Isolation, Screening, and Identification of Alkaline Protease-Producing Bacteria and Application of the Most Potent Enzyme from Bacillus sp. Mar64
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sampling and Isolation Procedure
2.3. Enzyme Production and Assay
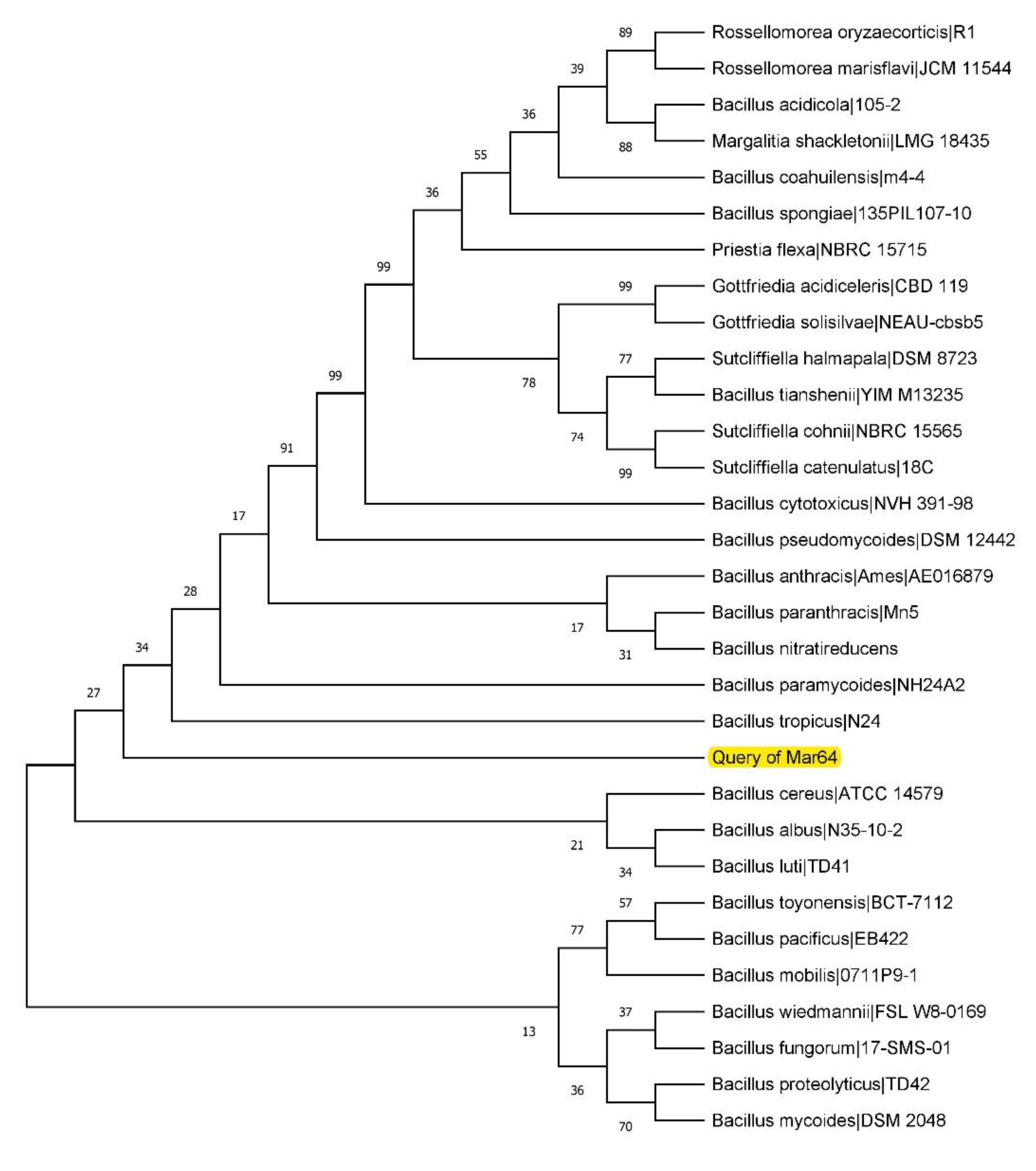
2.4. Molecular Characterization of the Most Potent Bacterial Isolates
2.5. Optimization of Production Conditions
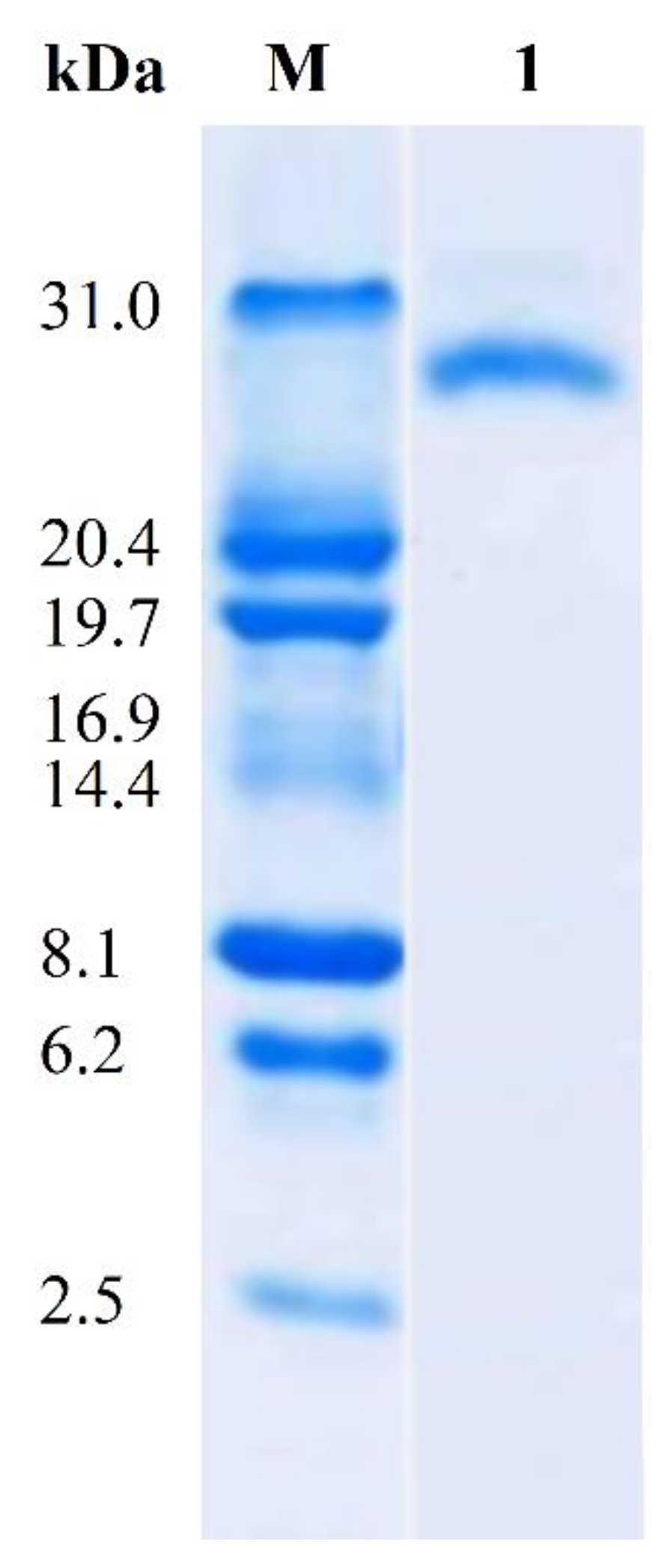
2.6. Enzyme Purification
2.7. Biochemical Properties of Enzyme
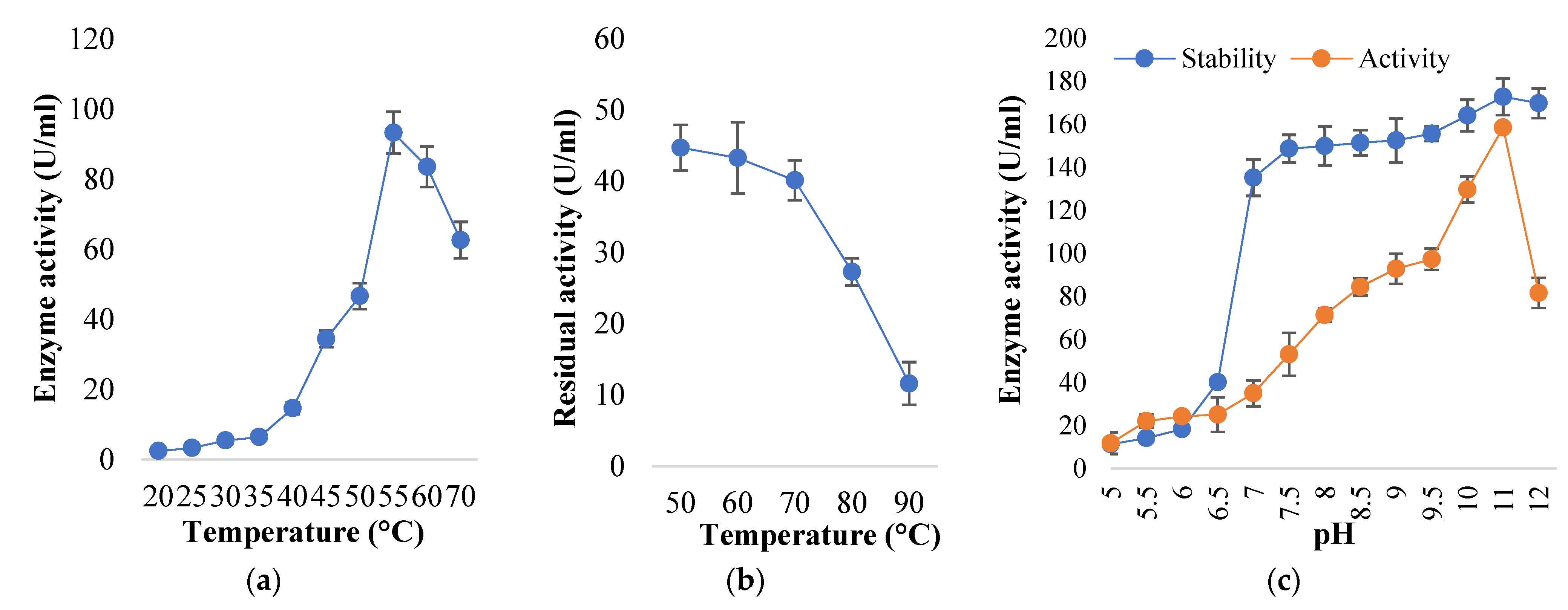
2.7.1. Effect of Temperature on Enzyme Activity and Stability
2.7.2. Effect of pH on Enzyme Activity and Stability
2.7.3. Effect of Metal Ions and Protease Inhibitors
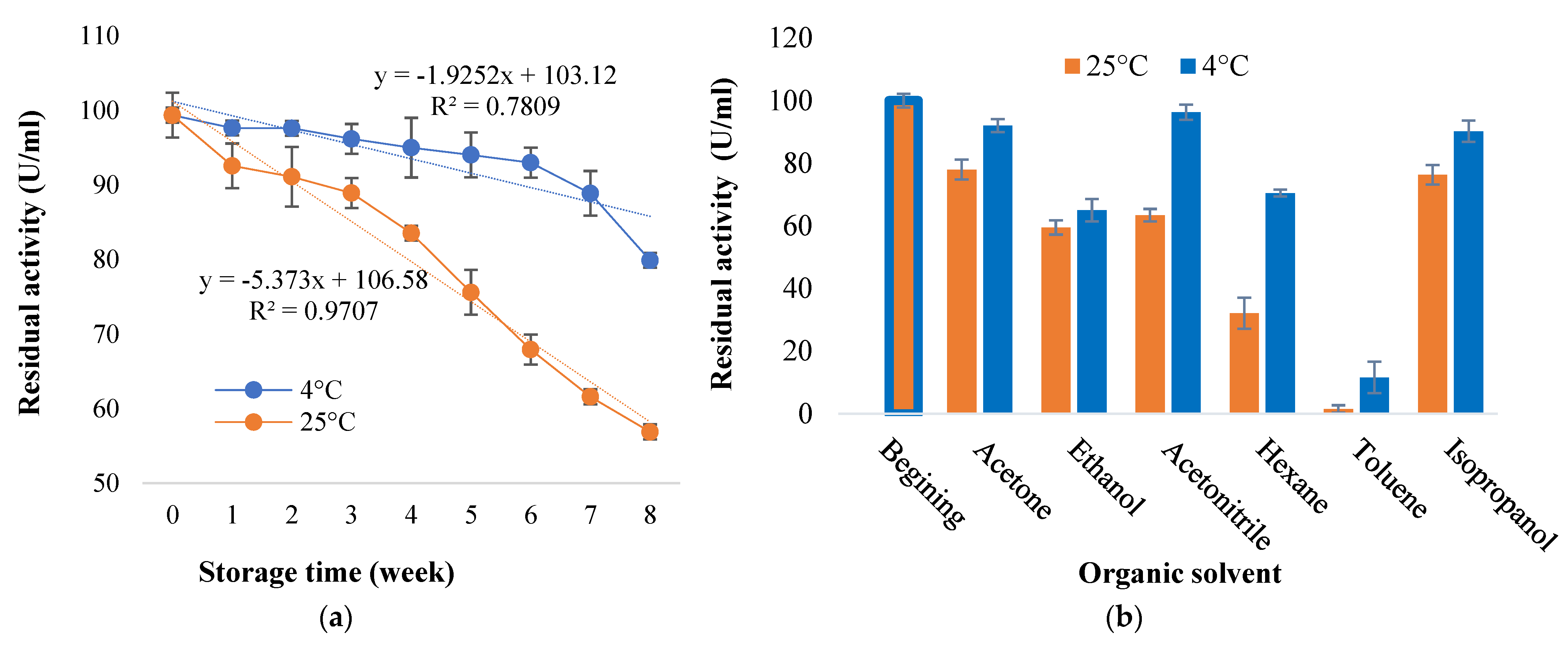
2.7.4. Effect of Storage Conditions and Organic Solvents
2.8. Detergent Compatibility
2.9. Destaining of Cotton Fabrics
2.10. Dehairing Activity
2.11. Hydrolysis of Gelatin and Recovery of Silver from Waste X-ray Films
2.12. Statistical Analysis
3. Results
3.1. Isolation of Alkaline Protease-Producing Bacteria
3.2. Optimization of Alkaline Protease Productivity
3.3. Enzyme Purification
3.4. Biochemical Properties of Alkaline Protease
3.5. Effect of Metal Ions and Protease Inhibitors
3.6. Storage Stability of Mar64P
3.7. Commercial Application of Mar64P
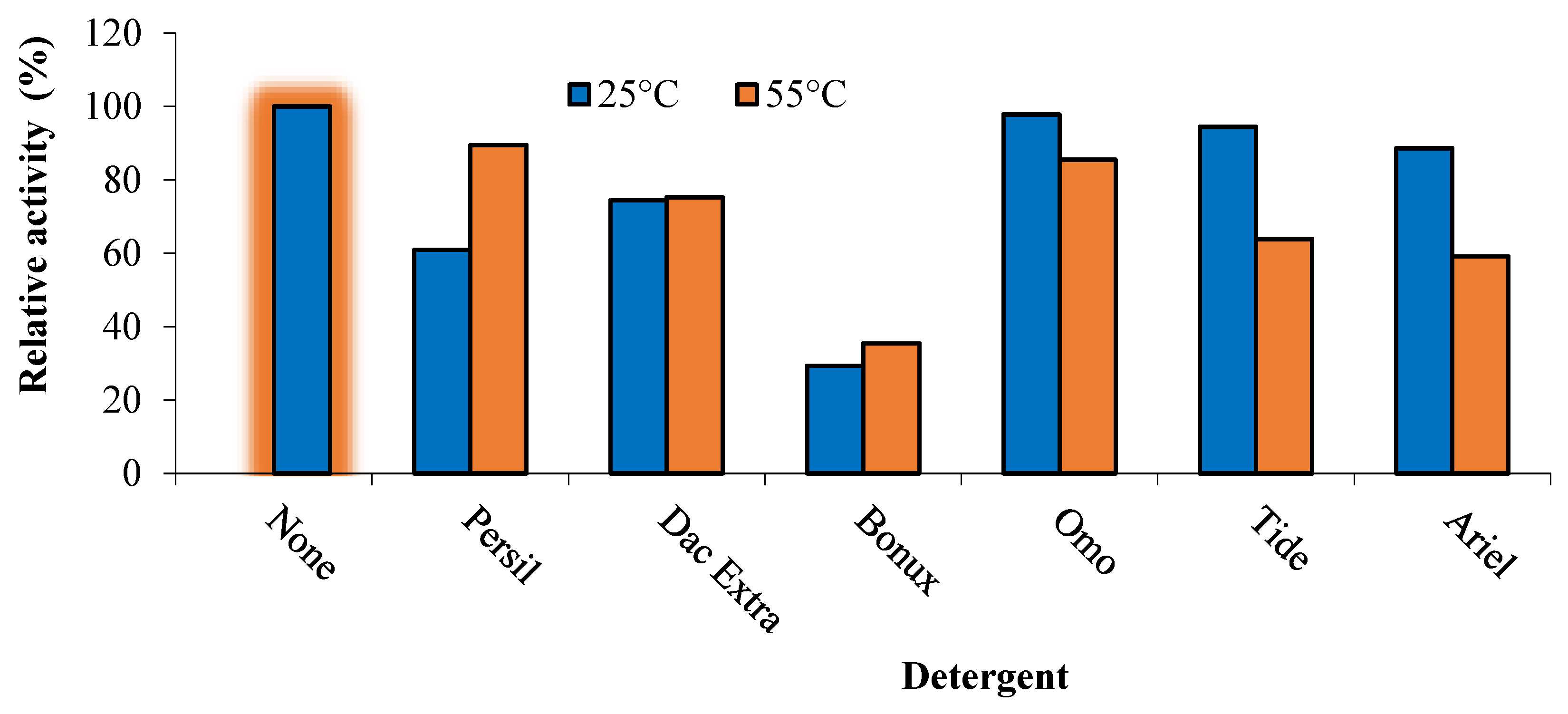
3.7.1. Detergent Compatibility
3.7.2. Destaining of Cotton Fabrics
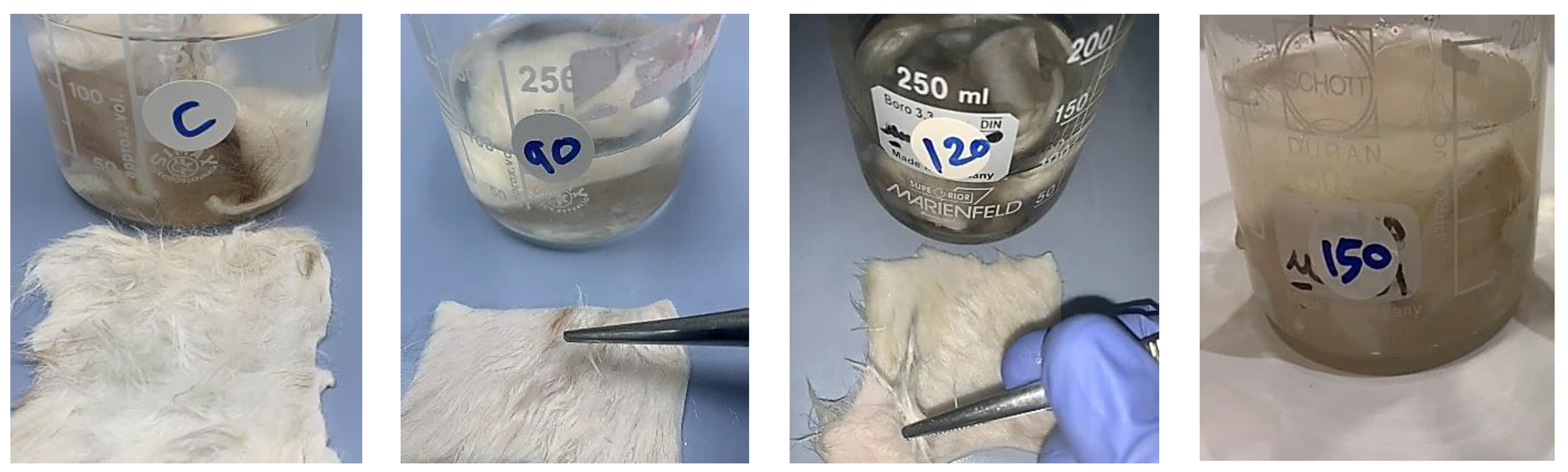
3.7.3. Dehairing Activity
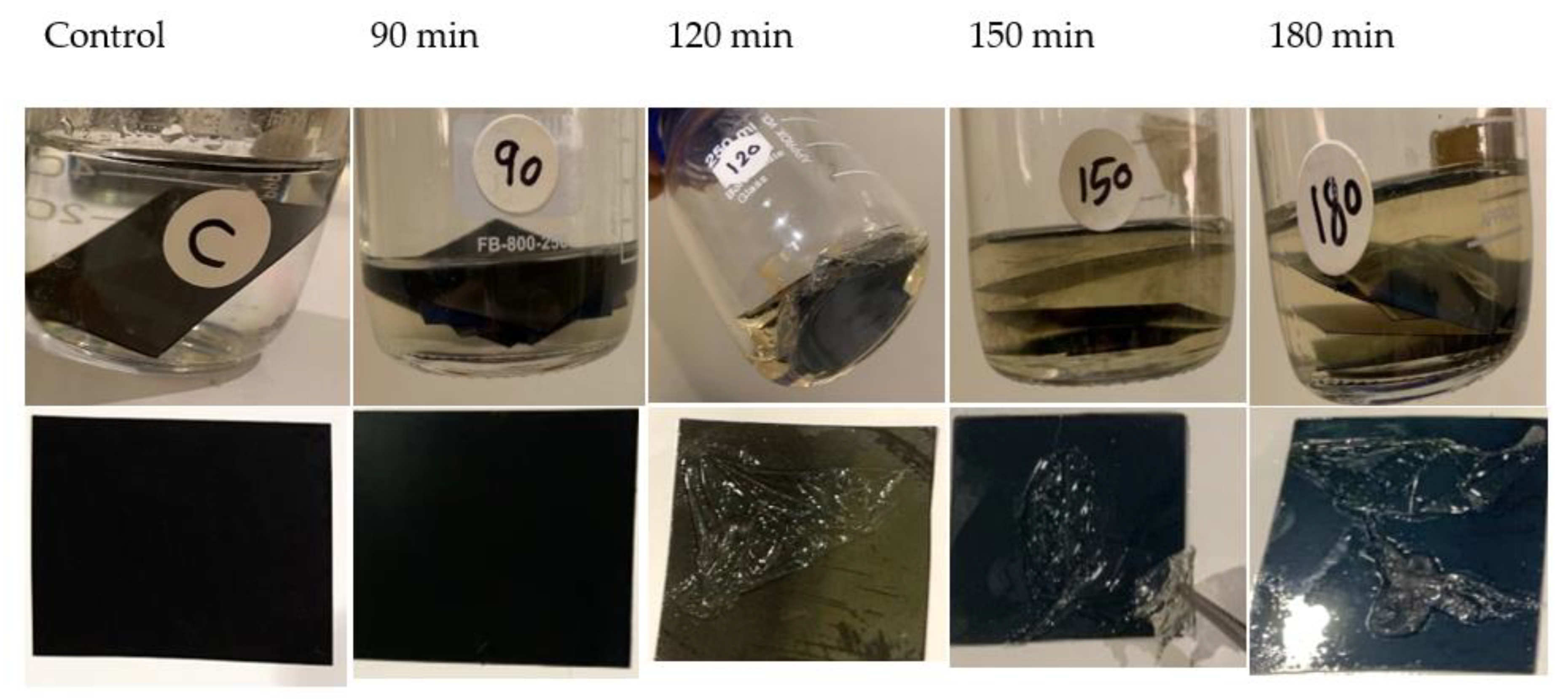
3.7.4. Degradation of Gelatin of Used X-ray Films
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Razzaq, A.; Shamsi, S.; Ali, A.; Ali, Q.; Sajjad, M.; Malik, A.; Ashraf, M. Microbial proteases applications. Front. Bioeng. Biotechnol. 2019, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, A.; Sivakumar, N.; Gujarathi, A.M.; Victor, R. Production of thermotolerant, detergent stable alkaline protease using the gut waste of Sardinella longiceps as a substrate: Optimization and characterization. Sci. Rep. 2018, 8, 12442. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Huang, X.; Ren, Q.; Wang, X. Purification and characterization of a H2O2-tolerant alkaline protease from Bacillus sp. ZJ1502, a newly isolated strain from fermented bean curd. Food Chem. 2019, 274, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kumar, S.; Kaur, R.; Kaur, R. Multipotential alkaline protease from a novel Pyxidicoccus sp. 252: Ecofriendly replacement to various chemical processes. Front. Microbiol. 2021, 12, 722719. [Google Scholar] [CrossRef]
- Shafique, T.; Shafique, J.; Zahid, S.; Kazi, M.; Alnemer, O.; Ahmad, A. Screening, selection and development of Bacillus subtilis apr- IBL04 for hyper production of macromolecule alkaline protease. Saudi J. Biol. Sci. 2020, 28, 1494–1501. [Google Scholar] [CrossRef]
- Zhou, C.; Zhou, H.; Zhang, H.; Lu, F. Optimization of alkaline protease production by rational deletion of sporulation related genes in Bacillus licheniformis. Microb. Cell Factories 2019, 18, 127. [Google Scholar] [CrossRef]
- Kotb, E.; El-Zawahry, Y.A.; Saleh, G.E. Isolation of a putative virulence agent, cytotoxic serine-elastase, from a newly isolated Pseudomonas aeruginosa ZuhP13. J. Biosci. 2019, 44, 7. [Google Scholar] [CrossRef]
- AlEraky, D.M.; Madi, M.; El Tantawi, M.; AlHumaid, J.; Fita, S.; AbdulAzeez, S.; Borgio, J.F.; Al-Harbi, F.A.; Alagl, A.S. Predominance of non-Streptococcus mutans bacteria in dental biofilm and its relation to caries progression. Saudi J. Biol. Sci. 2021, 28, 7390–7395. [Google Scholar] [CrossRef]
- AlShaikh-Mubarak, G.A.; Kotb, E.; Alabdalall, A.H.; Aldayel, M.F. A survey of elastase-producing bacteria and characteristics of the most potent producer, Priestia megaterium gasm32. PLoS ONE 2023, 18, e0282963. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Hassanein, W.A.; Kotb, E.; Awny, N.M.; El-Zawahry, Y.A. Fibrinolysis and anticoagulant potential of a metallo protease produced by Bacillus subtilis K42. J. Biosci. 2011, 36, 773–779. [Google Scholar] [CrossRef]
- Priya, J.D.A.; Divakar, K.; Prabha, M.S.; Selvam, G.P.; Gautam, P. Isolation, purification, and characterisation of an organic solvent-tolerant Ca2+-dependent protease from Bacillus megaterium AU02. Appl. Biochem. Biotechnol. 2014, 172, 910–932. [Google Scholar] [CrossRef]
- Mahakhan, P.; Aphaiso, P.; Srisunthorn, K.; Vichitphan, K.; Vichitphan, S.; Punyauppa-Path, S.; Sawaengkaew, J. Alkaline Protease Production from Bacillus gibsonii 6BS15-4 Using Dairy Effluent and Its Characterization as a Laundry Detergent Additive. J. Microbiol. Biotechnol. 2022, 33, 195–202. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Liu, H.; Lang, D.A.; Xu, H.; Zhu, H. The application of a halotolerant metalloprotease from marine bacterium Vibrio sp LA-05 in liquid detergent formulations. Int. Biodeterior. Biodegrad. 2019, 142, 18–25. [Google Scholar] [CrossRef]
- Ullah, N.; Rehman, M.U.; Sarwar, A.; Nadeem, M.; Nelofer, R.; Shakir, H.A.; Irfan, M.; Idrees, M.; Naz, S.; Nabi, G.; et al. Purification, characterization, and application of alkaline protease enzyme from a locally isolated Bacillus cereus strain. Fermentation. 2022, 8, 628. [Google Scholar] [CrossRef]
- Thomas, N.N.; Archana, V.; Shibina, S.; Edwin, B.T. Isolation and characterization of a protease from Bacillus sp. Mater. Today Proc. 2020, 41, 685–691. [Google Scholar] [CrossRef]
- Bhaskar, N.; Sudeepa, E.; Rashmi, H.; Tamilselvi, A. Partial purification and characterization of protease of Bacillus proteolyticus CFR3001 isolated from fish processing waste and its antibacterial activities. Bioresour. Technol. 2007, 98, 2758–2764. [Google Scholar] [CrossRef]
- Rao, C.S.; Sathish, T.; Pendyala, B.; Kumar, T.P.; Prakasham, R.S. Development of a mathematical model for Bacillus circulans growth and alkaline protease production kinetics. J. Chem. Technol. Biotechol. 2009, 84, 302–307. [Google Scholar]
- Shaikh, I.A.; Turakani, B.; Malpani, J.; Goudar, S.V.; Mahnashi, M.H.; Al-Serwi, R.H.; Ghoneim, M.M.; El-Sherbiny, M.; Mannasaheb, B.A.; Alsaikhan, F.; et al. Extracellular Protease Production, Optimization, and Partial Purification from Bacillus nakamurai PL4 and its Applications. J. King Saud Univ.-Sci. 2023, 35, 102429. [Google Scholar] [CrossRef]
- Joo, H.-S. Purification and Characterization of a Novel Alkaline Protease from Bacillus horikoshii. J. Microbiol. Biotechnol. 2012, 22, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, N.; Rajeswari, M.V.; Balasubramanian, T. Extraction, purification and application of thermostable and halostable alkaline protease from Bacillus alveayuensis CAS 5 using marine wastes. Food Bioprod. Process. 2014, 92, 335–342. [Google Scholar] [CrossRef]
- Kotb, E.; El-Nogoumy, B.A.; Alqahtani, H.A.; Ahmed, A.A.; Al-Shwyeh, H.A.; Algarudi, S.M.; Almahasheer, H. A putative cytotoxic serine protease from Salmonella typhimurium UcB5 recovered from undercooked burger. Sci. Rep. 2023, 13, 3926. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhuayan, I.; Kotb, E.; Alqosaibi, A.; Mahmoud, A. Histological Studies on a Newly Isolated Bacillus subtilis D10 Protease in the Debridement of Burn Wound Eschars Using Mouse Model. Pharmaceutics 2021, 13, 923. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.; Kotb, E.; Alqosaibi, A.I.; Al-Karmalawy, A.A.; Al-Dhuayan, I.S.; Alabkari, H. In vitro and in silico characterization of alkaline serine protease from Bacillus subtilis D9 recovered from Saudi Arabia. Heliyon 2021, 7, e08148. [Google Scholar] [CrossRef]
- Alqosaibi, A.I.; Mahmoud, A.; Kotb, E.; Huang, Y.; Al-Dhuayan, I.S.; Alhazmi, S.; Bahloul, A.A.; Okasha, S.T.; Otaibi, H.; AlYami, N.; et al. Saccharomyces cerevisiae OS303 expression of an alkaline protease from a newly isolated Bacillus subtilis D9. Braz. J. Biol. 2022, 82, 1–9. [Google Scholar] [CrossRef]
- Niyonzima, F.N.; More, S. Detergent-Compatible Proteases: Microbial Production, Properties, and Stain Removal Analysis. Prep. Biochem. Biotechnol. 2014, 45, 233–258. [Google Scholar] [CrossRef]
- Beg, Q.K.; Gupta, R. Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzym. Microb. Technol. 2003, 32, 294–304. [Google Scholar] [CrossRef]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef]
- Li, J.; Jiang, L.; Cao, X.; Wu, Y.; Lu, F.; Liu, F.; Li, Y.; Liu, Y. Improving the activity and stability of Bacillus clausii alkaline protease using directed evolution and molecular dynamics simulation. Enzym. Microb. Technol. 2021, 147, 109787. [Google Scholar] [CrossRef]
- Uttatree, S.; Kobtrakool, K.; Ketsuk, A.; Kaenngam, W.; Thakolprajak, P.; Charoenpanich, J. A novel metal-tolerant, solvent and surfactant stable protease from a new strain of Bacillus megaterium. Biocatal. Agric. Biotechnol. 2017, 12, 228–235. [Google Scholar] [CrossRef]
- Wang, S.L.; Yeh, P.Y. Production of a surfactant-and solvent stable alkaliphilic protease by bioconversion of shrimp shell wastes fermented by Bacillus subtilis TKU007. Process Biochem. 2006, 41, 1545–1552. [Google Scholar] [CrossRef]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Reddy, M.R.; Reddy, K.S.; Chouhan, Y.R.; Bee, H.; Reddy, G. Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Technol. 2017, 243, 254–263. [Google Scholar] [CrossRef]
- Ibrahim, A.S.S.; Elbadawi, Y.B.; El-Tayeb, M.A.; Al-Maary, K.S.; Maany, D.A.F.; Ibrahim, S.S.S.; Elagib, A.A. Alkaline serine protease from the new halotolerant alkaliphilic Salipaludibacillus agaradhaerens strain AK-R: Purification and properties. 3 Biotech 2019, 9, 391. [Google Scholar] [CrossRef]
- Jain, D.; Pancha, I.; Mishra, S.K.; Shrivastav, A.; Mishra, S. Purification and characterization of haloalkaline thermoactive, solvent stable and SDS-induced protease from Bacillus sp.: A potential additive for laundry detergents. Bioresour. Technol. 2011, 115, 228–236. [Google Scholar] [CrossRef]
- Masui, A.; Yasuda, M.; Fujiwara, N.; Ishikawa, H. Enzymatic hydrolysis of gelatin layers on used lith film using thermostable alkaline protease for recovery of silver and PET film. Biotechnol. Prog. 2004, 20, 1267–1269. [Google Scholar] [CrossRef]
- Sivasubramanian, S.; Murali Manohar, B.; Rajaram, A.; Puvanakrishnan, R. Ecofriendly lime and sulfide free enzymatic dehairing of skins and hides using a bacterial alkaline protease. Chemosphere 2008, 70, 1015–1024. [Google Scholar] [CrossRef]
- George, N.; Chauhan, P.S.; Kumar, V.; Puri, N.; Gupta, N. Approach to ecofriendly leather: Characterization and application of an alkaline protease for chemical free dehairing of skins and hides at pilot scale. J. Clean. Prod. 2014, 79, 249–257. [Google Scholar] [CrossRef]


| Isolate Number | Source Locality | Alkaline Protease Productivity (U/mL) | Strain Characterization | GenBank Accession Number |
|---|---|---|---|---|
| 9 | Saihat farm 1 | 142.8 ± 16.2 | Enterobacter sp. Mar9 | OP429622.1 |
| 64 | Saihat farm 2 | 159.9 ± 7.2 | Bacillus sp. Mar64 | OP429623.1 |
| 67 | Saihat farm 2 | 137.6 ± 7.2 | Bacillus sp. Mar67 | OP429624.1 |
| 77 | Dammam farm | 92.6 ± 7.6 | Bacillus sp. Mar77 | OP429625.1 |
| Parameter | Optimal Value | Maximal Productivity (U/mL) | Fold (×) |
|---|---|---|---|
| Unoptimized conditions | NA | 61.9 ± 2.2 | 1.0 |
| Incubation period | 60 h | 93.2 ± 3.0 | 1.5 |
| Initial pH | 9.0 | 110.1 ± 7.0 | 1.8 |
| Fermentation temperature | 45 °C | 155.3 ± 14.4 | 2.5 |
| Carbon source | 0.5% maltose | 162.1 ± 8.4 | 2.6 |
| Nitrogen source | 1% tyrosine | 172.4 ± 18.0 | 2.8 |
| Purification Step | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification (Fold) |
|---|---|---|---|---|
| Cell-free supernatant | 172,413.2 | 90.6 | 100.0 | 1.0 |
| 60% ammonium sulphate precipitation | 90,145.1 | 190.2 | 52.3 | 2.1 |
| Anion exchanger (DEAE-Sepharose CL-6B) | 53,312.0 | 571.8 | 30.9 | 6.3 |
| Gel permeation (Sephadex G-100 FF) | 21,368.6 | 769.1 | 12.4 | 8.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotb, E.; Alabdalall, A.H.; Alsayed, M.A.; Alghamdi, A.I.; Alkhaldi, E.; AbdulAzeez, S.; Borgio, J.F. Isolation, Screening, and Identification of Alkaline Protease-Producing Bacteria and Application of the Most Potent Enzyme from Bacillus sp. Mar64. Fermentation 2023, 9, 637. https://doi.org/10.3390/fermentation9070637
Kotb E, Alabdalall AH, Alsayed MA, Alghamdi AI, Alkhaldi E, AbdulAzeez S, Borgio JF. Isolation, Screening, and Identification of Alkaline Protease-Producing Bacteria and Application of the Most Potent Enzyme from Bacillus sp. Mar64. Fermentation. 2023; 9(7):637. https://doi.org/10.3390/fermentation9070637
Chicago/Turabian StyleKotb, Essam, Amira H. Alabdalall, Mariam A. Alsayed, Azzah I. Alghamdi, Eida Alkhaldi, Sayed AbdulAzeez, and J. Francis Borgio. 2023. "Isolation, Screening, and Identification of Alkaline Protease-Producing Bacteria and Application of the Most Potent Enzyme from Bacillus sp. Mar64" Fermentation 9, no. 7: 637. https://doi.org/10.3390/fermentation9070637
APA StyleKotb, E., Alabdalall, A. H., Alsayed, M. A., Alghamdi, A. I., Alkhaldi, E., AbdulAzeez, S., & Borgio, J. F. (2023). Isolation, Screening, and Identification of Alkaline Protease-Producing Bacteria and Application of the Most Potent Enzyme from Bacillus sp. Mar64. Fermentation, 9(7), 637. https://doi.org/10.3390/fermentation9070637








