. Anti-Photoaging Effect of Rhodiola rosea Fermented by Lactobacillus plantarum on UVA-Damaged Fibroblasts. Nutrients 2022, 14, 2324. [Google Scholar] [CrossRef]
- Liu, W.; Yan, F.; Xu, Z.; Chen, Q.; Ren, J.; Wang, Q.; Chen, L.; Ying, J.; Liu, Z.; Zhao, J.; et al. Urolithin A protects human dermal fibroblasts from UVA-induced photoaging through NRF2 activation and mitophagy. J. Photochem. Photobiol. B Biol. 2022, 232, 112462. [Google Scholar] [CrossRef]
- Jochum, W.; Passegué, E.; Wagner, E.F. AP-1 in mouse development and tumorigenesis. Oncogene 2001, 20, 2401–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenneisen, P.; Wenk, J.; Klotz, L.O.; Wlaschek, M.; Briviba, K.; Krieg, T.; Sies, H.; Scharffetter-Kochanek, K. Central role of Ferrous/Ferric iron in the ultraviolet B irradiation-mediated signaling pathway leading to increased interstitial collagenase (matrix-degrading metalloprotease (MMP)-1) and stromelysin-1 (MMP-3) mRNA levels in cultured human dermal fibroblasts. J. Biol. Chem. 1998, 273, 5279–5287. [Google Scholar] [CrossRef] [Green Version]
- Wenk, J.; Brenneisen, P.; Wlaschek, M.; Poswig, A.; Briviba, K.; Oberley, T.D.; Scharffetter-Kochanek, K. Stable overexpression of manganese superoxide dismutase in mitochondria identifies hydrogen peroxide as a major oxidant in the AP-1-mediated induction of matrix-degrading metalloprotease-1. J. Biol. Chem. 1999, 274, 25869–25876. [Google Scholar] [CrossRef]
- Lu, J.; Guo, J.H.; Tu, X.L.; Zhang, C.; Zhao, M.; Zhang, Q.W.; Gao, F.H. Tiron Inhibits UVB-Induced AP-1 Binding Sites Transcriptional Activation on MMP-1 and MMP-3 Promoters by MAPK Signaling Pathway in Human Dermal Fibroblasts. PLoS ONE 2016, 11, e0159998. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.Y.; Chuang, W.L. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed. Pharm. 2012, 66, 454–458. [Google Scholar] [CrossRef]
- Sengupta, P.; Xu, Y.; Wang, L.; Widom, R.; Smith, B.D. Collagen alpha1(I) gene (COL1A1) is repressed by RFX family. J. Biol. Chem. 2005, 280, 21004–21014. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Turmeric Milk | DPPH Free-Radical Scavenging Assay (mmole TE/g dw) | ABTs Free-Radical Scavenging Assay (mmole TE/g dw) | Ferric-Reducing Power Capacity Assay (mmole TE/g dw) |
|---|---|---|---|
| UF | 1.77 ± 0.01 cd | 3.98 ± 0.06 cd | 2.82 ± 0.08 ab |
| 14023 | 1.80 ± 0.01 c | 3.94 ± 0.06 d | 2.45 ± 0.11 c |
| 12195 | 1.86 ± 0.02 b | 4.15 ± 0.11 bc | 2.90 ± 0.02 ab |
| 12247 | 1.97 ± 0.02 a | 4.41 ± 0.05 a | 2.95 ± 0.03 a |
| 14634 | 1.74 ± 0.01 d | 4.28 ± 0.03 ab | 2.72 ± 0.04 b |
| 10696 | 1.86 ± 0.03 b | 4.18 ± 0.06 bc | 2.80 ± 0.09 ab |
| 14079 | 1.93 ± 0.02 cd | 4.26 ± 0.08 ab | 2.75 ± 0.07 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.-J.; Cheng, M.-C.; Khumsupan, D.; Hsieh, C.-C.; Hsieh, C.-W.; Cheng, K.-C. Evaluation of Fermented Turmeric Milk by Lactic Acid Bacteria to Prevent UV-Induced Oxidative Stress in Human Fibroblast Cells. Fermentation 2023, 9, 230. https://doi.org/10.3390/fermentation9030230
Lu J-J, Cheng M-C, Khumsupan D, Hsieh C-C, Hsieh C-W, Cheng K-C. Evaluation of Fermented Turmeric Milk by Lactic Acid Bacteria to Prevent UV-Induced Oxidative Stress in Human Fibroblast Cells. Fermentation. 2023; 9(3):230. https://doi.org/10.3390/fermentation9030230
Chicago/Turabian StyleLu, Jheng-Jhe, Meng-Chun Cheng, Darin Khumsupan, Chen-Che Hsieh, Chang-Wei Hsieh, and Kuan-Chen Cheng. 2023. "Evaluation of Fermented Turmeric Milk by Lactic Acid Bacteria to Prevent UV-Induced Oxidative Stress in Human Fibroblast Cells" Fermentation 9, no. 3: 230. https://doi.org/10.3390/fermentation9030230
APA StyleLu, J.-J., Cheng, M.-C., Khumsupan, D., Hsieh, C.-C., Hsieh, C.-W., & Cheng, K.-C. (2023). Evaluation of Fermented Turmeric Milk by Lactic Acid Bacteria to Prevent UV-Induced Oxidative Stress in Human Fibroblast Cells. Fermentation, 9(3), 230. https://doi.org/10.3390/fermentation9030230







