Fermentation of Date Pulp Residues Using Saccharomyces cerevisiae and Pichia kudriavzevii—Insights into Biological Activities, Phenolic and Volatile Compounds, Untargeted Metabolomics, and Carbohydrate Analysis Post In Vitro Digestion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Propagation, Proliferation, pH, and Titratable Acidity
2.2. Fermentation of Date Pulp Residues
2.3. Physicochemical Properties
2.3.1. Sugars and Organic Acids
2.3.2. Volatile Compounds
2.3.3. Quantification of Phenolic Compounds
2.4. In Vitro Digestion by INFOGEST2.0 and Bioaccessible Portion
2.5. Health-Promoting Properties
2.5.1. Inhibition of α-Amylase and α-Glucosidase
2.5.2. Antioxidant Capacity
2.5.3. Cytotoxicity Activities
2.5.4. OPA and TPC
2.5.5. ACE Inhibition
2.6. Untargeted Metabolomics and Carbohydrate Metabolomics Analyses
2.6.1. Untargeted Metabolite Analysis
2.6.2. Carbohydrate Metabolites Analysis
2.7. Data Processing
2.8. Statistical Analysis
3. Result and Discussion
3.1. Yeast Proliferation, pH, and Titratable Acidity
3.2. Physicochemical Properties
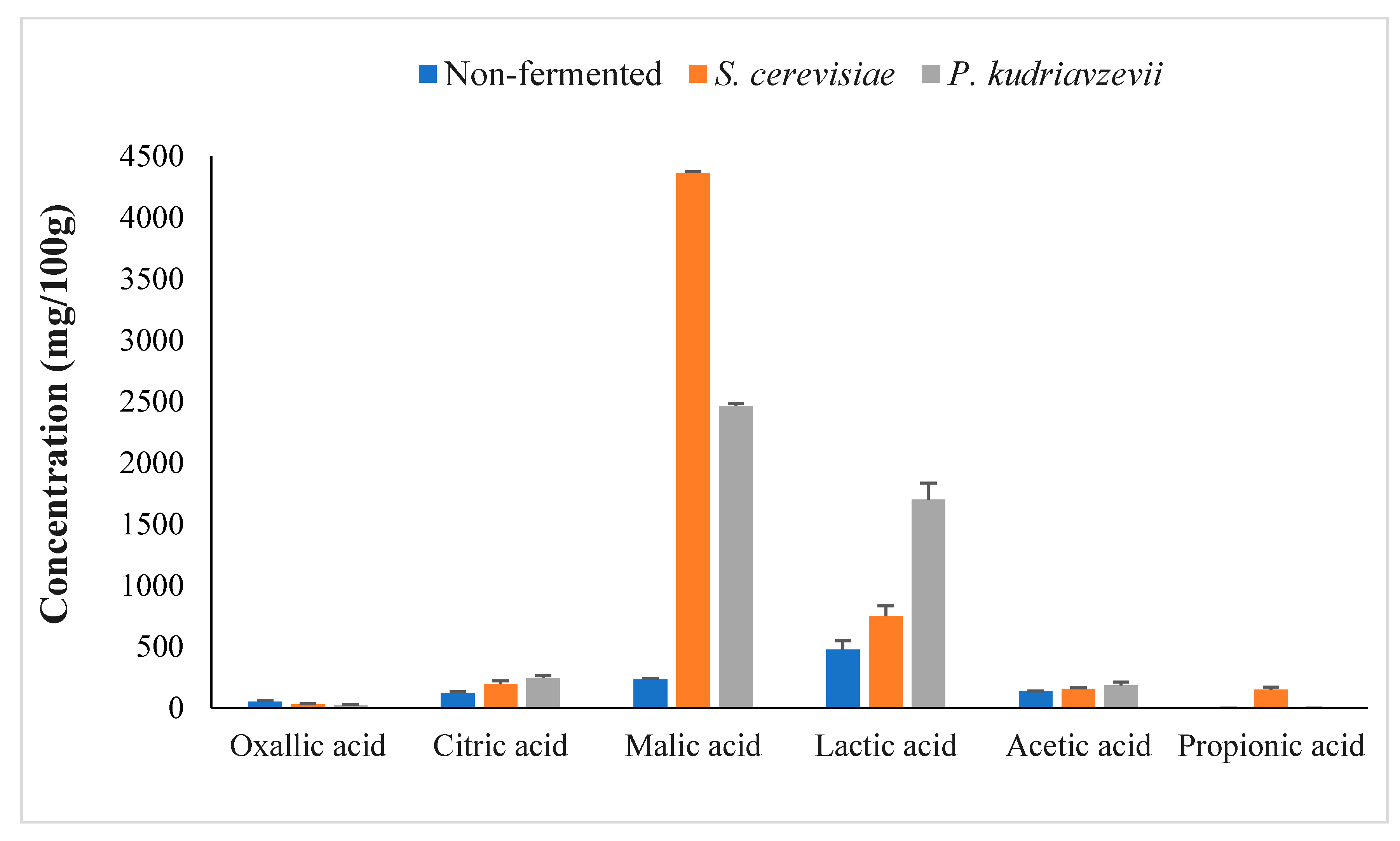
3.2.1. Evaluation of Sugars and Organic Acids
3.2.2. Evaluation of Volatile Compounds
3.2.3. Phenolic Compounds Profile
3.3. Health-Promoting Attributes
3.4. Untargeted and Carbohydrates Metabolomics Analyses
3.4.1. Untargeted Metabolite Analysis
3.4.2. Metabolic Pathway Analysis
3.4.3. Carbohydrate Metabolites Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capanoglu, E.; Tomás-Barberán, F.A. Introduction to Novel Approaches in the Valorization of Agricultural Wastes and Their Applications. J. Agric. Food Chem. 2022, 70, 6785–6786. [Google Scholar] [CrossRef] [PubMed]
- FAO. Food Wastage Footprint: Impacts on Natural Resources; FAO: Rome, Italy, 2013. [Google Scholar]
- Tropea, A. Food Waste Valorization. Fermentation 2022, 8, 168. [Google Scholar] [CrossRef]
- Patel, S.; Shukla, S. Chapter 30—Fermentation of Food Wastes for Generation of Nutraceuticals and Supplements. In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 707–734. [Google Scholar]
- Bilal, M.; Iqbal, H.M.N. Sustainable bioconversion of food waste into high-value products by immobilized enzymes to meet bio-economy challenges and opportunities—A review. Food Res. Int. 2019, 123, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Usmani, Z.; Gupta, V.K.; Bhat, R. Valorization of fruits and vegetable wastes and by-products to produce natural pigments. Crit. Rev. Biotechnol. 2021, 41, 535–563. [Google Scholar] [CrossRef]
- Krueger, R.R. Date Palm (Phoenix dactylifera L.) Biology and Utilization. In The Date Palm Genome, Vol. 1: Phylogeny, Biodiversity and Mapping; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 3–28. [Google Scholar]
- FAOSTAT. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 2 March 2023).
- Oladzad, S.; Fallah, N.; Mahboubi, A.; Afsham, N.; Taherzadeh, M.J. Date fruit processing waste and approaches to its valorization: A review. Bioresour. Technol. 2021, 340, 125625. [Google Scholar] [CrossRef]
- Barreveld, W. Date Palm Products; FAO: Rome, Italy, 1993. [Google Scholar]
- Siddiq, M.; Aleid, S.M.; Kader, A.A. Dates: Postharvest Science, Processing Technology and Health Benefits; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Majzoobi, M.; Karambakhsh, G.; Golmakani, M.; Mesbahi, G.; Farahnaky, A. Effects of level and particle size of date fruit press cake on batter rheological properties and physical and nutritional properties of cake. J. Agric. Sci. Technol. 2020, 22, 121–133. [Google Scholar]
- Cheok, C.Y.; Mohd Adzahan, N.; Abdul Rahman, R.; Zainal Abedin, N.H.; Hussain, N.; Sulaiman, R.; Chong, G.H. Current trends of tropical fruit waste utilization. Crit. Rev. Food Sci. Nutr. 2018, 58, 335–361. [Google Scholar] [CrossRef]
- Ahmad, A.; Banat, F.; Taher, H. Comparative study of lactic acid production from date pulp waste by batch and cyclic–mode dark fermentation. Waste Manag. 2021, 120, 585–593. [Google Scholar] [CrossRef]
- Seyed Reihani, S.F.; Khosravi-Darani, K. Mycoprotein Production from Date Waste Using Fusarium venenatum in a Submerged Culture. Appl. Food Biotechnol. 2019, 5, 243–352. [Google Scholar] [CrossRef]
- Rezazadeh Bari, M.; Alizadeh, M.; Farbeh, F. Optimizing endopectinase production from date pomace by Aspergillus niger PC5 using response surface methodology. Food Bioprod. Process. 2010, 88, 67–72. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Tan, X.; Show, P.L.; Rambabu, K.; Banat, F.; Veeramuthu, A.; Lau, B.F.; Ng, E.P.; Ling, T.C. Incorporating biowaste into circular bioeconomy: A critical review of current trend and scaling up feasibility. Environ. Technol. Innov. 2020, 19, 101034. [Google Scholar] [CrossRef]
- Struck, S.; Rohm, H. Chapter 1—Fruit processing by-products as food ingredients. In Valorization of Fruit Processing By-Products; Galanakis, C.M., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–16. [Google Scholar]
- Chandrasekaran, M. Valorization of Food Processing By-Products; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Gouw, V.P.; Jung, J.; Zhao, Y. Functional properties, bioactive compounds, and in vitro gastrointestinal digestion study of dried fruit pomace powders as functional food ingredients. LWT 2017, 80, 136–144. [Google Scholar] [CrossRef]
- Ribeiro, T.B.; Bonifácio-Lopes, T.; Morais, P.; Miranda, A.; Nunes, J.; Vicente, A.A.; Pintado, M. Incorporation of olive pomace ingredients into yoghurts as a source of fibre and hydroxytyrosol: Antioxidant activity and stability throughout gastrointestinal digestion. J. Food Eng. 2021, 297, 110476. [Google Scholar] [CrossRef]
- Coelho, M.C.; Ribeiro, T.B.; Oliveira, C.; Batista, P.; Castro, P.; Monforte, A.R.; Rodrigues, A.S.; Teixeira, J.; Pintado, M. In vitro gastrointestinal digestion impact on the bioaccessibility and antioxidant capacity of bioactive compounds from tomato flours obtained after conventional and ohmic heating extraction. Foods 2021, 10, 554. [Google Scholar] [CrossRef]
- Stafussa, A.P.; Maciel, G.M.; Bortolini, D.G.; Maroldi, W.V.; Ribeiro, V.R.; Fachi, M.M.; Pontarolo, R.; Bach, F.; Pedro, A.C.; Haminiuk, C.W.I. Bioactivity and bioaccessibility of phenolic compounds from Brazilian fruit purees. Future Foods 2021, 4, 100066. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Bahkali, A.H. Valorization of date palm (Phoenix dactylifera) fruit processing by-products and wastes using bioprocess technology—Review. Saudi J. Biol. Sci. 2013, 20, 105–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuvas-Limon, R.B.; Nobre, C.; Cruz, M.; Rodriguez-Jasso, R.M.; Ruíz, H.A.; Loredo-Treviño, A.; Texeira, J.A.; Belmares, R. Spontaneously fermented traditional beverages as a source of bioactive compounds: An overview. Crit. Rev. Food Sci. Nutr. 2020, 61, 2984–3006. [Google Scholar] [CrossRef]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Siriwardhana, N.; Kalupahana, N.S.; Cekanova, M.; LeMieux, M.; Greer, B.; Moustaid-Moussa, N. Modulation of adipose tissue inflammation by bioactive food compounds. J. Nutr. Biochem. 2013, 24, 613–623. [Google Scholar] [CrossRef]
- Munekata, P.E.S.; Domínguez, R.; Pateiro, M.; Nawaz, A.; Hano, C.; Walayat, N.; Lorenzo, J.M. Strategies to increase the value of pomaces with fermentation. Fermentation 2021, 7, 299. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-Waterhouse, D.; Waterhouse, G.I.N.; Cui, C.; Ruan, Z. Fermentation-enabled wellness foods: A fresh perspective. Food Sci. Hum. Wellness 2019, 8, 203–243. [Google Scholar] [CrossRef]
- Alkalbani, N.S.; Osaili, T.M.; Al-Nabulsi, A.A.; Obaid, R.S.; Olaimat, A.N.; Liu, S.Q.; Ayyash, M.M. In Vitro Characterization and Identification of Potential Probiotic Yeasts Isolated from Fermented Dairy and Non-Dairy Food Products. J. Fungi 2022, 8, 544. [Google Scholar] [CrossRef]
- Silva, P.D.; Cruz, R.; Casal, S. Sugars and artificial sweeteners in soft drinks: A decade of evolution in Portugal. Food Control 2021, 120, 107481. [Google Scholar] [CrossRef]
- Chidi, B.S.; Bauer, F.F.; Rossouw, D. Organic acid metabolism and the impact of fermentation practices on wine acidity—A review. S. Afr. J. Enol. Vitic. 2018, 39, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.Q.; Aung, M.T.; Lee, P.R.; Yu, B. Yeast and volatile evolution in cider co-fermentation with Saccharomyces cerevisiae and Williopsis saturnus. Ann. Microbiol. 2016, 66, 307–315. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Ayyash, M.; Abdalla, A.; Alhammadi, A.; Senaka Ranadheera, C.; Affan Baig, M.; Al-Ramadi, B.; Chen, G.; Kamal-Eldin, A.; Huppertz, T. Probiotic survival, biological functionality and untargeted metabolomics of the bioaccessible compounds in fermented camel and bovine milk after in vitro digestion. Food Chem. 2021, 363, 130243. [Google Scholar] [CrossRef]
- Ng, Z.X.; Samsuri, S.N.; Yong, P.H. The antioxidant index and chemometric analysis of tannin, flavonoid, and total phenolic extracted from medicinal plant foods with the solvents of different polarities. J. Food Process. Preserv. 2020, 44, e14680. [Google Scholar] [CrossRef]
- Ho, K.L.; Tan, C.G.; Yong, P.H.; Wang, C.W.; Lim, S.H.; Kuppusamy, U.R.; Ngo, C.T.; Massawe, F.; Ng, Z.X. Extraction of phytochemicals with health benefit from Peperomia pellucida (L.) Kunth through liquid-liquid partitioning. J. Appl. Res. Med. Aromat. Plants 2022, 30, 100392. [Google Scholar] [CrossRef]
- Ng, Z.X.; Kuppusamy, U.R. Effects of different heat treatments on the antioxidant activity and ascorbic acid content of bitter melon, Momordica charantia. Braz. J. Food Technol. 2019, 22. [Google Scholar] [CrossRef]
- Ayyash, M.; Abdalla, A.; Alameri, M.; Baig, M.A.; Kizhakkayil, J.; Chen, G.; Huppertz, T.; Kamal-Eldin, A. Biological activities of the bioaccessible compounds after in vitro digestion of low-fat Akawi cheese made from blends of bovine and camel milk. J. Dairy Sci. 2021, 104, 9450–9464. [Google Scholar] [CrossRef]
- Kashyap, P.; Riar, C.S.; Jindal, N. Effect of extraction methods and simulated in vitro gastrointestinal digestion on phenolic compound profile, bio-accessibility, and antioxidant activity of Meghalayan cherry (Prunus nepalensis) pomace extracts. LWT 2022, 153, 112570. [Google Scholar] [CrossRef]
- Liu, C.; Fang, L.; Min, W.; Liu, J.; Li, H. Exploration of the molecular interactions between angiotensin-I-converting enzyme (ACE) and the inhibitory peptides derived from hazelnut (Corylus heterophylla Fisch.). Food Chem. 2018, 245, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Flowers, J.M.; Hazzouri, K.M.; Lemansour, A.; Capote, T.; Gros-Balthazard, M.; Ferrand, S.; Lebrun, M.; Amiri, K.M.A.; Purugganan, M.D. Patterns of Volatile Diversity Yield Insights Into the Genetics and Biochemistry of the Date Palm Fruit Volatilome. Front. Plant Sci. 2022, 13, 408. [Google Scholar] [CrossRef] [PubMed]
- Ghnimi, S.; Al-Shibli, M.; Al-Yammahi, H.R.; Al-Dhaheri, A.; Al-Jaberi, F.; Jobe, B.; Kamal-Eldin, A. Reducing sugars, organic acids, size, color, and texture of 21 Emirati date fruit varieties (Phoenix dactylifera L.). NFS J. 2018, 12, 1–10. [Google Scholar] [CrossRef]
- Elshibli, S.; Korpelainen, H. Biodiversity of date palms (Phoenix dactylifera L.) in Sudan: Chemical, morphological and DNA polymorphisms of selected cultivars. Plant Genet. Resour. Characterisation Util. 2009, 7, 194–203. [Google Scholar] [CrossRef]
- Kamal-Eldin, A.; Ghnimi, S. Classification of date fruit (Phoenix dactylifera L.) based on chemometric analysis with multivariate approach. J. Food Meas. Charact. 2018, 12, 1020–1027. [Google Scholar] [CrossRef]
- Farag, M.A.; Mohsen, M.; Heinke, R.; Wessjohann, L.A. Metabolomic fingerprints of 21 date palm fruit varieties from Egypt using UPLC/PDA/ESI–qTOF-MS and GC–MS analyzed by chemometrics. Food Res. Int. 2014, 64, 218–226. [Google Scholar] [CrossRef]
- Zelle, R.M.; De Hulster, E.; Van Winden, W.A.; De Waard, P.; Dijkema, C.; Winkler, A.A.; Geertman, J.M.A.; Van Dijken, J.P.; Pronk, J.T.; Van Maris, A.J.A. Malic acid production by Saccharomyces cerevisiae: Engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 2008, 74, 2766–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Xu, Y.; Xu, Q.; Cao, W.; Huang, H.; Liu, H. Microbial Biosynthesis of L-Malic Acid and Related Metabolic Engineering Strategies: Advances and Prospects. Front. Bioeng. Biotechnol. 2021, 9, 901. [Google Scholar] [CrossRef] [PubMed]
- Al-Farsi, M.; Alasalvar, C.; Morris, A.; Baron, M.; Shahidi, F. Compositional and sensory characteristics of three native sun-dried date (Phoenix dactylifera L.) varieties grown in Oman. J. Agric. Food Chem. 2005, 53, 7586–7591. [Google Scholar] [CrossRef] [PubMed]
- Alahyane, A.; Harrak, H.; Elateri, I.; Ayour, J.; Ait-Oubahou, A.; Benichou, M.; Abderrazik, M.E. Evaluation of some nutritional quality criteria of seventeen moroccan dates varieties and clones, fruits of date palm (Phoenix dactylifera L.). Braz. J. Biol. 2022, 82, e236471. [Google Scholar] [CrossRef]
- Cherif, S.; Le Bourvellec, C.; Bureau, S.; Benabda, J. Effect of storage conditions on ‘Deglet Nour’ date palm fruit organoleptic and nutritional quality. LWT 2021, 137, 110343. [Google Scholar] [CrossRef]
- Lee, L.W.; Tay, G.Y.; Cheong, M.W.; Curran, P.; Yu, B.; Liu, S.Q. Modulation of the volatile and non-volatile profiles of coffee fermented with Yarrowia lipolytica: I. Green coffee. LWT 2017, 77, 225–232. [Google Scholar] [CrossRef]
- Canonico, L.; Solomon, M.; Comitini, F.; Ciani, M.; Varela, C. Volatile profile of reduced alcohol wines fermented with selected non-Saccharomyces yeasts under different aeration conditions. Food Microbiol. 2019, 84, 103247. [Google Scholar] [CrossRef]
- Torrens, J.; Urpí, P.; Riu-Aumatell, M.; Vichi, S.; López-Tamames, E.; Buxaderas, S. Different commercial yeast strains affecting the volatile and sensory profile of cava base wine. Int. J. Food Microbiol. 2008, 124, 48–57. [Google Scholar] [CrossRef]
- Lakshmi, N.M.; Binod, P.; Sindhu, R.; Awasthi, M.K.; Pandey, A. Microbial engineering for the production of isobutanol: Current status and future directions. Bioengineered 2021, 12, 12308–12321. [Google Scholar] [CrossRef]
- Ravasio, D.; Wendland, J.; Walther, A. Major contribution of the Ehrlich pathway for 2-phenylethanol/rose flavor production in Ashbya gossypii. FEMS Yeast Res. 2014, 14, 833–844. [Google Scholar] [CrossRef] [Green Version]
- Abe, F.; Horikoshi, K.J.C.; Letters, M.B. Enhanced production of isoamyl alcohol and isoamyl acetate by ubiquitination-deficient Saccharomyces cerevisiae mutants. Cell. Mol. Biol. Lett. 2005, 10, 383. [Google Scholar] [PubMed]
- Shalit, M.; Katzir, N.; Tadmor, Y.; Larkov, O.; Burger, Y.; Shalekhet, F.; Lastochkin, E.; Ravid, U.; Amar, O.; Edelstein, M.; et al. Acetyl-CoA: Alcohol acetyltransferase activity and aroma formation in ripening melon fruits. J. Agric. Food Chem. 2001, 49, 794–799. [Google Scholar] [CrossRef] [PubMed]
- Yahyaoui, F.E.L.; Wongs-Aree, C.; Latché, A.; Hackett, R.; Grierson, D.; Pech, J.C. Molecular and biochemical characteristics of a gene encoding an alcohol acyl-transferase involved in the generation of aroma volatile esters during melon ripening. Eur. J. Biochem. 2002, 269, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Van Laere, S.D.M.; Vanderhaegen, B.M.P.; Derdelinckx, G.; Dufour, J.P.; Pretorius, I.S.; Winderickx, J.; Thevelein, J.M.; Delvaux, F.R. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl. Environ. Microbiol. 2003, 69, 5228–5237. [Google Scholar] [CrossRef] [Green Version]
- Saerens, S.M.G.; Delvaux, F.; Verstrepen, K.J.; Van Dijck, P.; Thevelein, J.M. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl. Environ. Microbiol. 2008, 74, 454–461. [Google Scholar] [CrossRef] [Green Version]
- Siddeeg, A.; Zeng, X.A.; Ammar, A.F.; Han, Z. Sugar profile, volatile compounds, composition and antioxidant activity of Sukkari date palm fruit. J. Food Sci. Technol. 2019, 56, 754–762. [Google Scholar] [CrossRef]
- Saafi, E.B.; Arem, A.E.; Chahdoura, H.; Flamini, G.; Lachheb, B.; Ferchichi, A.; Hammami, M.; Achour, L. Nutritional properties, aromatic compounds and in vitro antioxidant activity of ten date palm fruit (Phoenix dactylifera L.) varieties grown in Tunisia. Braz. J. Pharm. Sci. 2022, 58. [Google Scholar] [CrossRef]
- Rashmi, H.B.; Negi, P.S. Phenolic acids from vegetables: A review on processing stability and health benefits. Food Res. Int. 2020, 136, 109298. [Google Scholar] [CrossRef]
- Leonard, W.; Zhang, P.; Ying, D.; Adhikari, B.; Fang, Z. Fermentation transforms the phenolic profiles and bioactivities of plant-based foods. Biotechnol. Adv. 2021, 49, 107763. [Google Scholar] [CrossRef]
- Zhang, P.; Ma, W.; Meng, Y.; Zhang, Y.; Jin, G.; Fang, Z. Wine phenolic profile altered by yeast: Mechanisms and influences. Compr. Rev. Food Sci. Food Saf. 2021, 20, 3579–3619. [Google Scholar] [CrossRef]
- Alam, M.Z.; Ramachandran, T.; Antony, A.; Hamed, F.; Ayyash, M.; Kamal-Eldin, A. Melanin is a plenteous bioactive phenolic compound in date fruits (Phoenix dactylifera L.). Sci. Rep. 2022, 12, 6614. [Google Scholar] [CrossRef] [PubMed]
- Hammouda, H.; Chérif, J.K.; Trabelsi-Ayadi, M.; Baron, A.; Guyot, S. Detailed polyphenol and tannin composition and its variability in Tunisian dates (Phoenix dactylifera L.) at different maturity stages. J. Agric. Food Chem. 2013, 61, 3252–3263. [Google Scholar] [CrossRef] [PubMed]
- Tassoult, M.; Kati, D.E.; Bachir-bey, M.; Benouadah, A.; Rodriguez-Gutiérrez, G. Valorization of date palm biodiversity: Physico-chemical composition, phenolic profile, antioxidant activity, and sensory evaluation of date pastes. J. Food Meas. Charact. 2021, 15, 2601–2612. [Google Scholar] [CrossRef]
- Lasanta, C.; Durán-Guerrero, E.; Díaz, A.B.; Castro, R. Influence of fermentation temperature and yeast type on the chemical and sensory profile of handcrafted beers. J. Sci. Food Agric. 2021, 101, 1174–1181. [Google Scholar] [CrossRef]
- Liu, H.; Tian, Y.; Zhou, Y.; Kan, Y.; Wu, T.; Xiao, W.; Luo, Y. Multi-modular engineering of Saccharomyces cerevisiae for high-titre production of tyrosol and salidroside. Microb. Biotechnol. 2021, 14, 2605–2616. [Google Scholar] [CrossRef]
- Alshwyeh, H.A. Phenolic profiling and antibacterial potential of Saudi Arabian native date palm (Phoenix dactylifera) cultivars. Int. J. Food Prop. 2020, 23, 627–638. [Google Scholar] [CrossRef] [Green Version]
- Amira, E.A.; Behija, S.E.; Beligh, M.; Lamia, L.; Manel, I.; Mohamed, H.; Lotfi, A. Effects of the ripening stage on phenolic profile, phytochemical composition and antioxidant activity of date palm fruit. J. Agric. Food Chem. 2012, 60, 10896–10902. [Google Scholar] [CrossRef]
- Zihad, S.M.N.K.; Uddin, S.J.; Sifat, N.; Lovely, F.; Rouf, R.; Shilpi, J.A.; Sheikh, B.Y.; Göransson, U. Antioxidant properties and phenolic profiling by UPLC-QTOF-MS of Ajwah, Safawy and Sukkari cultivars of date palm. Biochem. Biophys. Rep. 2021, 25, 100909. [Google Scholar] [CrossRef]
- Barros, R.G.C.; Pereira, U.C.; Andrade, J.K.S.; de Oliveira, C.S.; Vasconcelos, S.V.; Narain, N. In vitro gastrointestinal digestion and probiotics fermentation impact on bioaccessbility of phenolics compounds and antioxidant capacity of some native and exotic fruit residues with potential antidiabetic effects. Food Res. Int. 2020, 136, 109614. [Google Scholar] [CrossRef]
- Ketnawa, S.; Reginio, F.C., Jr.; Thuengtung, S.; Ogawa, Y. Changes in bioactive compounds and antioxidant activity of plant-based foods by gastrointestinal digestion: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 4684–4705. [Google Scholar] [CrossRef]
- Jakobek, L. Interactions of polyphenols with carbohydrates, lipids and proteins. Food Chem. 2015, 175, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Sęczyk, Ł.; Gawlik-Dziki, U.; Świeca, M. Influence of phenolic-food matrix interactions on in vitro bioaccessibility of selected phenolic compounds and nutrients digestibility in fortified white bean paste. Antioxidants 2021, 10, 1825. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein–phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Seraglio, S.K.T.; Valese, A.C.; Daguer, H.; Bergamo, G.; Azevedo, M.S.; Nehring, P.; Gonzaga, L.V.; Fett, R.; Costa, A.C.O. Effect of in vitro gastrointestinal digestion on the bioaccessibility of phenolic compounds, minerals, and antioxidant capacity of Mimosa scabrella Bentham honeydew honeys. Food Res. Int. 2017, 99, 670–678. [Google Scholar] [CrossRef]
- Shahidi, F.; Naczk, M. Phenolics in Food and Nutraceuticals; CRC Press: Boca Raton, FL, USA, 2003; Volume 1. [Google Scholar]
- Andrade Barreto, S.M.; Martins da Silva, A.B.; Prudêncio Dutra, M.d.C.; Costa Bastos, D.; de Brito Araújo Carvalho, A.J.; Cardoso Viana, A.; Narain, N.; dos Santos Lima, M. Effect of commercial yeasts (Saccharomyces cerevisiae) on fermentation metabolites, phenolic compounds, and bioaccessibility of Brazilian fermented oranges. Food Chem. 2023, 408, 135121. [Google Scholar] [CrossRef]
- Djaoudene, O.; Mansinhos, I.; Gonçalves, S.; Jara-Palacios, M.J.; Romano, A. Phenolic profile, antioxidant activity and enzyme inhibitory capacities of fruit and seed extracts from different Algerian cultivars of date (Phoenix dactylifera L.) were affected by in vitro simulated gastrointestinal digestion. S. Afr. J. Bot. 2021, 137, 133–148. [Google Scholar] [CrossRef]
- Sharma, R.; Garg, P.; Kumar, P.; Bhatia, S.K.; Kulshrestha, S. Microbial fermentation and its role in quality improvement of fermented foods. Fermentation 2020, 6, 106. [Google Scholar] [CrossRef]
- Bambouskova, M.; Gorvel, L.; Lampropoulou, V.; Sergushichev, A.; Loginicheva, E.; Johnson, K.; Korenfeld, D.; Mathyer, M.E.; Kim, H.; Huang, L.H.; et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature 2018, 556, 501–504. [Google Scholar] [CrossRef]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.H.; Higgins, M.; Hams, E.; et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Z.; Li, Z.; Wang, K.; Kong, B.; Chen, Q. l-glycine and l-glutamic acid protect Pediococcus pentosaceus R1 against oxidative damage induced by hydrogen peroxide. Food Microbiol. 2022, 101, 103897. [Google Scholar] [CrossRef]
- Zhou, Y.; Xie, Z.; Zhang, Z.; Yang, J.; Chen, M.; Chen, F.; Ma, Y.; Chen, C.; Peng, Q.; Zou, L.; et al. Plasma metabolites changes in male heroin addicts during acute and protracted withdrawal. Aging 2021, 13, 18669–18688. [Google Scholar] [CrossRef]
- Masyita, A.; Mustika Sari, R.; Dwi Astuti, A.; Yasir, B.; Rahma Rumata, N.; Emran, T.B.; Nainu, F.; Simal-Gandara, J. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X 2022, 13, 100217. [Google Scholar] [CrossRef]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Akihisa, T.; Ogihara, J.; Kato, J.; Yasukawa, K.; Ukiya, M.; Yamanouchi, S.; Oishi, K. Inhibitory effects of triterpenoids and sterols on human immunodeficiency virus-1 reverse transcriptase. Lipids 2001, 36, 507–512. [Google Scholar] [CrossRef]
- Szakiel, A.; Pączkowski, C.; Pensec, F.; Bertsch, C. Fruit cuticular waxes as a source of biologically active triterpenoids. Phytochem. Rev. 2012, 11, 263–284. [Google Scholar] [CrossRef] [Green Version]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkalbani, N.S.; Alam, M.Z.; Al-Nabulsi, A.; Osaili, T.M.; Olaimat, A.; Liu, S.-Q.; Kamal-Eldin, A.; Ayyash, M. Fermentation of Date Pulp Residues Using Saccharomyces cerevisiae and Pichia kudriavzevii—Insights into Biological Activities, Phenolic and Volatile Compounds, Untargeted Metabolomics, and Carbohydrate Analysis Post In Vitro Digestion. Fermentation 2023, 9, 561. https://doi.org/10.3390/fermentation9060561
Alkalbani NS, Alam MZ, Al-Nabulsi A, Osaili TM, Olaimat A, Liu S-Q, Kamal-Eldin A, Ayyash M. Fermentation of Date Pulp Residues Using Saccharomyces cerevisiae and Pichia kudriavzevii—Insights into Biological Activities, Phenolic and Volatile Compounds, Untargeted Metabolomics, and Carbohydrate Analysis Post In Vitro Digestion. Fermentation. 2023; 9(6):561. https://doi.org/10.3390/fermentation9060561
Chicago/Turabian StyleAlkalbani, Nadia S., Muneeba Zubair Alam, Anas Al-Nabulsi, Tareq M. Osaili, Amin Olaimat, Shao-Quan Liu, Afaf Kamal-Eldin, and Mutamed Ayyash. 2023. "Fermentation of Date Pulp Residues Using Saccharomyces cerevisiae and Pichia kudriavzevii—Insights into Biological Activities, Phenolic and Volatile Compounds, Untargeted Metabolomics, and Carbohydrate Analysis Post In Vitro Digestion" Fermentation 9, no. 6: 561. https://doi.org/10.3390/fermentation9060561
APA StyleAlkalbani, N. S., Alam, M. Z., Al-Nabulsi, A., Osaili, T. M., Olaimat, A., Liu, S.-Q., Kamal-Eldin, A., & Ayyash, M. (2023). Fermentation of Date Pulp Residues Using Saccharomyces cerevisiae and Pichia kudriavzevii—Insights into Biological Activities, Phenolic and Volatile Compounds, Untargeted Metabolomics, and Carbohydrate Analysis Post In Vitro Digestion. Fermentation, 9(6), 561. https://doi.org/10.3390/fermentation9060561






