High Salt Concentration Affects the Microbial Diversity of Cassava during Fermentation, as Revealed by 16S rRNA Gene Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Enumeration and Isolation of Microorganisms during Cassava Fermentation
2.3. Molecular Characterization and Identification of Bacterial Isolates
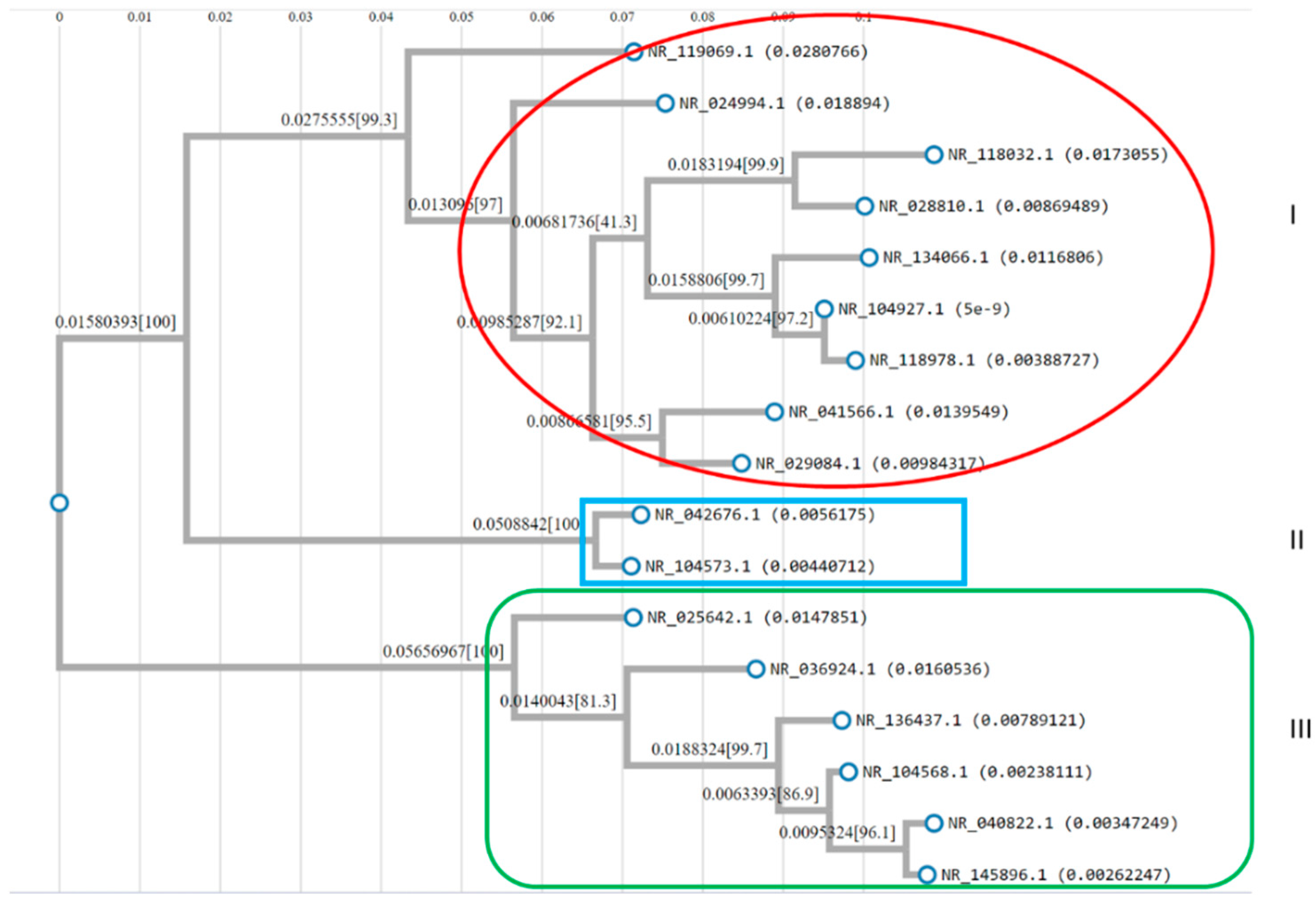
2.4. Construction of Phylogenetic Tree Construction
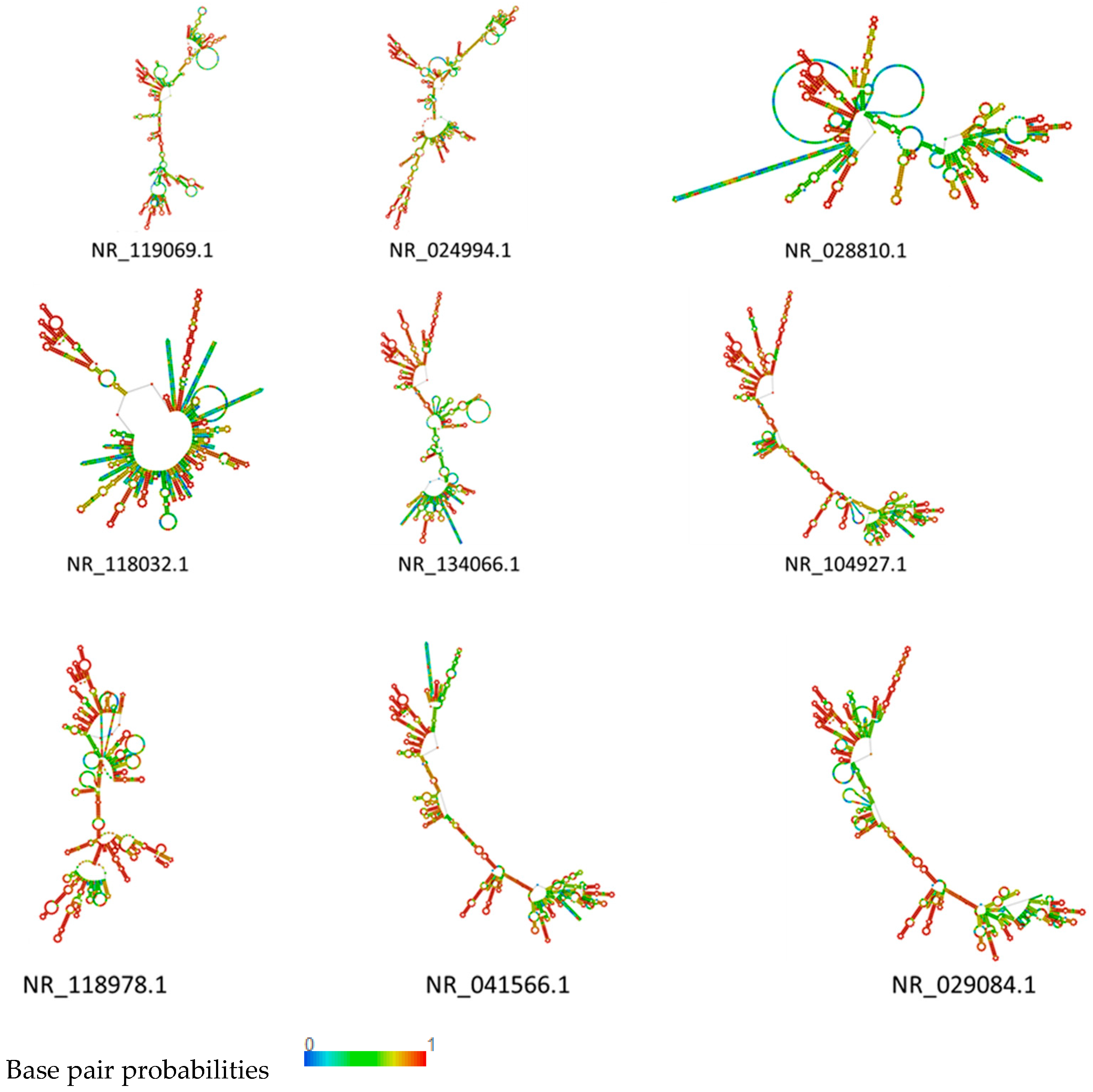
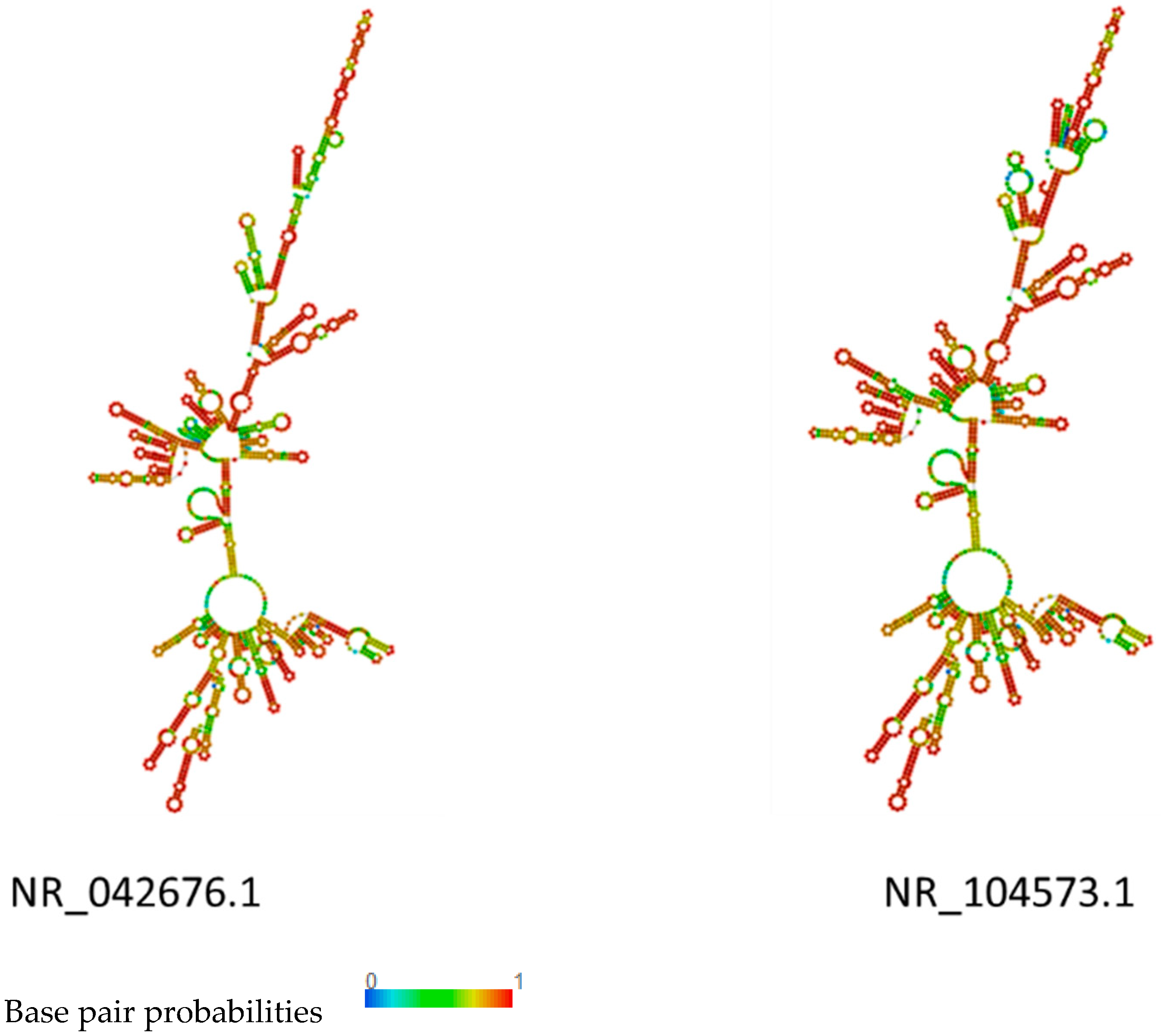
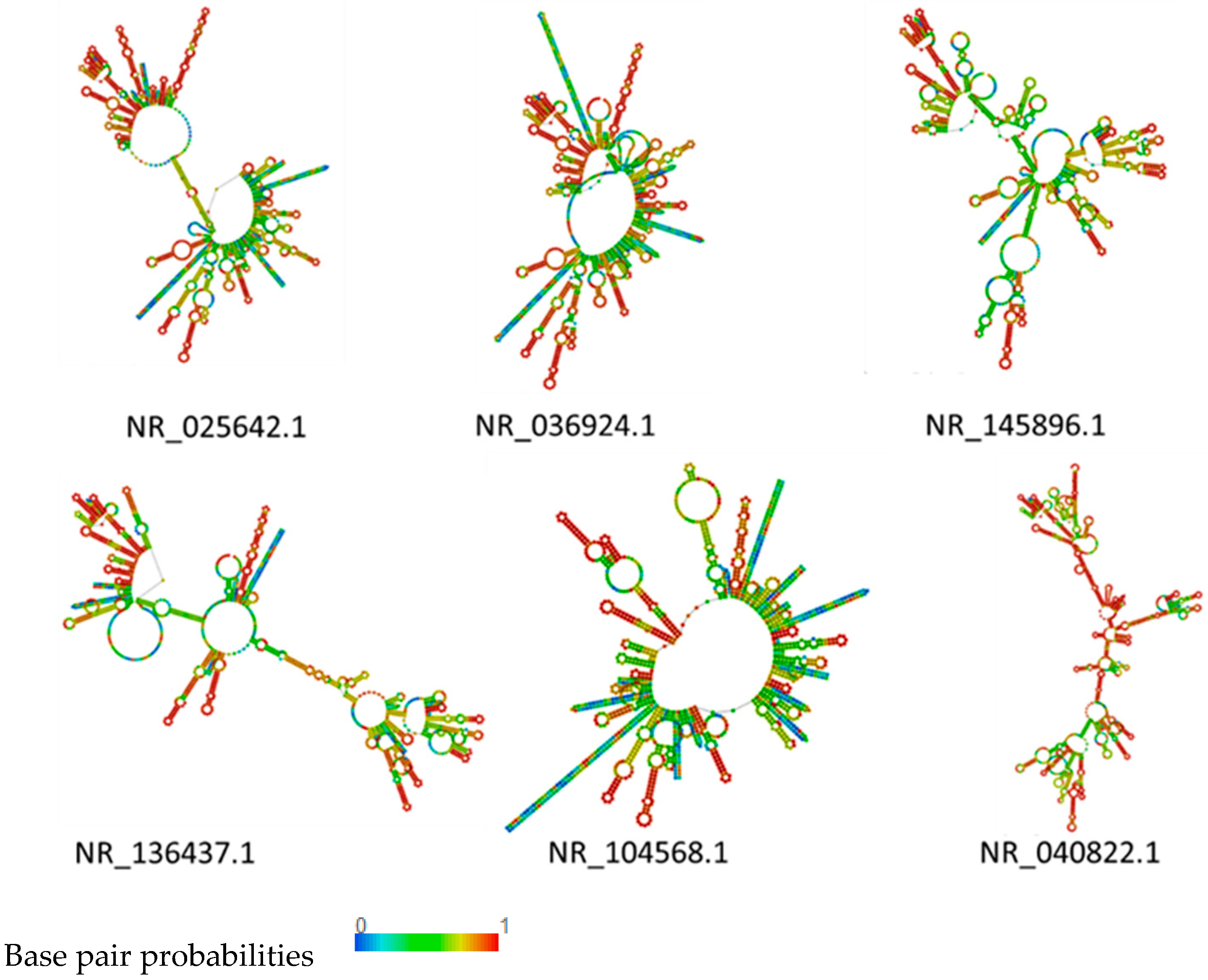
2.5. RNA Structure Prediction
2.6. Similarity Analysis of 16S rRNA Sequences
3. Results
3.1. Colony Sequencing and Similarity Detection Analysis
3.2. Colony Classification Based on Sequence Similarity
3.3. Strain Identification
3.4. rRNA Structure and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falade, K.O.; Akingbala, J.O. Utilization of cassava for food. Food Rev. Int. 2010, 27, 51–83. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations (FAO); FAOSTAT: Rome, Italy, 2020. [Google Scholar]
- Awoyale, W.; Alamu, E.O.; Chijioke, U.; Tran, T.; Takam Tchuente, H.N.; Ndjouenkeu, R.; Kegah, N.; Maziya-Dixon, B. A review of cassava semolina (gari and eba) end-user preferences and implications for varietal trait evaluation. Int. J. Food Sci. Technol. 2021, 56, 1206–1222. [Google Scholar] [CrossRef] [PubMed]
- Komen, J.; Tripathi, L.; Mkoko, B.; Ofosu, D.O.; Oloka, H.; Wangari, D. Biosafety regulatory reviews and leeway to operate: Case studies from sub-Sahara Africa. Front. Plant Sci. 2020, 11, 130. [Google Scholar] [PubMed] [Green Version]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented foods: Definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, M.E.; O’Donovan, C.M.; de Ullivarri, M.F.; Cotter, P.D. Microorganisms present in artisanal fermented food from South America. Front. Microbiol. 2022, 13, 941866. [Google Scholar] [CrossRef]
- Owusu-Kwarteng, J.; Agyei, D.; Akabanda, F.; Atuna, R.A.; Amagloh, F.K. Plant-Based Alkaline Fermented Foods as Sustainable Sources of Nutrients and Health-Promoting Bioactive Compounds. Front. Sustain. Food Syst. 2022, 6, 885328. [Google Scholar] [CrossRef]
- Aryee, A.N.A.; Owusu-Kwarteng, J.; Senwo, Z.; Alvarez, M.N. Characterizing fermented habanero pepper (Capsicum chinense L). Food Chem. Adv. 2022, 1, 100137. [Google Scholar] [CrossRef]
- Hawashi, M.; Widjaja, T.; Gunawan, S. Solid-State Fermentation of Cassava Products for Degradation of Anti-Nutritional Value and Enrichment of Nutritional Value. In New Advances on Fermentation Processes; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef] [Green Version]
- Seesaard, T.; Wongchoosuk, C. Recent Progress in Electronic Noses for Fermented Foods and Beverages Applications. Fermentation 2022, 8, 302. [Google Scholar] [CrossRef]
- Theron, C. The Advantages and Disadvantages of Spontaneous Fermentation—Wineland Media; Wineland Media: Paarl, South Africa, 2021. [Google Scholar]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. Toxins in fermented foods: Prevalence and preventions—A mini review. Toxins 2018, 11, 4. [Google Scholar] [CrossRef] [Green Version]
- El Sheikha, A.F.; Ray, R.C. Bioprocessing of horticultural wastes by solid-state fermentation into value-added/innovative bioproducts: A review. Food Rev. Int. 2022, 39, 1–57. [Google Scholar]
- Boonnop, K.; Wanapat, M.; Nontaso, N.; Wanapat, S. Enriching nutritive value of cassava root by yeast fermentation. Sci. Agric. 2009, 66, 629–633. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarska, K.; Taylor, M.; Piyasiri, U.; Frank, D. Flavor and Metabolite Profiles of Meat, Meat Substitutes, and Traditional Plant-Based High-Protein Food Products Available in Australia. Foods 2021, 10, 801. [Google Scholar] [CrossRef] [PubMed]
- Frediansyar, A.; Kurniadi, M. Comparative influence of salinity and temperature on cassava flour product by Lactobacillus plantarum and Lactobacillus acidophilus during single culture fermentation. Nusant. Biosci. 2016, 8, 207–214. [Google Scholar] [CrossRef]
- Khanna, S. DigitalCommons@UMaine Effects of Salt Concentration on the Physicochemical Properties and Microbial Safety of Spontaneously Fermented Cabbage; The University of Maine: Orono, ME, USA, 2018. [Google Scholar]
- Barcenilla, C.; Álvarez-Ordóñez, A.; López, M.; Alvseike, O.; Prieto, M. Microbiological safety and shelf-life of low-salt meat products—A Review. Foods 2022, 11, 2331. [Google Scholar] [CrossRef] [PubMed]
- Meryandini, A.; Melani, V.; Sunarti, T.C. Addition of cellulolytic bacteria to improved the quality of fermented cassava flour. Afr. J. Food Sci. Technol. 2011, 2, 30–35. [Google Scholar]
- Anal, A.K. Quality ingredients and safety concerns for traditional fermented foods and beverages from Asia: A review. Fermentation 2019, 5, 8. [Google Scholar] [CrossRef] [Green Version]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhang, L.; Zhang, Q.; Zi, X.; Lv, R.; Tang, J.; Zhou, H. Impacts of citric acid and malic acid on fermentation quality and bacterial community of cassava foliage silage. Front. Microbiol. 2020, 11, 595622. [Google Scholar] [CrossRef]
- Balogun, O.B.; Adeleke, B.S.; Owoseni, I. Characterization of bacteria isolates from fermented cassava steeping water. Int. J. Appl. Biol. 2021, 5, 190–199. [Google Scholar]
- Flibert, G.; Abel, T.; Aly, S. African cassava Traditional Fermented Food: The Microorganism’s Contribution to their Nutritional and Safety Values—A Review. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 664–687. [Google Scholar] [CrossRef] [Green Version]
- Rapoo, S.M.; Budeli, P.; Thaoge, M.L. Recovery of Potential Starter Cultures and Probiotics from Fermented Sorghum (Ting) Slurries. Microorganisms 2023, 11, 715. [Google Scholar] [PubMed]
- Oyeyinka, S.A.; Adeloye, A.A.; Olaomo, O.O.; Kayitesi, E. Effect of fermentation time on physicochemical properties of starch extracted from cassava root. Food Biosci. 2020, 33, 100485. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [Green Version]
- Koch, C.; Kroppenstedt, R.M.; Stackebrandt, E. Intrageneric relationships of the actinomycete genus Micromonospora. Int. J. Syst. Evol. Microbiol. 1996, 46, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Chruszcz, M.; Kapingidza, A.B.; Dolamore, C.; Kowal, K. A robust method for the estimation and visualization of IgE cross-reactivity likelihood between allergens belonging to the same protein family. PLoS ONE 2018, 13, e0208276. [Google Scholar]
- Björkroth, J.; Holzapfel, W. Genera Leuconostoc, Oenococcus and Weissella. Prokaryotes 2006, 4, 267–319. [Google Scholar]
- Adesulu-Dahunsi, A.T.; Dahunsi, S.O.; Ajayeoba, T.A. Co-occurrence of Lactobacillus Species During Fermentation of African Indigenous Foods: Impact on Food Safety and Shelf-Life Extension. Front. Microbiol. 2022, 13, 684730. [Google Scholar] [PubMed]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yap, P.-C.; MatRahim, N.-A.; AbuBakar, S.; Lee, H.Y. Antilisterial potential of lactic acid bacteria in eliminating Listeria monocytogenes in host and ready-to-eat food application. Microbiol. Res. 2021, 12, 234–257. [Google Scholar]
- Orike, E.L.; Adeyemo, S.M.; Omafuvbe, B.O. Probiotic potentials of lactic acid bacteria isolated from fermenting cassava. Int. J. Probiot. Prebiot. 2018, 13, 69–76. [Google Scholar]
- Pang, H.; Xin, X.; He, J.; Cui, B.; Guo, D.; Liu, S.; Yan, Z.; Liu, C.; Wang, X.; Nan, J. Effect of NaCl Concentration on Microbiological Properties in NaCl Assistant Anaerobic Fermentation: Hydrolase Activity and Microbial Community Distribution. Front. Microbiol. 2020, 11, 589222. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Rosicka-Kaczmarek, J.; Ryngajłło, M.; Czarnecka-Chrebelska, K.H. Characterization and Protective Properties of Lactic Acid Bacteria Intended to Be Used in Probiotic Preparation for Honeybees (Apis mellifera L.)—An In Vitro Study. Animals 2023, 13, 1059. [Google Scholar] [CrossRef]
- Ali, M.S.; Lee, E.-B.; Lim, S.-K.; Suk, K.; Park, S.-C. Isolation and Identification of Limosilactobacillus reuteri PSC102 and Evaluation of Its Potential Probiotic, Antioxidant, and Antibacterial Properties. Antioxidants 2023, 12, 238. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Pan, X.; Xu, T.; Zhang, Z.; Zi, X.; Jiang, Y. Cassava foliage affects the microbial diversity of Chinese indigenous geese caecum using 16S rRNA sequencing. Sci. Rep. 2017, 7, 45697. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, J.; Ge, L.; Huang, L.; Peng, Z.; Liang, Y.; Zhang, X.; Fan, S. Probiotic Lacticaseibacillus rhamnosus GR-1 and Limosilactobacillus reuteri RC-14 as an Adjunctive Treatment for Bacterial Vaginosis Do Not Increase the Cure Rate in a Chinese Cohort: A Prospective, Parallel-Group, Randomized, Controlled Study. Front. Cell. Infect. Microbiol. 2021, 11, 669901. [Google Scholar] [CrossRef]
- Sharma, N.; Gupta, D.; Park, Y.-S. Genome analysis revealed a repertoire of oligosaccharide utilizing CAZymes in Weissella confusa CCK931 and Weissella cibaria YRK005. Food Sci. Biotechnol. 2023, 32, 553–564. [Google Scholar] [CrossRef]
- Nachtigall, C.; Hassler, V.; Wefers, D.; Rohm, H.; Jaros, D. Dextrans of Weissella cibaria DSM14295: Microbial production, structure and functionality. Int. J. Biol. Macromol. 2023, 246, 125631. [Google Scholar] [CrossRef] [PubMed]
- Kiran, F.; Demirhan, H.K.; Haliscelik, O.; Zatari, D. Metabolic profiles of Weissella spp. postbiotics with anti-microbial and anti-oxidant effects. J. Infect. Dev. Ctries. 2023, 17, 507–517. [Google Scholar] [PubMed]

| Treatment Code | Salt Concentration (%) | Accession # | Species | BLAST Max Score | Species Rate (%) | Genus Rate (%) |
|---|---|---|---|---|---|---|
| FCS0 (control) | 0 | NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 10 | 100 |
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 10 | |||
| NR_104573.1 | Lactiplantibacillus plantarum | 2656 | 58 | |||
| NR_042676.1 | Lactiplantibacillus fabifermentans | 2591 | 12 | |||
| NR_042254.1 | Lactiplantibacillus argentoratensis | 2617 | 10 | |||
| FCS20 | 5 | NR_113339.1 | Lactiplantibacillus fabifermentans | 2591 | 4 | 80 |
| NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 8 | |||
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 8 | |||
| NR_104573.1 | Lactiplantibacillus plantarum | 2656 | 48 | |||
| NR_042676.1 | Lactiplantibacillus fabifermentans | 2591 | 4 | |||
| NR_042254.1 | Lactiplantibacillus argentoratensis | 2617 | 8 | |||
| NR_041566.1 | Limosilactobacillus equigenerosi | 2398 | 2 | 20 | ||
| NR_113335.1 | Limosilactobacillus fermentum | 2763 | 2 | |||
| NR_029084.1 | Limosilactobacillus gastricus | 2431 | 2 | |||
| NR_118032.1 | Limosilactobacillus alvi | 2344 | 2 | |||
| NR_104927.1 | Limosilactobacillus fermentum | 2760 | 4 | |||
| NR_134066.1 | Limosilactobacillus gorillae | 2634 | 2 | |||
| NR_028810.1 | Limosilactobacillus ingluviei | 2401 | 2 | |||
| NR_024994.1 | Limosilactobacillus mucosae | 2390 | 2 | |||
| NR_119069.1 | Limosilactobacillus reuteri | 2278 | 2 | |||
| FCS40 | 10 | NR_042676.1 | Lactiplantibacillus fabifermentans | 2591 | 10 | 100 |
| NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 10 | |||
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 10 | |||
| NR_104573.1 | Lactiplantibacillus plantarum | 2656 | 60 | |||
| NR_042254.1 | Lactiplantibacillus argentoratensis | 2617 | 10 | |||
| FCS60 | 15 | NR_113339.1 | Lactiplantibacillus fabifermentans | 2591 | 12 | 100 |
| NR_115605.1 | Lactiplantibacillus plantarum | 2660 | 58 | |||
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 10 | |||
| NR_042254.1 | Lactiplantibacillus argentoratensi | 2617 | 10 | |||
| NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 10 | |||
| FCS80 | 20 | NR_042676.1 | Lactiplantibacillus fabifermentans | 2591 | 2 | 32 |
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 2 | |||
| NR_115605.1 | Lactiplantibacillus plantarum | 2660 | 12 | |||
| NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 16 | |||
| NR_136437.1 | Weissella bombi | 2368 | 2 | 68 | ||
| NR_036924.1 | Weissella cibaria | 2183 | 8 | |||
| NR_113258.1 | Weissella confusa | 2211 | 8 | |||
| NR_118771.1 | Weissella hellenica | 2398 | 16 | |||
| NR_113775.1 | Weissella hellenica | 2398 | 8 | |||
| NR_145896.1 | Weissella jogaejeotgali | 2198 | 6 | |||
| NR_104568.1 | Weissella paramesenteroides | 2483 | 8 | |||
| NR_040822.1 | Weissella thailandensis | 2226 | 8 | |||
| NR_025642.1 | Weissella soli | 2370 | 4 | |||
| FCS100 | 25 | NR_042676.1 | Lactiplantibacillus fabifermentans | 2591 | 6 | 60 |
| NR_025447.1 | Lactiplantibacillus paraplantarum | 2643 | 6 | |||
| NR_029133.1 | Lactiplantibacillus pentosus | 2658 | 6 | |||
| NR_115605.1 | Lactiplantibacillus plantarum | 2660 | 36 | |||
| NR_042254.1 | Lactiplantibacillus argentoratensis | 2617 | 6 | |||
| NR_136437.1 | Weissella bombi | 2368 | 4 | 40 | ||
| NR_036924.1 | Weissella cibaria | 2183 | 4 | |||
| NR_113258.1 | Weissella confusa | 2211 | 8 | |||
| NR_118771.1 | Weissella hellenica | 2398 | 12 | |||
| NR_145896.1 | Weissella jogaejeotgali | 2198 | 2 | |||
| NR_025642.1 | Weissella soli | 2370 | 2 | |||
| NR_040822.1 | Weissella thailandensis | 2226 | 4 | |||
| NR_104568.1 | Weissella paramesenteroides | 2483 | 4 |
| Group | Accession # | Strains | RNA Type | Partial Sequence |
|---|---|---|---|---|
| Group A | NR_042676.1 | Lactiplantibacillus fabifermentans LMG 24284 | 16S rRNA | Partial sequence |
| NR_104573.1 | Lactiplantibacillus plantarum CIP 103151 | 16S rRNA | Partial sequence | |
| Group B | NR_025642.1 | Weissella soli Mi268 | 16S rRNA | Partial sequence |
| Group C | NR_036924.1 | Weissella cibaria II-I-59 | 16S rRNA | Partial sequence |
| NR_040822.1 | Weissella thailandensis FS61-1 | 16S rRNA | Partial sequence | |
| NR_104568.1 | Weissella paramesenteroides NRIC 1542 | 16S rRNA | Partial sequence | |
| NR_136437.1 | Weissella bombi R-53094 | 16S rRNA | Partial sequence | |
| NR_145896.1 | Weissella jogaejeotgali FOL01 | 16S rRNA | Partial sequence | |
| Group D | NR_024994.1 | Limosilactobacillus mucosae S32 | 16S rRNA | Partial sequence |
| NR_028810.1 | Limosilactobacillus ingluviei KR3 | 16S rRNA | Partial sequence | |
| NR_029084.1 | Limosilactobacillus gastricus Kx156A7 | 16S rRNA | Partial sequence | |
| NR_041566.1 | Limosilactobacillus equigenerosi NRIC 0697 | 16S rRNA | Partial sequence | |
| NR_104927.1 | Limosilactobacillus fermentum CIP 102980 | 16S rRNA | Partial sequence | |
| NR_118032.1 | Limosilactobacillus alvi R54 | 16S rRNA | Partial sequence | |
| NR_118978.1 | Limosilactobacillus fermentum NCDO 1750 | 16S rRNA | Partial sequence | |
| NR_119069.1 | Limosilactobacillus reuteri subsp. Reuteri DSM 20016 | 16S rRNA | Partial sequence | |
| NR_134066.1 | Limosilactobacillus orilla KZ01 | 16S rRNA | Partial sequence |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, W.; Ananga, A.; Ukuku, D.O.; Aryee, A.N.A. High Salt Concentration Affects the Microbial Diversity of Cassava during Fermentation, as Revealed by 16S rRNA Gene Sequencing. Fermentation 2023, 9, 727. https://doi.org/10.3390/fermentation9080727
Zhou W, Ananga A, Ukuku DO, Aryee ANA. High Salt Concentration Affects the Microbial Diversity of Cassava during Fermentation, as Revealed by 16S rRNA Gene Sequencing. Fermentation. 2023; 9(8):727. https://doi.org/10.3390/fermentation9080727
Chicago/Turabian StyleZhou, Wei, Anthony Ananga, Dike O. Ukuku, and Alberta N. A. Aryee. 2023. "High Salt Concentration Affects the Microbial Diversity of Cassava during Fermentation, as Revealed by 16S rRNA Gene Sequencing" Fermentation 9, no. 8: 727. https://doi.org/10.3390/fermentation9080727
APA StyleZhou, W., Ananga, A., Ukuku, D. O., & Aryee, A. N. A. (2023). High Salt Concentration Affects the Microbial Diversity of Cassava during Fermentation, as Revealed by 16S rRNA Gene Sequencing. Fermentation, 9(8), 727. https://doi.org/10.3390/fermentation9080727





