Pyruvic Acid Production from Sucrose by Escherichia coli Pyruvate Dehydrogenase Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Genetic Modifications
2.2. Media
2.3. Shake Flask Experiments
2.4. Batch Processes
2.5. Fed-Batch Processes
2.6. Analytical Methods
3. Results
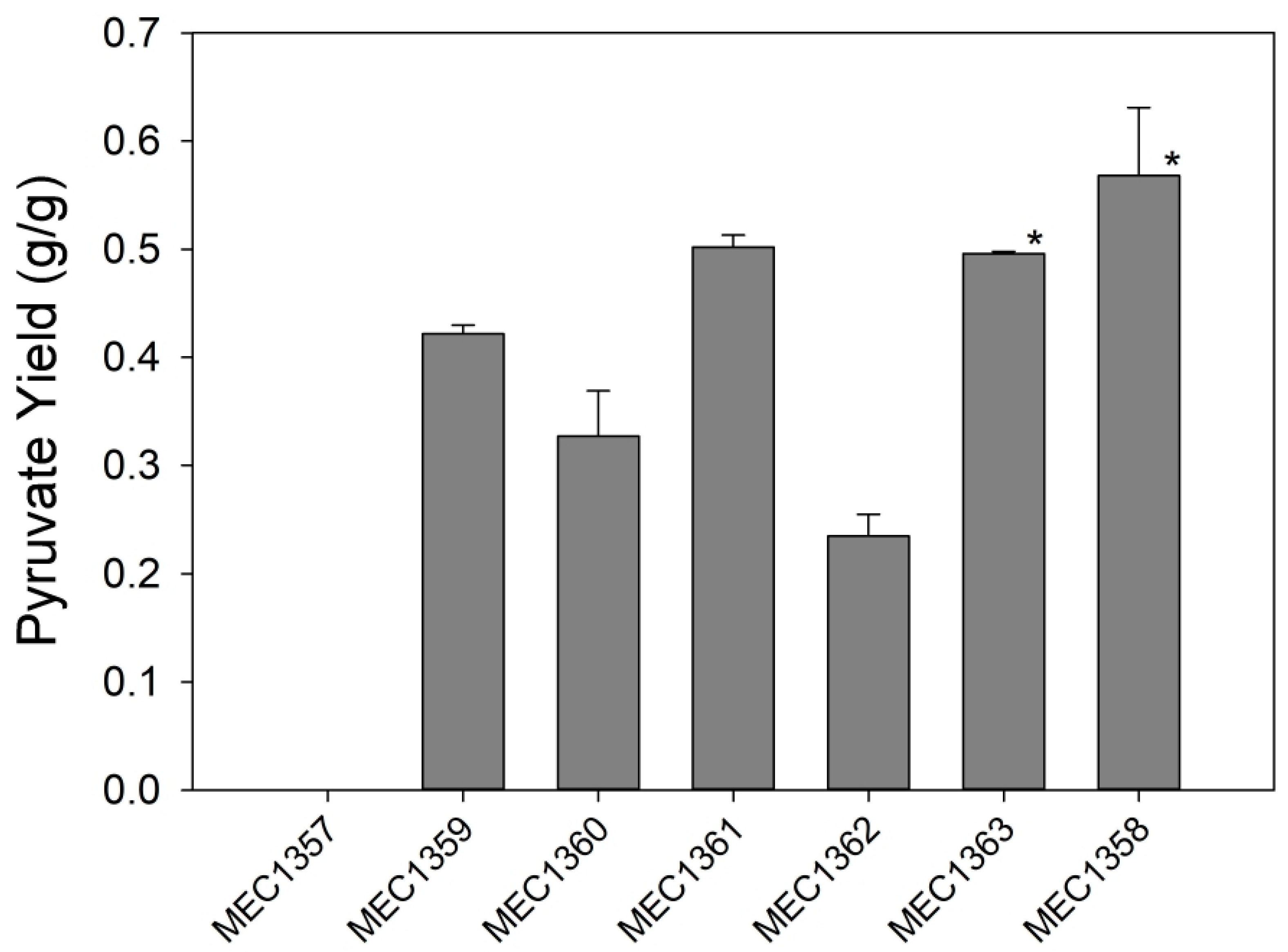
3.1. Variant Strain Screening for Pyruvate Yield
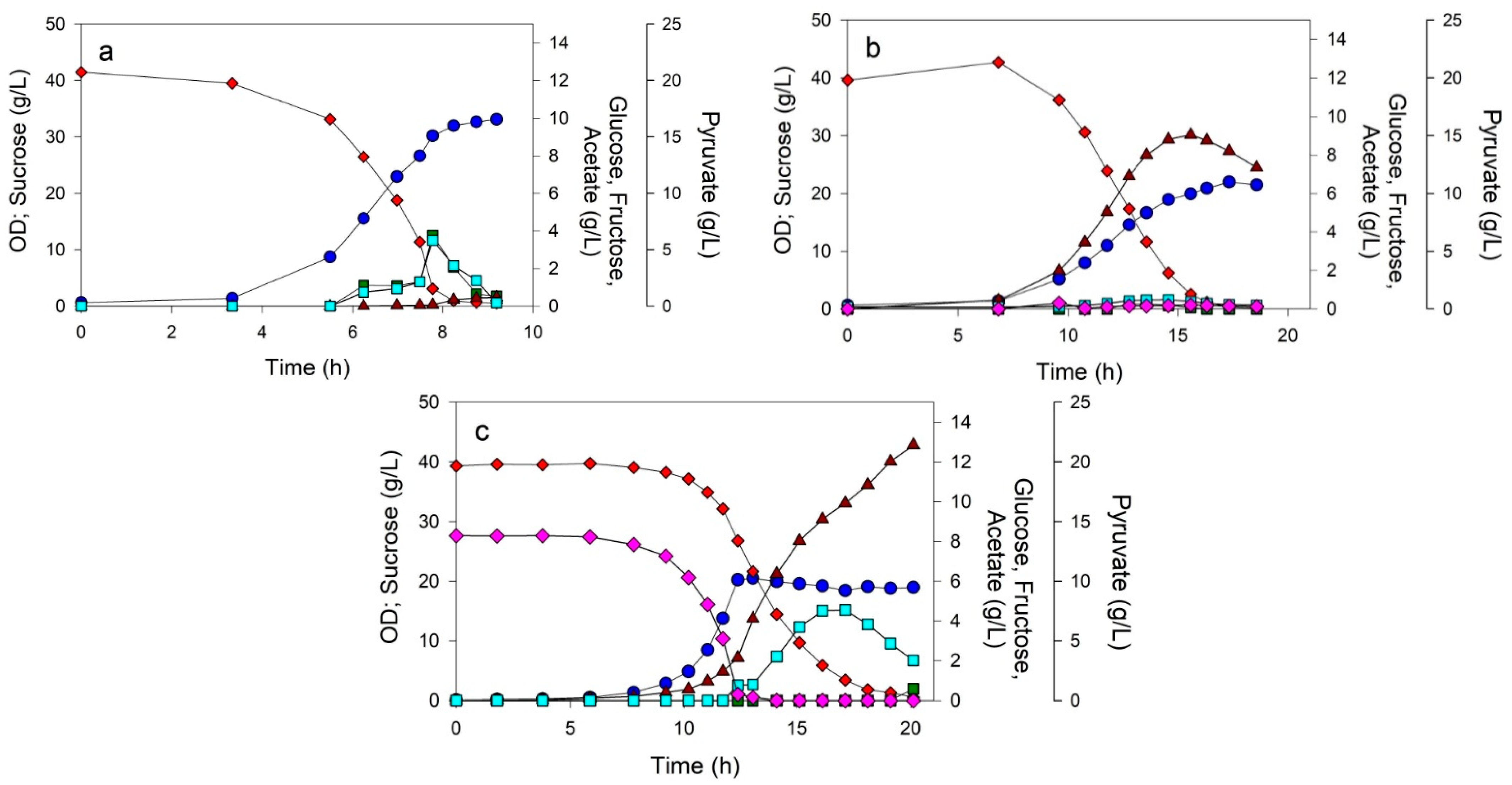
3.2. Controlled Batch Processes
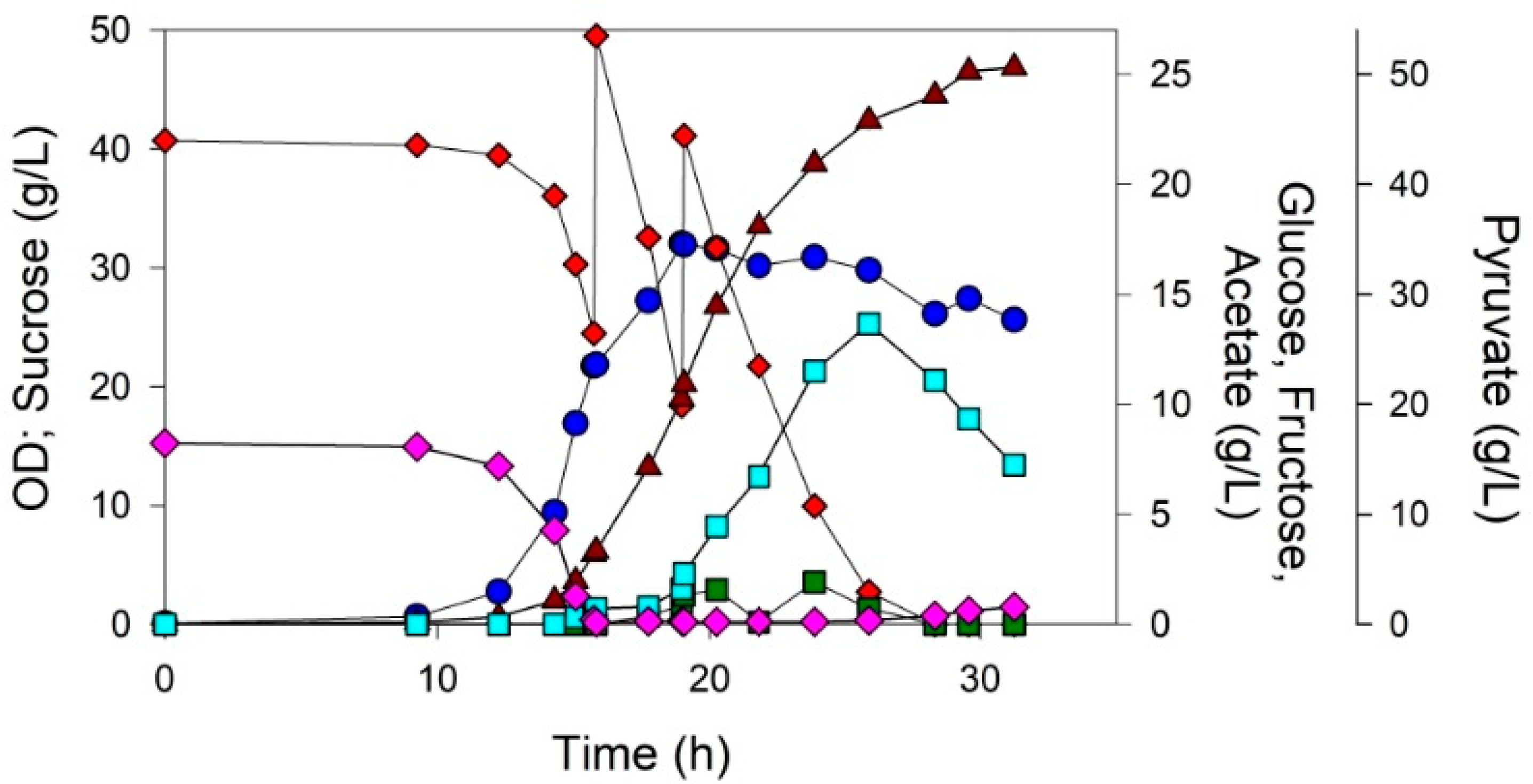
3.3. Fed-Batch Process
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kunamneni, A.; Singh, S. Response surface optimization of enzymatic hydrolysis of maize starch for higher glucose production. Biochem. Eng. J. 2005, 27, 179–190. [Google Scholar] [CrossRef]
- van der Poel, P.; Schiweck, H.; Schwartz, T. Sugar Technology, Beet and Cane Sugar Manufacture, Sugar Technology; Verlag Dr. Albert Bartens KG: Berlin, Germany, 1998. [Google Scholar] [CrossRef]
- Yim, H.; Haselbeck, R.; Niu, W.; Pujol-Baxley, C.; Burgard, A.; Boldt, J.; Khandurina, J.; Trawick, J.D.; Osterhout, R.E.; Stephen, R.; et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar] [CrossRef]
- Bockmann, J.; Heuel, H.; Lengeler, J.W. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. 1992, 235, 22–32. [Google Scholar] [CrossRef]
- Sabri, S.; Nielsen, L.K.; Vickers, C.E. Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Appl. Environ. Microbiol. 2013, 79, 478–487. [Google Scholar] [CrossRef]
- Tsunekawa, H.; Azuma, S.; Okabe, M.; Okamoto, R.; Aiba, S. Acquisition of a sucrose utilization system in Escherichia coli K-12 derivatives and its application to industry. Appl. Environ. Microbiol. 1992, 58, 2081–2088. [Google Scholar] [CrossRef]
- Arifin, Y.; Sabri, S.; Sugiarto, H.; Krömer, J.O.; Vickers, C.E.; Nielsen, L.K. Deletion of cscR in Escherichia coli W improves growth and poly-3-hydroxybutyrate (PHB) production from sucrose in fed batch culture. J. Biotechnol. 2011, 156, 275–278. [Google Scholar] [CrossRef]
- Erian, A.M.; Gibisch, M.; Pflügl, S. Engineered E. coli W enables efficient 2,3-butanediol production from glucose and sugar beet molasses using defined minimal medium as economic basis. Microb. Cell Fact. 2018, 17, 190. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, S.; Park, J.H.; Vickers, C.E.; Nielsen, L.K.; Lee, S.Y. Development of sucrose-utilizing Escherichia coli K-12 strain by cloning β-fructofuranosidases and its application for L-threonine production. Appl. Microbiol. Biotechnol. 2010, 88, 905–913. [Google Scholar] [CrossRef]
- Shukla, V.B.; Zhou, S.; Yomano, L.P.; Shanmugam, K.T.; Preston, J.F.; Ingram, L.O. Production of d(−)-lactate from sucrose and molasses. Biotechnol. Lett. 2004, 26, 689–693. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, J.; Bennett, G.N.; San, K.Y. Succinate production from sucrose by metabolic engineered Escherichia coli strains under aerobic conditions. Biotechnol. Prog. 2011, 27, 1242–1247. [Google Scholar] [CrossRef] [PubMed]
- Arifin, Y.; Archer, C.; Lim, S.A.; Quek, L.E.; Sugiarto, H.; Marcellin, E.; Vickers, C.E.; Krömer, J.O.; Nielsen, L.K. Escherichia coli W shows fast, highly oxidative sucrose metabolism and low acetate formation. Appl. Microbiol. Biotechnol. 2014, 98, 9033–9044. [Google Scholar] [CrossRef] [PubMed]
- Archer, C.T.; Kim, J.F.; Jeong, H.; Park, J.H.; Vickers, C.E.; Lee, S.Y.; Nielsen, L.K. The genome sequence of E. coli W (ATCC 9637): Comparative genome analysis and an improved genome-scale reconstruction of E. coli. BMC Genom. 2011, 12, 9. [Google Scholar] [CrossRef]
- Steen, J.A.; Bohlke, N.; Vickers, C.E.; Nielsen, L.K. The Trehalose Phosphotransferase System (PTS) in E. coli W can transport low levels of sucrose that are sufficient to facilitate induction of the csc sucrose catabolism operon. PLoS ONE 2014, 9, e88688. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Chang, H.N. High cell density cultivation of Escherichia coli W using sucrose as a carbon source. Biotechnol. Lett. 1993, 15, 971–974. [Google Scholar] [CrossRef]
- Maleki, N.; Eiteman, M.A. Recent progress on the microbial production of pyruvic acid. Fermentation 2017, 3, 8. [Google Scholar] [CrossRef]
- Blombach, B.; Schreiner, M.E.; Holátko, J.; Bartek, T.; Oldiges, M.; Eikmanns, B.J. L-Valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl. Environ. Microbiol. 2007, 73, 2079–2084. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, M.A.; Ferraz, C.A.; Scrutton, N.S. Alternative metabolic pathways and strategies to high-titre terpenoid production in Escherichia coli. Nat. Prod. Rep. 2022, 39, 90–118. [Google Scholar] [CrossRef]
- Rolf, J.; Julsing, M.K.; Rosenthal, K.; Lütz, S. A gram-scale limonene production process with engineered Escherichia coli. Molecules 2020, 25, 1881. [Google Scholar] [CrossRef]
- Smith, G.M.; Lee, S.A.; Reilly, K.C.; Eiteman, M.A.; Altman, E. Fed-batch two-phase production of alanine by a metabolically engineered Escherichia coli. Biotechnol. Lett. 2006, 28, 1695–1700. [Google Scholar] [CrossRef]
- Zhu, Y.; Eiteman, M.A.; Altman, R.; Altman, E. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl. Environ. Microbiol. 2008, 74, 6649–6655. [Google Scholar] [CrossRef]
- Tomar, A.; Eiteman, M.A.; Altman, E. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl. Microbiol. Biotechnol. 2003, 62, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Zelić, B.; Gerharz, T.; Bott, M.; Vasić-Rački, Đ.; Wandrey, C.; Takors, R. Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 Strain. Eng. Life Sci. 2003, 3, 299–305. [Google Scholar] [CrossRef]
- Ziegler, M.; Hägele, L.; Gäbele, T.; Takors, R. CRISPRi enables fast growth followed by stable aerobic pyruvate formation in Escherichia coli without auxotrophy. Eng. Life Sci. 2021, 22, 70–84. [Google Scholar] [CrossRef]
- Moxley, W.C.; Eiteman, M.A. Pyruvate production by Escherichia coli by use of pyruvate dehydrogenase variants. Appl. Environ. Microbiol. 2021, 87, e00487-21. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Moxley, W.C.; Brown, R.E.; Eiteman, M.A. Escherichia coli aceE variants coding pyruvate dehydrogenase improve the generation of pyruvate-derived acetoin. Eng. Life Sci. 2023, 23, 2200054. [Google Scholar] [CrossRef]
- Eiteman, M.A.; Chastain, M.J. Optimization of the ion-exchange analysis of organic acids from fermentation. Anal. Chim. Acta 1997, 338, 69–75. [Google Scholar] [CrossRef]
- Kale, S.; Arjunan, P.; Furey, W.; Jordan, F. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component. J. Biol. Chem. 2007, 282, 28106–28116. [Google Scholar] [CrossRef]
- Arjunan, P.; Nemeria, N.; Brunskill, A.; Chandrasekhar, K.; Sax, M.; Yan, Y.; Jordan, F.; Guest, J.R.; Furey, W. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 2002, 41, 5213–5221. [Google Scholar] [CrossRef]
- Park, Y.H.; Wei, W.; Zhou, L.; Nemeria, N.; Jordan, F. Amino-terminal residues 1–45 of the Escherichia coli pyruvate dehydrogenase complex E1 subunit interact with the E2 subunit and are required for activity of the complex but not for reductive acetylation of the E2 subunit. Biochemistry 2004, 43, 14037–14046. [Google Scholar] [CrossRef]
- Jeckelmann, J.M.; Erni, B. Carbohydrate transport by group translocation: The bacterial phosphoenolpyruvate: Sugar phosphotransferase system, in: Subcellular. Biochemistry 2019, 92, 223–274. [Google Scholar] [CrossRef]
- Postma, P.W.; Lengeler, J.W.; Jacobson, G.R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 1993, 57, 543–594. [Google Scholar] [CrossRef]
- Emmerling, M.; Dauner, M.; Ponti, A.; Fiaux, J.; Hochuli, M.; Szyperski, T.; Wüthrich, K.; Bailey, J.E.; Sauer, U. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 2002, 184, 152–164. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP—Pyruvate—Oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005, 29, 765–794. [Google Scholar] [CrossRef]
- Löwe, H.; Schmauder, L.; Hobmeier, K.; Kremling, A.; Pflüger-Grau, K. Metabolic engineering to expand the substrate spectrum of Pseudomonas putida toward sucrose. Microbiolgyopen 2017, 6, e00473. [Google Scholar] [CrossRef]
- Sahin-Tóth, M.; Lengyel, Z.; Tsunekawa, H. Cloning, sequencing, and expression of cscA invertase from Escherichia coli B-62. Can. J. Microbiol. 1999, 45, 418–422. [Google Scholar] [CrossRef] [PubMed]
| Strain | Relevant Characteristics | Reference |
|---|---|---|
| ATCC 9637 | Escherichia coli W | Wild-type |
| MEC1319 | ATCC 9637 ΔldhA ΔpoxB ΔppsA | [27] |
| MEC1320 | MEC1319 ΔaceE::cam-sacB | [27] |
| MEC1322 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE | [27] |
| MEC1332 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE::aceE[H106V] | [27] |
| MEC1339 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE::aceE[N276S;R465C;V668A;Y696N] | [27] |
| MEC1340 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE::aceE[V169A;P190Q;F532L] | [27] |
| MEC1341 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE::aceE[H106M] | [27] |
| MEC1342 | ATCC 9637 ΔldhA ΔpoxB ΔppsA ΔaceE::aceE[H106M;E401A] | [27] |
| MEC1357 | MEC1319 ΔcscR::Kan | This study |
| MEC1358 | MEC1322 ΔcscR::Kan | This study |
| MEC1359 | MEC1332 ΔcscR::Kan | This study |
| MEC1360 | MEC1339 ΔcscR::Kan | This study |
| MEC1361 | MEC1340 ΔcscR::Kan | This study |
| MEC1362 | MEC1341 ΔcscR::Kan | This study |
| MEC1363 | MEC1342 ΔcscR::Kan | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moxley, W.C.; Eiteman, M.A. Pyruvic Acid Production from Sucrose by Escherichia coli Pyruvate Dehydrogenase Variants. Fermentation 2023, 9, 478. https://doi.org/10.3390/fermentation9050478
Moxley WC, Eiteman MA. Pyruvic Acid Production from Sucrose by Escherichia coli Pyruvate Dehydrogenase Variants. Fermentation. 2023; 9(5):478. https://doi.org/10.3390/fermentation9050478
Chicago/Turabian StyleMoxley, W. Chris, and Mark A. Eiteman. 2023. "Pyruvic Acid Production from Sucrose by Escherichia coli Pyruvate Dehydrogenase Variants" Fermentation 9, no. 5: 478. https://doi.org/10.3390/fermentation9050478
APA StyleMoxley, W. C., & Eiteman, M. A. (2023). Pyruvic Acid Production from Sucrose by Escherichia coli Pyruvate Dehydrogenase Variants. Fermentation, 9(5), 478. https://doi.org/10.3390/fermentation9050478






